A Deep Exon Cryptic Splice Site Promotes Aberrant Intron Retention in a Von Willebrand Disease Patient
Abstract
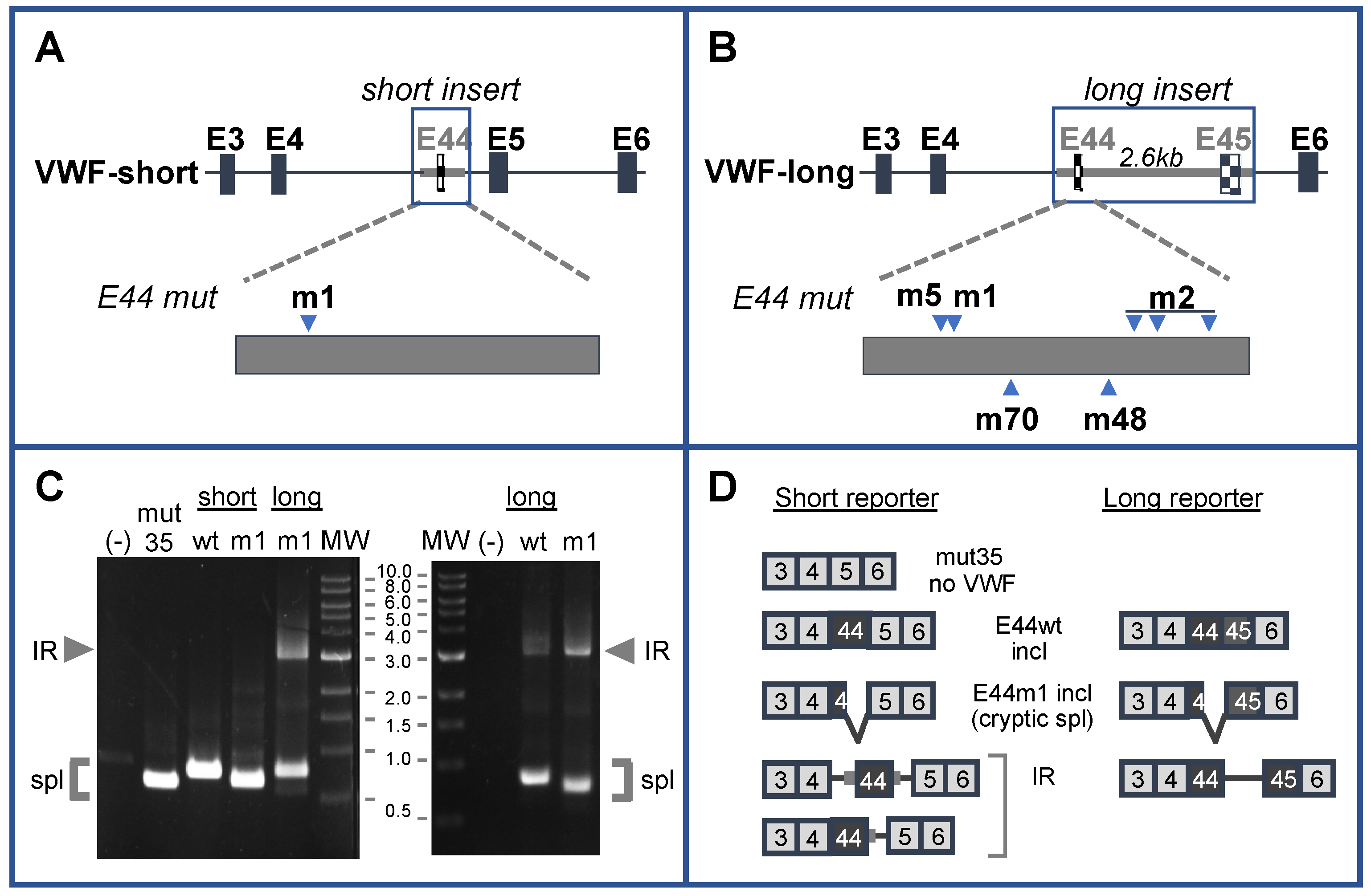
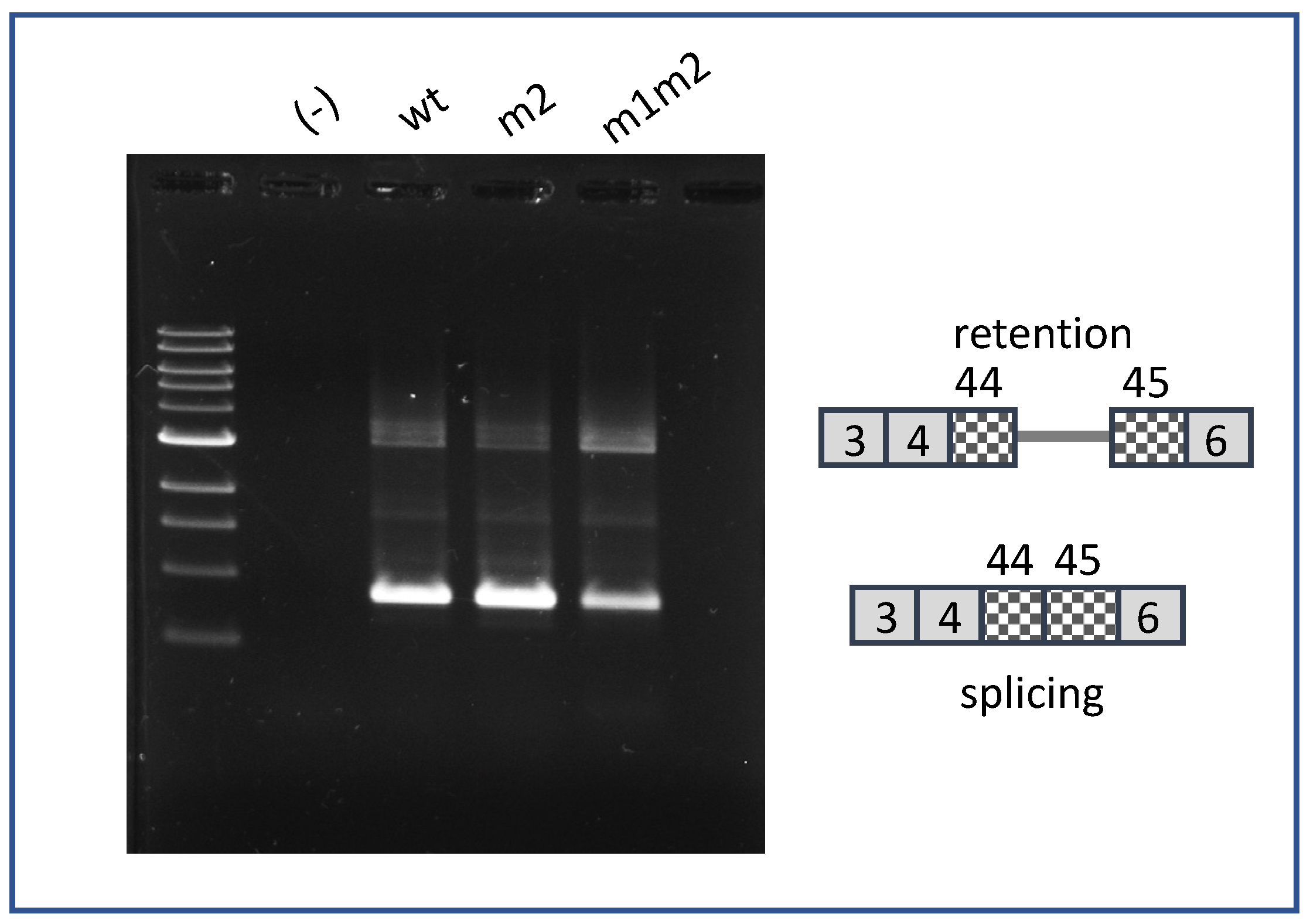
1. Introduction
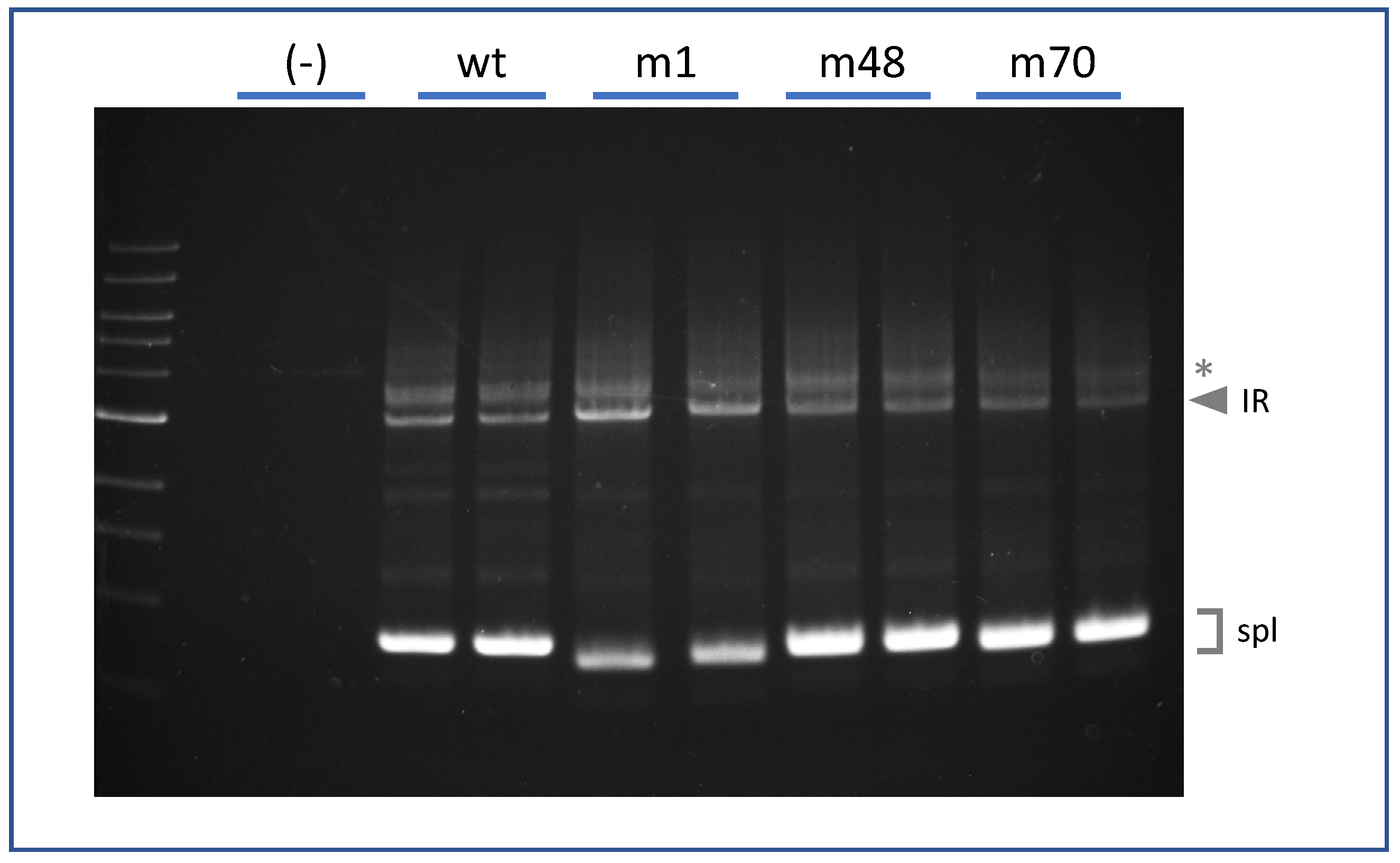
2. Results
3. Discussion
4. Materials and Methods
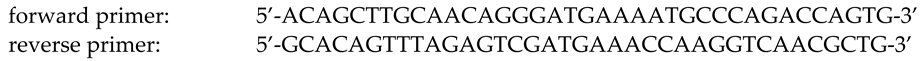


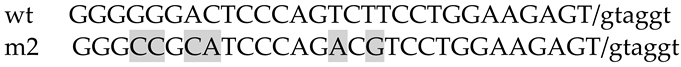
Funding
Acknowledgments
Conflicts of Interest
References
- Krawczak, M.; Reiss, J.; Cooper, D.N. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: Causes and consequences. Hum. Genet. 1992, 90, 41–54. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Bigas, N.; Audit, B.; Ouzounis, C.; Parra, G.; Guigo, R. Are splicing mutations the most frequent cause of hereditary disease? FEBS Lett. 2005, 579, 1900–1903. [Google Scholar] [CrossRef]
- Lord, J.; Baralle, D. Splicing in the Diagnosis of Rare Disease: Advances and Challenges. Front. Genet. 2021, 12, 689892. [Google Scholar] [CrossRef] [PubMed]
- Casadei, S.; Gulsuner, S.; Shirts, B.H.; Mandell, J.B.; Kortbawi, H.M.; Norquist, B.S.; Swisher, E.M.; Lee, M.K.; Goldberg, Y.; O’Connor, R.; et al. Characterization of splice-altering mutations in inherited predisposition to cancer. Proc. Natl. Acad. Sci. USA 2019, 116, 26798–26807. [Google Scholar] [CrossRef]
- Cartegni, L.; Chew, S.L.; Krainer, A.R. Listening to silence and understanding nonsense: Exonic mutations that affect splicing. Nat. Rev. Genet. 2002, 3, 285–298. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Rio, D.C. Mechanisms and Regulation of Alternative Pre-mRNA Splicing. Annu. Rev. Biochem. 2015, 84, 291–323. [Google Scholar] [CrossRef]
- Hershberger, C.E.; Moyer, D.C.; Adema, V.; Kerr, C.M.; Walter, W.; Hutter, S.; Meggendorfer, M.; Baer, C.; Kern, W.; Nadarajah, N.; et al. Complex landscape of alternative splicing in myeloid neoplasms. Leukemia 2021, 35, 1108–1120. [Google Scholar] [CrossRef]
