The SR Splicing Factors: Providing Perspectives on Their Evolution, Expression, Alternative Splicing, and Function in Populus trichocarpa
Abstract
:1. Introduction
2. Results
2.1. Identification of PtSR Family Genes and Their Characteristics
2.2. Phylogenetic and Architectural Analysis of PtSR Family Genes
2.3. The Expansion History of the PtSR Gene Family in P. trichocarpa
2.4. GO Term Enrichment and Promoter Cis-element Analysis of PtSR Genes
2.5. Constitutive and Abundant Expression Patterns of PtSR Genes in P. trichocarpa
2.6. Perturbation in PtSR Gene Expressions by Hormones and Abiotic Stresses
2.7. Conserved Alternative Splicing of PtSR Genes in P. trichocarpa Tissues and Their Perturbance by Hormones and Abiotic Stresses
2.8. Overexpression of PtSCL30 Decreased the Freezing Tolerance of Arabidopsis
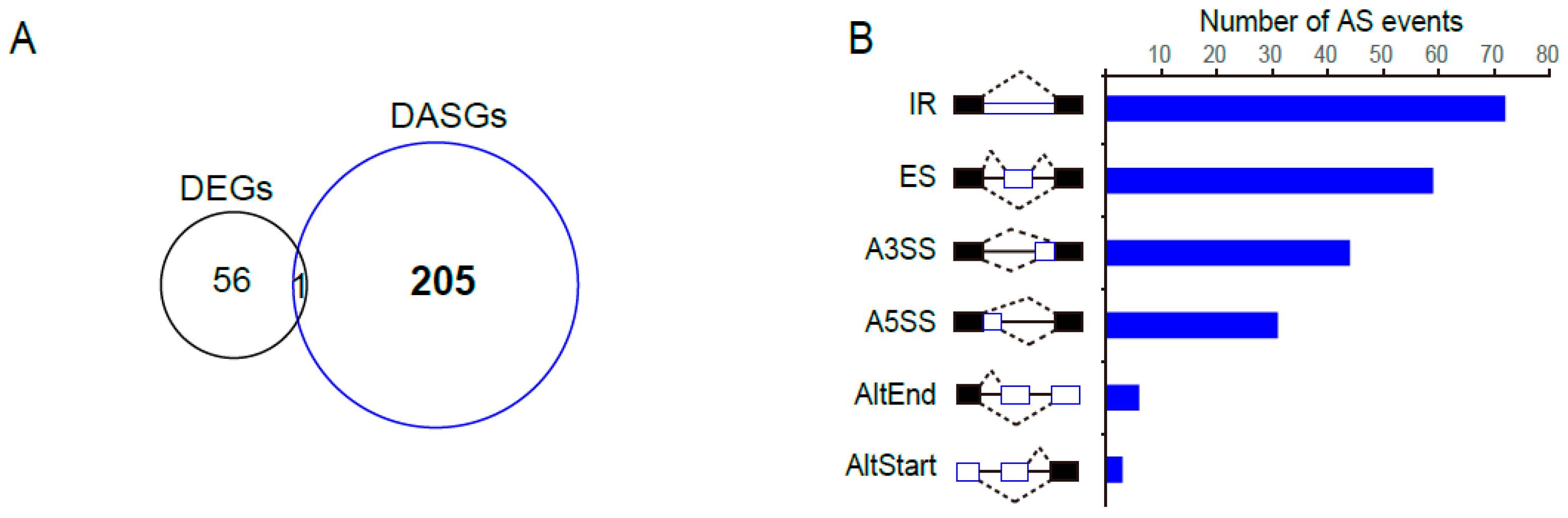
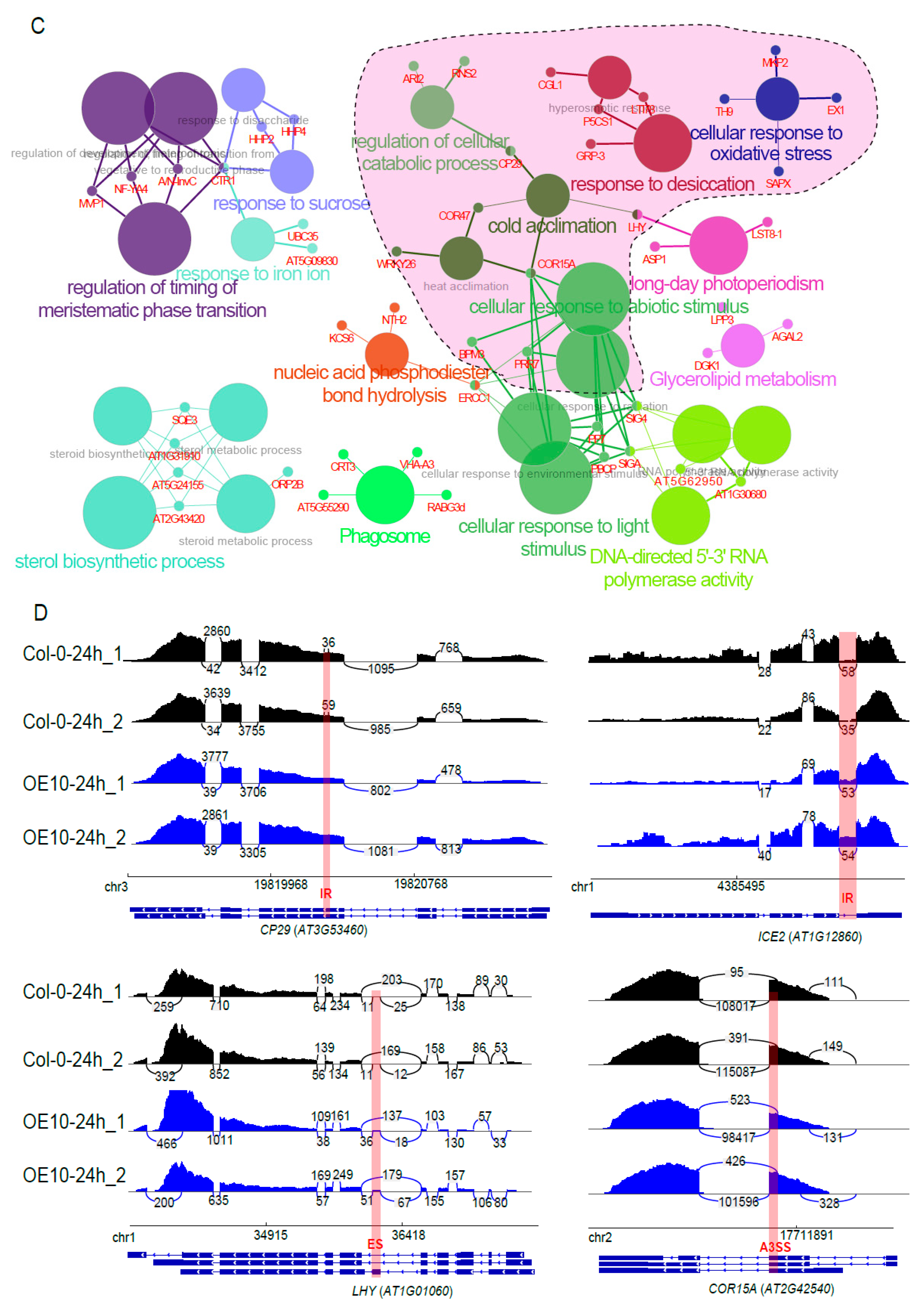
2.9. PtSCL30 Overexpression Affected the Alternative Splicing of Hundreds of Genes, including Cold-Responsive Genes
2.10. PtSCL30 Overexpression Were Hypersensitive to Salt Stress
3. Discussion
4. Materials and Methods
4.1. Identification and Basic Features of PtSR Family Genes
4.2. Phylogenetic Analysis, Domain Identification, and Architecture Visualization of PtSR Genes
4.3. Chromosomal Localization and Expansion History of PtSR Genes
4.4. GO-Term Enrichment and Promoter Cis-Element Analysis of PtSR Genes
4.5. Expression Profile Analysis of PtSR Genes
4.6. Quantitative RT-PCR (qRT-PCR) Analysis
4.7. Alternative Splicing (AS) Pattern Analysis of PtSR Genes
4.8. Plasmid Construction and Plant Transformation
4.9. Abiotic Tolerance and Seed Germination Assays
4.10. High-Throughput mRNA Sequencing and Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| SR | serine/arginine-rich |
| AS | alternative splicing |
| GO | gene ontology |
| snRNP | small nuclear ribonucleoprotein particle |
| CDS | coding sequences |
| GFF | genomic structure information |
| pI | protein isoelectric point |
| ML | maximum likelihood |
| HMM | hidden Markov model |
| Ks | synonymous substitutions rate |
| EF | enrichment factor |
| MW | molecular weight |
| ZF | zinc finger |
| WGD | whole-genome duplication |
| ABA | abscisic acid |
| MeJA | methyl jasmonate |
| SA | salicylic acid |
| GA | gibberellin |
| MS | Murashige and Skoog |
| DEGs | differentially expressed genes |
| DASGs | differentially alternatively spliced genes |
References
- Reddy, A.S. Plant serine/arginine-rich proteins and their role in pre-mRNA splicing. Trends Plant Sci. 2004, 9, 541–547. [Google Scholar] [CrossRef]
- Reddy, A.S.; Marquez, Y.; Kalyna, M.; Barta, A. Complexity of the alternative splicing landscape in plants. Plant Cell 2013, 25, 3657–3683. [Google Scholar] [CrossRef] [Green Version]
