Instability of the NS1 Glycoprotein from La Reunion 2018 Dengue 2 Virus (Cosmopolitan-1 Genotype) in Huh7 Cells Is Due to Lysine Residues on Positions 272 and 324
Abstract
1. Introduction
2. Results
2.1. Expression of Recombinant SWIO DENV-2 NS1 Proteins in HEK-293T Cells
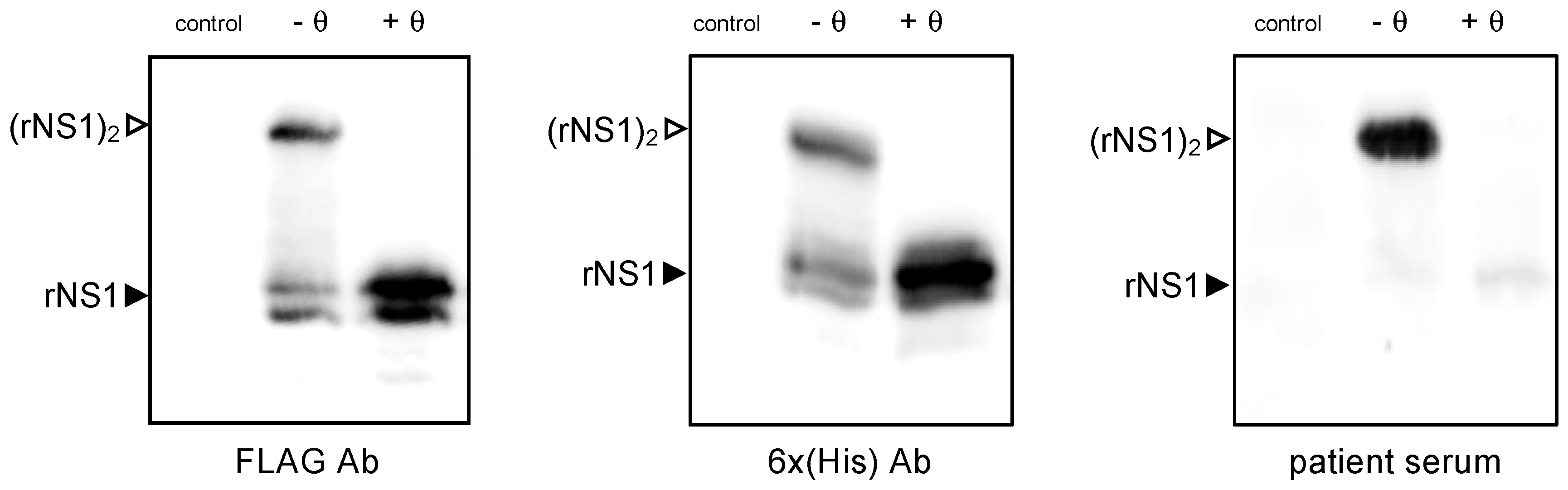