- Bonnal, S.C.; Lopez-Oreja, I.; Valcarcel, J. Roles and mechanisms of alternative splicing in cancer—Implications for care. Nat. Rev. Clin. Oncol. 2020, 17, 457–474. [Google Scholar] [CrossRef]
- Shepard, P.J.; Hertel, K.J. Conserved RNA secondary structures promote alternative splicing. RNA 2008, 14, 1463–1469. [Google Scholar] [CrossRef] [PubMed]
- Grover, A.; Houlden, H.; Baker, M.; Adamson, J.; Lewis, J.; Prihar, G.; Pickering-Brown, S.; Duff, K.; Hutton, M. 5’ splice site mutations in tau associated with the inherited dementia FTDP-17 affect a stem-loop structure that regulates alternative splicing of exon 10. J. Biol. Chem. 1999, 274, 15134–15143. [Google Scholar] [CrossRef]
- Varani, L.; Hasegawa, M.; Spillantini, M.G.; Smith, M.J.; Murrell, J.R.; Ghetti, B.; Klug, A.; Goedert, M.; Varani, G. Structure of tau exon 10 splicing regulatory element RNA and destabilization by mutations of frontotemporal dementia and parkinsonism linked to chromosome 17. Proc. Natl. Acad. Sci. USA 1999, 96, 8229–8234. [Google Scholar] [CrossRef] [PubMed]
- Bartys, N.; Kierzek, R.; Lisowiec-Wachnicka, J. The regulation properties of RNA secondary structure in alternative splicing. Biochim. Biophys. Acta. Gene. Regul. Mech. 2019, 1862, 194401. [Google Scholar] [CrossRef] [PubMed]
- Shilo, A.; Tosto, F.A.; Rausch, J.W.; Le Grice, S.F.J.; Misteli, T. Interplay of primary sequence, position and secondary RNA structure determines alternative splicing of LMNA in a pre-mature aging syndrome. Nucleic Acids Res. 2019, 47, 5922–5935. [Google Scholar] [CrossRef]
- Taliaferro, J.M.; Lambert, N.J.; Sudmant, P.H.; Dominguez, D.; Merkin, J.J.; Alexis, M.S.; Bazile, C.; Burge, C.B. RNA Sequence Context Effects Measured In Vitro Predict In Vivo Protein Binding and Regulation. Mol. Cell 2016, 64, 294–306. [Google Scholar] [CrossRef]
- Jacob, A.G.; Smith, C.W.J. Intron retention as a component of regulated gene expression programs. Hum. Genet. 2017, 136, 1043–1057. [Google Scholar] [CrossRef] [PubMed]