- Wang, E.T.; Sandberg, R.; Luo, S.; Khrebtukova, I.; Zhang, L.; Mayr, C.; Kingsmore, S.F.; Schroth, G.P.; Burge, C.B. Alternative isoform regulation in human tissue transcriptomes. Nature 2008, 456, 470–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nilsen, T.W.; Graveley, B.R. Expansion of the eukaryotic proteome by alternative splicing. Nature 2010, 463, 457–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, F.Y.; Chen, M.X.; Ye, N.H.; Shi, L.; Ma, K.L.; Yang, J.F.; Cao, Y.Y.; Zhang, Y.; Yoshida, T.; Fernie, A.R.; et al. Proteogenomic analysis reveals alternative splicing and translation as part of the abscisic acid response in Arabidopsis seedlings. Plant J. 2017, 91, 518–533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, M.X.; Zhu, F.Y.; Gao, B.; Ma, K.L.; Zhang, Y.; Fernie, A.R.; Chen, X.; Dai, L.; Ye, N.H.; Zhang, X.; et al. Full-Length Transcript-Based Proteogenomics of Rice Improves Its Genome and Proteome Annotation. Plant Physiol. 2020, 182, 1510–1526. [Google Scholar] [CrossRef] [PubMed]
- Matlin, A.J.; Clark, F.; Smith, C.W. Understanding alternative splicing: Towards a cellular code. Nat. Rev. Mol. Cell Biol. 2005, 6, 386–398. [Google Scholar] [CrossRef] [PubMed]
- Ner-Gaon, H.; Halachmi, R.; Savaldi-Goldstein, S.; Rubin, E.; Ophir, R.; Fluhr, R. Intron retention is a major phenomenon in alternative splicing in Arabidopsis. Plant J. 2004, 39, 877–885. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.; Shai, O.; Lee, L.J.; Frey, B.J.; Blencowe, B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008, 40, 1413–1415. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Zhou, Z.; Wang, Z.; Li, W.; Fang, C.; Wu, M.; Ma, Y.; Liu, T.; Kong, L.A.; Peng, D.L.; et al. Global dissection of alternative splicing in paleopolyploid soybean. Plant Cell 2014, 26, 996–1008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kornblihtt, A.R.; Schor, I.E.; Allo, M.; Dujardin, G.; Petrillo, E.; Munoz, M.J. Alternative splicing: A pivotal step between eukaryotic transcription and translation. Nat. Rev. Mol. Cell Biol. 2013, 14, 153–165. [Google Scholar] [CrossRef]
- Kim, H.K.; Pham, M.H.C.; Ko, K.S.; Rhee, B.D.; Han, J. Alternative splicing isoforms in health and disease. Pflugers Arch. 2018, 470, 995–1016. [Google Scholar] [CrossRef]
- Mastrangelo, A.M.; Marone, D.; Laido, G.; De Leonardis, A.M.; De Vita, P. Alternative splicing: Enhancing ability to cope with stress via transcriptome plasticity. Plant Sci. 2012, 185–186, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Calixto, C.P.G.; Guo, W.; James, A.B.; Tzioutziou, N.A.; Entizne, J.C.; Panter, P.E.; Knight, H.; Nimmo, H.G.; Zhang, R.; Brown, J.W.S. Rapid and Dynamic Alternative Splicing Impacts the Arabidopsis Cold Response Transcriptome. Plant Cell 2018, 30, 1424–1444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, J.; Li, J.; Gao, Z.; Lu, Y.; Yu, J.; Zheng, Q.; Yan, S.; Zhang, W.; He, H.; Ma, L.; et al. SKIP Confers Osmotic Tolerance during Salt Stress by Controlling Alternative Gene Splicing in Arabidopsis. Mol. Plant 2015, 8, 1038–1052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, J.; Liu, X.; Liu, C.; Liu, G.; Li, S.; Wang, L. Integrating Omics and Alternative Splicing Reveals Insights into Grape Response to High Temperature. Plant Physiol. 2017, 173, 1502–1518. [Google Scholar] [CrossRef] [Green Version]
- Reddy, A.S. Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu. Rev. Plant Biol. 2007, 58, 267–294. [Google Scholar] [CrossRef] [Green Version]
- Sanchez, S.E.; Petrillo, E.; Beckwith, E.J.; Zhang, X.; Rugnone, M.L.; Hernando, C.E.; Cuevas, J.C.; Godoy Herz, M.A.; Depetris-Chauvin, A.; Simpson, C.G.; et al. A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature 2010, 468, 112–116. [Google Scholar] [CrossRef] [Green Version]
- Cui, Z.; Tong, A.; Huo, Y.; Yan, Z.; Yang, W.; Yang, X.; Wang, X.X. SKIP controls flowering time via the alternative splicing of SEF pre-mRNA in Arabidopsis. BMC Biol. 2017, 15, 80. [Google Scholar] [CrossRef] [Green Version]
- Martin-Trillo, M.; Grandio, E.G.; Serra, F.; Marcel, F.; Rodriguez-Buey, M.L.; Schmitz, G.; Theres, K.; Bendahmane, A.; Dopazo, H.; Cubas, P. Role of tomato BRANCHED1-like genes in the control of shoot branching. Plant J. 2011, 67, 701–714. [Google Scholar] [CrossRef]
- Will, C.L.; Luhrmann, R. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 2011, 3, a003707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelemen, O.; Convertini, P.; Zhang, Z.; Wen, Y.; Shen, M.; Falaleeva, M.; Stamm, S. Function of alternative splicing. Gene 2013, 514, 1–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Long, J.C.; Caceres, J.F. The SR protein family of splicing factors: Master regulators of gene expression. Biochem. J. 2009, 417, 15–27. [Google Scholar] [CrossRef] [Green Version]
- Luo, C.; Cheng, Y.; Liu, Y.; Chen, L.; Liu, L.; Wei, N.; Xie, Z.; Wu, W.; Feng, Y. SRSF2 Regulates Alternative Splicing to Drive Hepatocellular Carcinoma Development. Cancer Res. 2017, 77, 1168–1178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barta, A.; Kalyna, M.; Reddy, A.S. Implementing a rational and consistent nomenclature for serine/arginine-rich protein splicing factors (SR proteins) in plants. Plant Cell 2010, 22, 2926–2929. [Google Scholar] [CrossRef] [PubMed]
- Iida, K.; Go, M. Survey of conserved alternative splicing events of mRNAs encoding SR proteins in land plants. Mol. Biol. Evol. 2006, 23, 1085–1094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butt, H.; Piatek, A.; Li, L.; Reddy, A.S.N.; Mahfouz, M.M. Multiplex CRISPR Mutagenesis of the Serine/Arginine-Rich (SR) Gene Family in Rice. Genes 2019, 10, 596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Huang, S.; Jiang, M.; Chen, Y.; XuHan, X.; Zhang, Z.; Lin, Y.; Lai, Z. Genome-wide identification and expression analysis of the SR gene family in longan (Dimocarpus longan Lour.). PLoS ONE 2020, 15, e0238032. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Li, J.; Liu, Y.; Li, H. Genome-Wide Analysis of Serine/Arginine-Rich Protein Family in Wheat and Brachypodium distachyon. Plants 2019, 8, 188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barta, A.; Kalyna, M.; Lorkovic, Z.J. Plant SR proteins and their functions. Curr. Top. Microbiol. Immunol. 2008, 326, 83–102. [Google Scholar]