- Monteuuis, G.; Wong, J.J.L.; Bailey, C.G.; Schmitz, U.; Rasko, J.E.J. The changing paradigm of intron retention: Regulation, ramifications and recipes. Nucleic Acids Res. 2019, 47, 11497–11513. [Google Scholar] [CrossRef]
- Grabski, D.F.; Broseus, L.; Kumari, B.; Rekosh, D.; Hammarskjold, M.L.; Ritchie, W. Intron retention and its impact on gene expression and protein diversity: A review and a practical guide. Wiley Interdiscip. Rev. RNA 2021, 12, e1631. [Google Scholar] [CrossRef]
- Yao, J.; Ding, D.; Li, X.; Shen, T.; Fu, H.; Zhong, H.; Wei, G.; Ni, T. Prevalent intron retention fine-tunes gene expression and contributes to cellular senescence. Aging Cell 2020, 19, e13276. [Google Scholar] [CrossRef]
- Boothby, T.C.; Zipper, R.S.; van der Weele, C.M.; Wolniak, S.M. Removal of retained introns regulates translation in the rapidly developing gametophyte of Marsilea vestita. Dev. Cell 2013, 24, 517–529. [Google Scholar] [CrossRef]
- Mauger, O.; Scheiffele, P. Beyond proteome diversity: Alternative splicing as a regulator of neuronal transcript dynamics. Curr. Opin. Neurobiol. 2017, 45, 162–168. [Google Scholar] [CrossRef]
- Ninomiya, K.; Kataoka, N.; Hagiwara, M. Stress-responsive maturation of Clk1/4 pre-mRNAs promotes phosphorylation of SR splicing factor. J. Cell Biol. 2011, 195, 27–40. [Google Scholar] [CrossRef] [PubMed]
- Dvinge, H.; Bradley, R.K. Widespread intron retention diversifies most cancer transcriptomes. Genome Med. 2015, 7, 45. [Google Scholar] [CrossRef]
- Jung, H.; Lee, D.; Lee, J.; Park, D.; Kim, Y.J.; Park, W.-Y.; Hong, D.; Park, P.J.; Lee, E. Intron retention is a widespread mechanism of tumor-suppressor inactivation. Nat. Genet. 2015, 47, 1242–1248. [Google Scholar] [CrossRef] [PubMed]
- Sznajder, L.J.; Thomas, J.D.; Carrell, E.M.; Reid, T.; McFarland, K.N.; Cleary, J.D.; Oliveira, R.; Nutter, C.A.; Bhatt, K.; Sobczak, K.; et al. Intron retention induced by microsatellite expansions as a disease biomarker. Proc. Natl. Acad. Sci. USA 2018, 115, 4234–4239. [Google Scholar] [CrossRef]