- Morton, M.; AlTamimi, N.; Butt, H.; Reddy, A.S.N.; Mahfouz, M. Serine/Arginine-rich protein family of splicing regulators: New approaches to study splice isoform functions. Plant Sci. 2019, 283, 127–134. [Google Scholar] [CrossRef]
- Lopato, S.; Kalyna, M.; Dorner, S.; Kobayashi, R.; Krainer, A.R.; Barta, A. atSRp30, one of two SF2/ASF-like proteins from Arabidopsis thaliana, regulates splicing of specific plant genes. Genes Dev. 1999, 13, 987–1001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, Q.; Xia, X.; Sun, Z.; Fang, Y. Depletion of Arabidopsis SC35 and SC35-like serine/arginine-rich proteins affects the transcription and splicing of a subset of genes. PLoS Genet. 2017, 13, e1006663. [Google Scholar] [CrossRef] [PubMed]
- Isshiki, M.; Tsumoto, A.; Shimamoto, K. The serine/arginine-rich protein family in rice plays important roles in constitutive and alternative splicing of pre-mRNA. Plant Cell 2006, 18, 146–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Filichkin, S.A.; Priest, H.D.; Givan, S.A.; Shen, R.; Bryant, D.W.; Fox, S.E.; Wong, W.K.; Mockler, T.C. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 2010, 20, 45–58. [Google Scholar] [CrossRef] [Green Version]
- Jansson, S.; Douglas, C.J. Populus: A model system for plant biology. Annu. Rev. Plant Biol. 2007, 58, 435–458. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [Green Version]
- Reddy, A.S.; Shad Ali, G. Plant serine/arginine-rich proteins: Roles in precursor messenger RNA splicing, plant development, and stress responses. Wiley Interdiscip. Rev. RNA 2011, 2, 875–889. [Google Scholar] [CrossRef]
- Duque, P. A role for SR proteins in plant stress responses. Plant Signal. Behav. 2011, 6, 49–54. [Google Scholar] [CrossRef] [Green Version]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Ren, R.; Wang, H.; Guo, C.; Zhang, N.; Zeng, L.; Chen, Y.; Ma, H.; Qi, J. Widespread Whole Genome Duplications Contribute to Genome Complexity and Species Diversity in Angiosperms. Mol. Plant 2018, 11, 414–428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tuskan, G.A.; Difazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, N.; Ralph, S.; Rombauts, S.; Salamov, A.; et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 2006, 313, 1596–1604. [Google Scholar] [PubMed] [Green Version]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Rauch, H.B.; Patrick, T.L.; Klusman, K.M.; Battistuzzi, F.U.; Mei, W.; Brendel, V.P.; Lal, S.K. Discovery and expression analysis of alternative splicing events conserved among plant SR proteins. Mol. Biol. Evol. 2014, 31, 605–613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Y.; Ding, Y.; Yang, S. Molecular Regulation of CBF Signaling in Cold Acclimation. Trends Plant Sci. 2018, 23, 623–637. [Google Scholar] [CrossRef] [PubMed]