- Inoue, D.; Polaski, J.T.; Taylor, J.; Castel, P.; Chen, S.; Kobayashi, S.; Hogg, S.J.; Hayashi, Y.; Pineda, J.M.B.; El Marabti, E.; et al. Minor intron retention drives clonal hematopoietic disorders and diverse cancer predisposition. Nat. Genet. 2021, 53, 707–718. [Google Scholar] [CrossRef]
- Wang, Q.; Conlon, E.G.; Manley, J.L.; Rio, D.C. Widespread intron retention impairs protein homeostasis in C9orf72 ALS brains. Genome Res. 2020, 30, 1705–1715. [Google Scholar] [CrossRef] [PubMed]
- Li, H.D.; Funk, C.C.; McFarland, K.; Dammer, E.B.; Allen, M.; Carrasquillo, M.M.; Levites, Y.; Chakrabarty, P.; Burgess, J.D.; Wang, X.; et al. Integrative functional genomic analysis of intron retention in human and mouse brain with Alzheimer’s disease. Alzheimers Dement. 2021, 17, 984–1004. [Google Scholar] [CrossRef] [PubMed]
- Luisier, R.; Tyzack, G.E.; Hall, C.E.; Mitchell, J.S.; Devine, H.; Taha, D.M.; Malik, B.; Meyer, I.; Greensmith, L.; Newcombe, J.; et al. Intron retention and nuclear loss of SFPQ are molecular hallmarks of ALS. Nat. Commun. 2018, 9, 2010. [Google Scholar] [CrossRef] [PubMed]
- Jangi, M.; Sharp, P.A. Building robust transcriptomes with master splicing factors. Cell 2014, 159, 487–498. [Google Scholar] [CrossRef]
- Humphrey, J.; Birsa, N.; Milioto, C.; McLaughlin, M.; Ule, A.M.; Robaldo, D.; Eberle, A.B.; Krauchi, R.; Bentham, M.; Brown, A.L.; et al. FUS ALS-causative mutations impair FUS autoregulation and splicing factor networks through intron retention. Nucleic Acids Res. 2020, 48, 6889–6905. [Google Scholar] [CrossRef] [PubMed]
- Yadegari, H.; Biswas, A.; Akhter, M.S.; Driesen, J.; Ivaskevicius, V.; Marquardt, N.; Oldenburg, J. Intron retention resulting from a silent mutation in the VWF gene that structurally influences the 5′ splice site. Blood 2016, 128, 2144–2152. [Google Scholar] [CrossRef]
- Parra, M.; Booth, B.W.; Weiszmann, R.; Yee, B.; Yeo, G.W.; Brown, J.B.; Celniker, S.E.; Conboy, J.G. An important class of intron retention events in human erythroblasts is regulated by cryptic exons proposed to function as splicing decoys. RNA 2018, 24, 1255–1265. [Google Scholar] [CrossRef] [PubMed]
- Pirnie, S.P.; Osman, A.; Zhu, Y.; Carmichael, G.G. An Ultraconserved Element (UCE) controls homeostatic splicing of ARGLU1 mRNA. Nucleic Acids Res. 2017, 45, 3473–3486. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Siebel, C.W.; Fresco, L.D.; Rio, D.C. The mechanism of somatic inhibition of Drosophila P-element pre-mRNA splicing: Multiprotein complexes at an exon pseudo-5’ splice site control U1 snRNP binding. Genes. Dev. 1992, 6, 1386–1401. [Google Scholar] [CrossRef] [PubMed]
- Yeo, G.; Burge, C.B. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004, 11, 377–394. [Google Scholar] [CrossRef]
- Yu, Y.; Maroney, P.A.; Denker, J.A.; Zhang, X.H.; Dybkov, O.; Luhrmann, R.; Jankowsky, E.; Chasin, L.A.; Nilsen, T.W. Dynamic regulation of alternative splicing by silencers that modulate 5’ splice site competition. Cell 2008, 135, 1224–1236. [Google Scholar] [CrossRef] [PubMed]
- Roca, X.; Krainer, A.R.; Eperon, I.C. Pick one, but be quick: 5’ splice sites and the problems of too many choices. Genes. Dev. 2013, 27, 129–144. [Google Scholar] [CrossRef] [PubMed]
- Borras, N.; Orriols, G.; Batlle, J.; Perez-Rodriguez, A.; Fidalgo, T.; Martinho, P.; Lopez-Fernandez, M.F.; Rodriguez-Trillo, A.; Loures, E.; Parra, R.; et al. Unraveling the effect of silent, intronic and missense mutations on VWF splicing: Contribution of next generation sequencing in the study of mRNA. Haematologica 2019, 104, 587–598. [Google Scholar] [CrossRef]
- de Jong, A.; Eikenboom, J. Von Willebrand disease mutation spectrum and associated mutation mechanisms. Thromb. Res. 2017, 159, 65–75. [Google Scholar] [CrossRef]
- Bao, C.; Zhu, M.; Nykonchuk, I.; Wakabayashi, H.; Mathews, D.H.; Ermolenko, D.N. Specific length and structure rather than high thermodynamic stability enable regulatory mRNA stem-loops to pause translation. bioRxiv 2021. [Google Scholar] [CrossRef]