- Lazar, G.; Schaal, T.; Maniatis, T.; Goodman, H.M. Identification of a plant serine-arginine-rich protein similar to the mammalian splicing factor SF2/ASF. Proc. Natl. Acad. Sci. USA 1995, 92, 7672–7676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palusa, S.G.; Ali, G.S.; Reddy, A.S. Alternative splicing of pre-mRNAs of Arabidopsis serine/arginine-rich proteins: Regulation by hormones and stresses. Plant J. 2007, 49, 1091–1107. [Google Scholar] [CrossRef]
- Tanabe, N.; Yoshimura, K.; Kimura, A.; Yabuta, Y.; Shigeoka, S. Differential expression of alternatively spliced mRNAs of Arabidopsis SR protein homologs, atSR30 and atSR45a, in response to environmental stress. Plant Cell Physiol. 2007, 48, 1036–1049. [Google Scholar] [CrossRef] [Green Version]
- Cruz, T.M.; Carvalho, R.F.; Richardson, D.N.; Duque, P. Abscisic acid (ABA) regulation of Arabidopsis SR protein gene expression. Int. J. Mol. Sci. 2014, 15, 17541–17564. [Google Scholar] [CrossRef] [Green Version]
- Day, I.S.; Golovkin, M.; Palusa, S.G.; Link, A.; Ali, G.S.; Thomas, J.; Richardson, D.N.; Reddy, A.S. Interactions of SR45, an SR-like protein, with spliceosomal proteins and an intronic sequence: Insights into regulated splicing. Plant J. 2012, 71, 936–947. [Google Scholar] [CrossRef]
- Albaqami, M.; Laluk, K.; Reddy, A.S.N. The Arabidopsis splicing regulator SR45 confers salt tolerance in a splice isoform-dependent manner. Plant Mol. Biol. 2019, 100, 379–390. [Google Scholar] [CrossRef]
- Rausin, G.; Tillemans, V.; Stankovic, N.; Hanikenne, M.; Motte, P. Dynamic nucleocytoplasmic shuttling of an Arabidopsis SR splicing factor: Role of the RNA-binding domains. Plant Physiol. 2010, 153, 273–284. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Du, B.; Liu, D.; Qi, X. Splicing factor SR34b mutation reduces cadmium tolerance in Arabidopsis by regulating iron-regulated transporter 1 gene. Biochem. Biophys. Res. Commun. 2014, 455, 312–317. [Google Scholar] [CrossRef]
- Chen, T.; Cui, P.; Chen, H.; Ali, S.; Zhang, S.; Xiong, L. A KH-domain RNA-binding protein interacts with FIERY2/CTD phosphatase-like 1 and splicing factors and is important for pre-mRNA splicing in Arabidopsis. PLoS Genet 2013, 9, e1003875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, J.; Ma, S.; Zhang, Y.; Wang, D.; Gao, S.; Zhang, Z.Y. Genome-Wide Identification of Cassava Serine/Arginine-Rich Proteins: Insights into Alternative Splicing of Pre-mRNAs and Response to Abiotic Stress. Plant Cell Physiol. 2020, 61, 178–191. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Sun, J.; Xu, P.; Zhang, R.; Li, L. Intron-mediated alternative splicing of WOOD-ASSOCIATED NAC TRANSCRIPTION FACTOR1B regulates cell wall thickening during fiber development in Populus species. Plant Physiol. 