- Horan, L.; Yasuhara, J.C.; Kohlstaedt, L.A.; Rio, D.C. Biochemical identification of new proteins involved in splicing repression at the Drosophila P-element exonic splicing silencer. Genes. Dev. 2015, 29, 2298–2311. [Google Scholar] [CrossRef] [PubMed]
- Labourier, E.; Adams, M.D.; Rio, D.C. Modulation of P-element pre-mRNA splicing by a direct interaction between PSI and U1 snRNP 70K protein. Mol. Cell 2001, 8, 363–373. [Google Scholar] [CrossRef]
- Siebel, C.W.; Kanaar, R.; Rio, D.C. Regulation of tissue-specific P-element pre-mRNA splicing requires the RNA-binding protein PSI. Genes Dev. 1994, 8, 1713–1725. [Google Scholar] [CrossRef]
- Nilsen, T.W.; Graveley, B.R. Expansion of the eukaryotic proteome by alternative splicing. Nature 2010, 463, 457–463. [Google Scholar] [CrossRef] [PubMed]
- Ohe, K.; Miyajima, S.; Tanaka, T.; Hamaguchi, Y.; Harada, Y.; Horita, Y.; Beppu, Y.; Ito, F.; Yamasaki, T.; Terai, H.; et al. HMGA1a Induces Alternative Splicing of the Estrogen Receptor-alphalpha Gene by Trapping U1 snRNP to an Upstream Pseudo-5’ Splice Site. Front. Mol. Biosci. 2018, 5, 52. [Google Scholar] [CrossRef] [PubMed]
- Shao, C.; Yang, B.; Wu, T.; Huang, J.; Tang, P.; Zhou, Y.; Zhou, J.; Qiu, J.; Jiang, L.; Li, H.; et al. Mechanisms for U2AF to define 3’ splice sites and regulate alternative splicing in the human genome. Nat. Struct. Mol. Biol. 2014, 21, 997–1005. [Google Scholar] [CrossRef]
- De Conti, L.; Baralle, M.; Buratti, E. Exon and intron definition in pre-mRNA splicing. Wiley Interdiscip. Rev. RNA 2013, 4, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Shiraishi, Y.; Kataoka, K.; Chiba, K.; Okada, A.; Kogure, Y.; Tanaka, H.; Ogawa, S.; Miyano, S. A comprehensive characterization of cis-acting splicing-associated variants in human cancer. Genome Res. 2018, 28, 1111–1125. [Google Scholar] [CrossRef]
- Pimentel, H.; Conboy, J.G.; Pacher, L. Keep Me Around: Intron Retention Detection and Analysis. arXiv 2015, arXiv:1510.00696. [Google Scholar]