2014, 164, 765–776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forment, J.; Naranjo, M.A.; Roldan, M.; Serrano, R.; Vicente, O. Expression of Arabidopsis SR-like splicing proteins confers salt tolerance to yeast and transgenic plants. Plant J. 2002, 30, 511–519. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Huala, E.; Dickerman, A.W.; Garcia-Hernandez, M.; Weems, D.; Reiser, L.; LaFond, F.; Hanley, D.; Kiphart, D.; Zhuang, M.; Huang, W.; et al. The Arabidopsis Information Resource (TAIR): A comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res. 2001, 29, 102–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [Green Version]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A Toolkit Incorporating Gamma-Series Methods and Sliding Window Strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef] [Green Version]
- Huerta-Cepas, J.; Forslund, K.; Coelho, L.P.; Szklarczyk, D.; Jensen, L.J.; von Mering, C.; Bork, P. Fast Genome-Wide Functional Annotation through Orthology Assignment by eggNOG-Mapper. Mol. Biol. Evol. 2017, 34, 2115–2122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Filichkin, S.A.; Hamilton, M.; Dharmawardhana, P.D.; Singh, S.K.; Sullivan, C.; Ben-Hur, A.; Reddy, A.S.N.; Jaiswal, P. Abiotic Stresses Modulate Landscape of Poplar Transcriptome via Alternative Splicing, Differential Intron Retention, and Isoform Ratio Switching. Front. Plant Sci. 2018, 9, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Ding, Y.; Shi, Y.; Zhang, X.; Gong, Z.; Yang, S. The cbfs triple mutants reveal the essential functions of CBFs in cold acclimation and allow the definition of CBF regulons in Arabidopsis. New Phytol. 2016, 212, 345–353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Wu, W.; Zong, J.; Wei, N.; Cheng, J.; Zhou, X.; Cheng, Y.; Chen, D.; Guo, Q.; Zhang, B.; Feng, Y. CASH: A constructing comprehensive splice site method for detecting alternative splicing events. Brief. Bioinform. 2017, 19, 905–917. [Google Scholar] [CrossRef]








Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, X.; Tan, L.; Wang, S.; Shen, Y.; Guo, L.; Ye, X.; Liu, S.; Feng, Y.; Wu, W. The SR Splicing Factors: Providing Perspectives on Their Evolution, Expression, Alternative Splicing, and Function in Populus trichocarpa. Int. J. Mol. Sci. 2021, 22, 11369. https://doi.org/10.3390/ijms222111369
Zhao X, Tan L, Wang S, Shen Y, Guo L, Ye X, Liu S, Feng Y, Wu W. The SR Splicing Factors: Providing Perspectives on Their Evolution, Expression, Alternative Splicing, and Function in Populus trichocarpa. International Journal of Molecular Sciences. 2021; 22(21):11369. https://doi.org/10.3390/ijms222111369
Chicago/Turabian StyleZhao, Xijuan, Lingling Tan, Shuo Wang, Yirong Shen, Liangyu Guo, Xiaoxue Ye, Shenkui Liu, Ying Feng, and Wenwu Wu. 2021. "The SR Splicing Factors: Providing Perspectives on Their Evolution, Expression, Alternative Splicing, and Function in Populus trichocarpa" International Journal of Molecular Sciences 22, no. 21: 11369. https://doi.org/10.3390/ijms222111369