- Middleton, R.; Gao, D.; Thomas, A.; Singh, B.; Au, A.; Wong, J.J.; Bomane, A.; Cosson, B.; Eyras, E.; Rasko, J.E.; et al. IRFinder: Assessing the impact of intron retention on mammalian gene expression. Genome Biol. 2017, 18, 51. [Google Scholar] [CrossRef]
- Ni, T.; Yang, W.; Han, M.; Zhang, Y.; Shen, T.; Nie, H.; Zhou, Z.; Dai, Y.; Yang, Y.; Liu, P.; et al. Global intron retention mediated gene regulation during CD4+ T cell activation. Nucleic Acids Res. 2016, 44, 6817–6829. [Google Scholar] [CrossRef] [PubMed]
- Tang, A.D.; Soulette, C.M.; van Baren, M.J.; Hart, K.; Hrabeta-Robinson, E.; Wu, C.J.; Brooks, A.N. Full-length transcript characterization of SF3B1 mutation in chronic lymphocytic leukemia reveals downregulation of retained introns. Nat. Commun. 2020, 11, 1438. [Google Scholar] [CrossRef] [PubMed]
- Varela, P.; Caldas, M.M.; Pesquero, J.B. Novel GLA Mutation Promotes Intron Inclusion Leading to Fabry Disease. Front. Genet. 2019, 10, 783. [Google Scholar] [CrossRef]
- Gibson, D.G. Enzymatic assembly of overlapping DNA fragments. Methods Enzymol. 2011, 498, 349–361. [Google Scholar] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Conboy, J.G. A Deep Exon Cryptic Splice Site Promotes Aberrant Intron Retention in a Von Willebrand Disease Patient. Int. J. Mol. Sci. 2021, 22, 13248. https://doi.org/10.3390/ijms222413248
Conboy JG. A Deep Exon Cryptic Splice Site Promotes Aberrant Intron Retention in a Von Willebrand Disease Patient. International Journal of Molecular Sciences. 2021; 22(24):13248. https://doi.org/10.3390/ijms222413248
Chicago/Turabian StyleConboy, John G. 2021. "A Deep Exon Cryptic Splice Site Promotes Aberrant Intron Retention in a Von Willebrand Disease Patient" International Journal of Molecular Sciences 22, no. 24: 13248. https://doi.org/10.3390/ijms222413248
APA StyleConboy, J. G. (2021). A Deep Exon Cryptic Splice Site Promotes Aberrant Intron Retention in a Von Willebrand Disease Patient. International Journal of Molecular Sciences, 22(24), 13248. https://doi.org/10.3390/ijms222413248