2.1.1. Antigenic Reactivity of SWIO DENV-2 rNS1 Protein
2.1.2. Expression of SWIO DENV-2 NS1 Monomeric and Dimeric Forms
2.2. Expression of Recombinant SWIO DENV-2 NS1 Proteins in Huh7 Cells
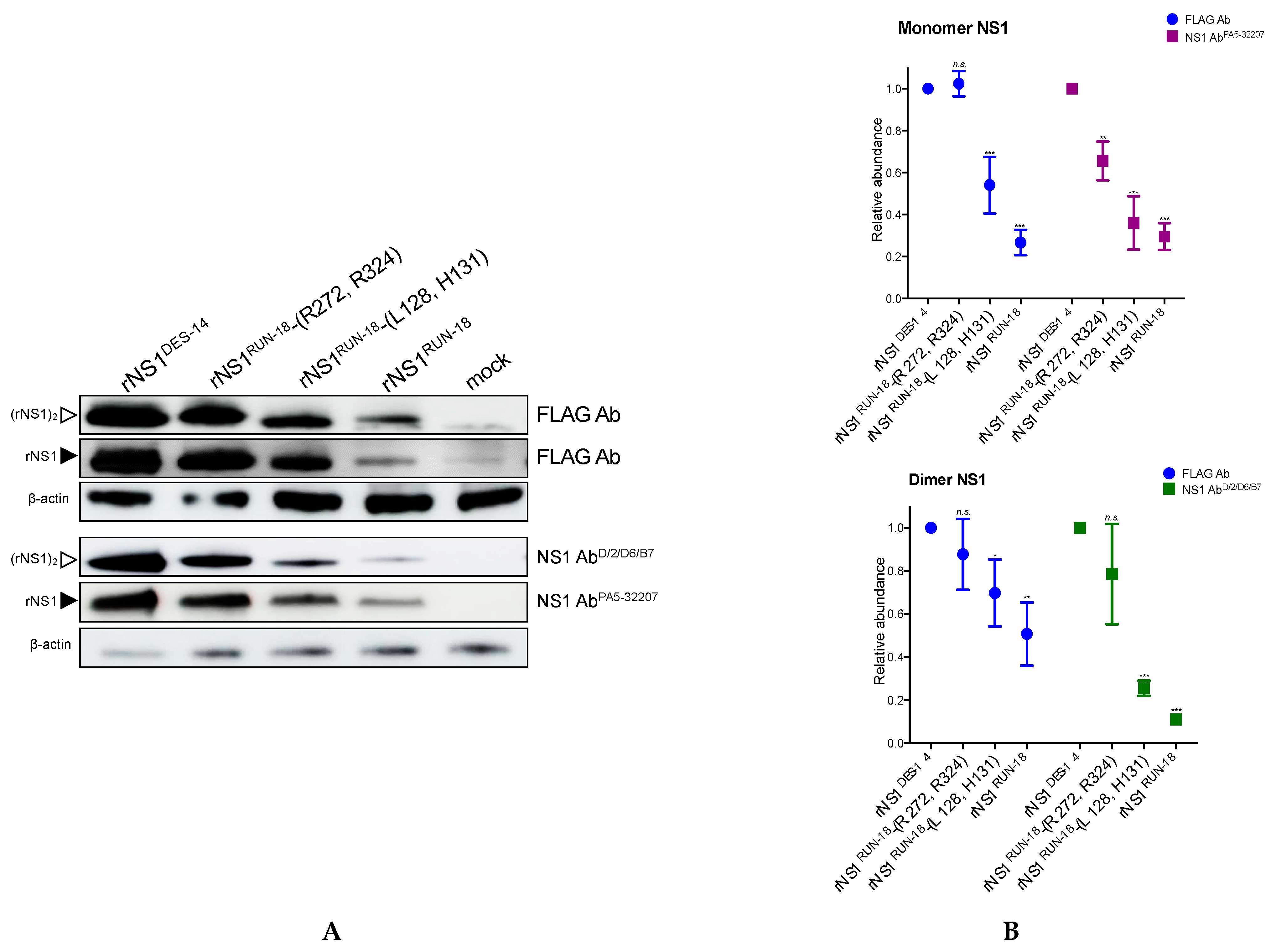
2.2.1. Expression of DES-14 rNS1 and RUN-18 rNS1 Proteins
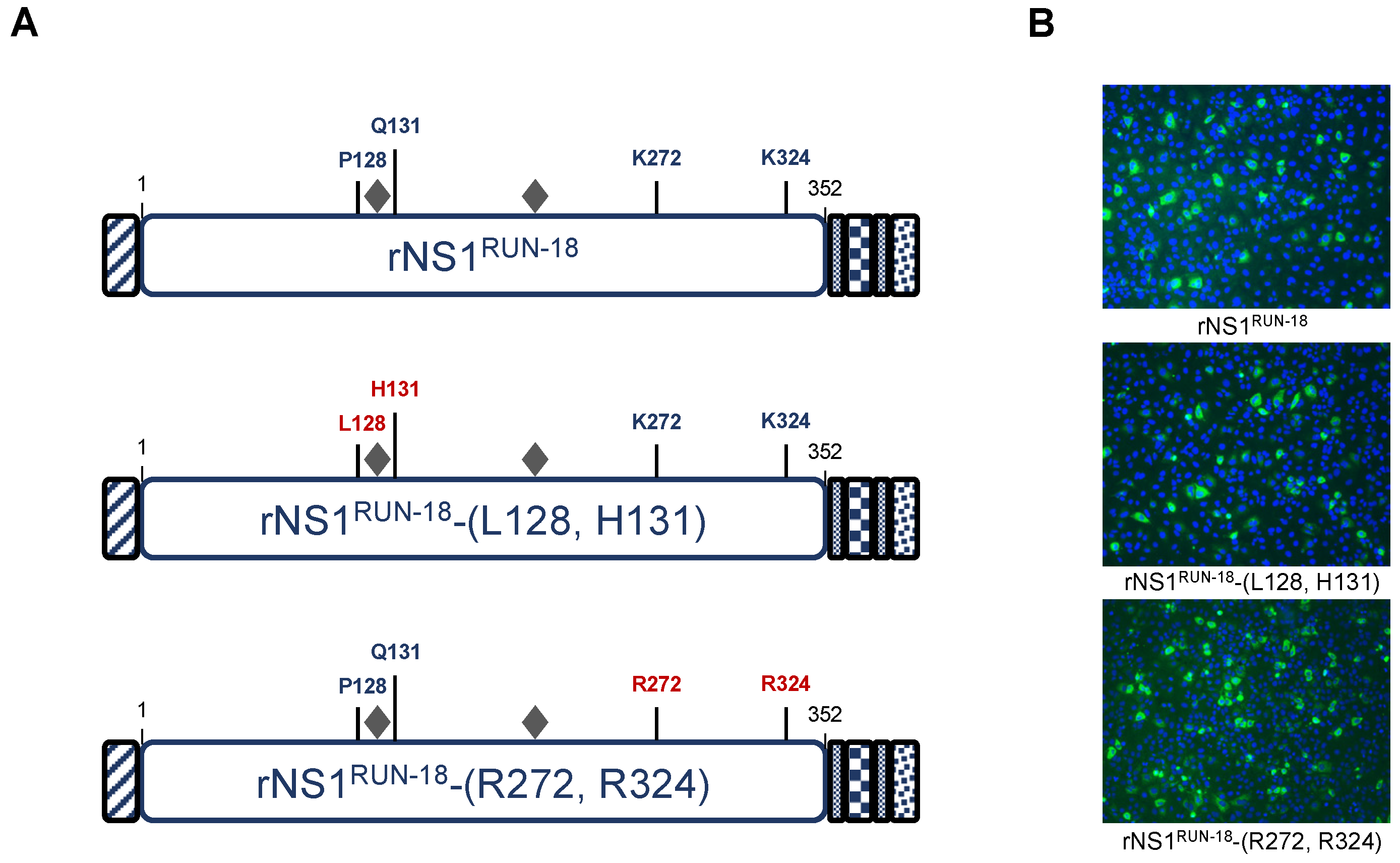
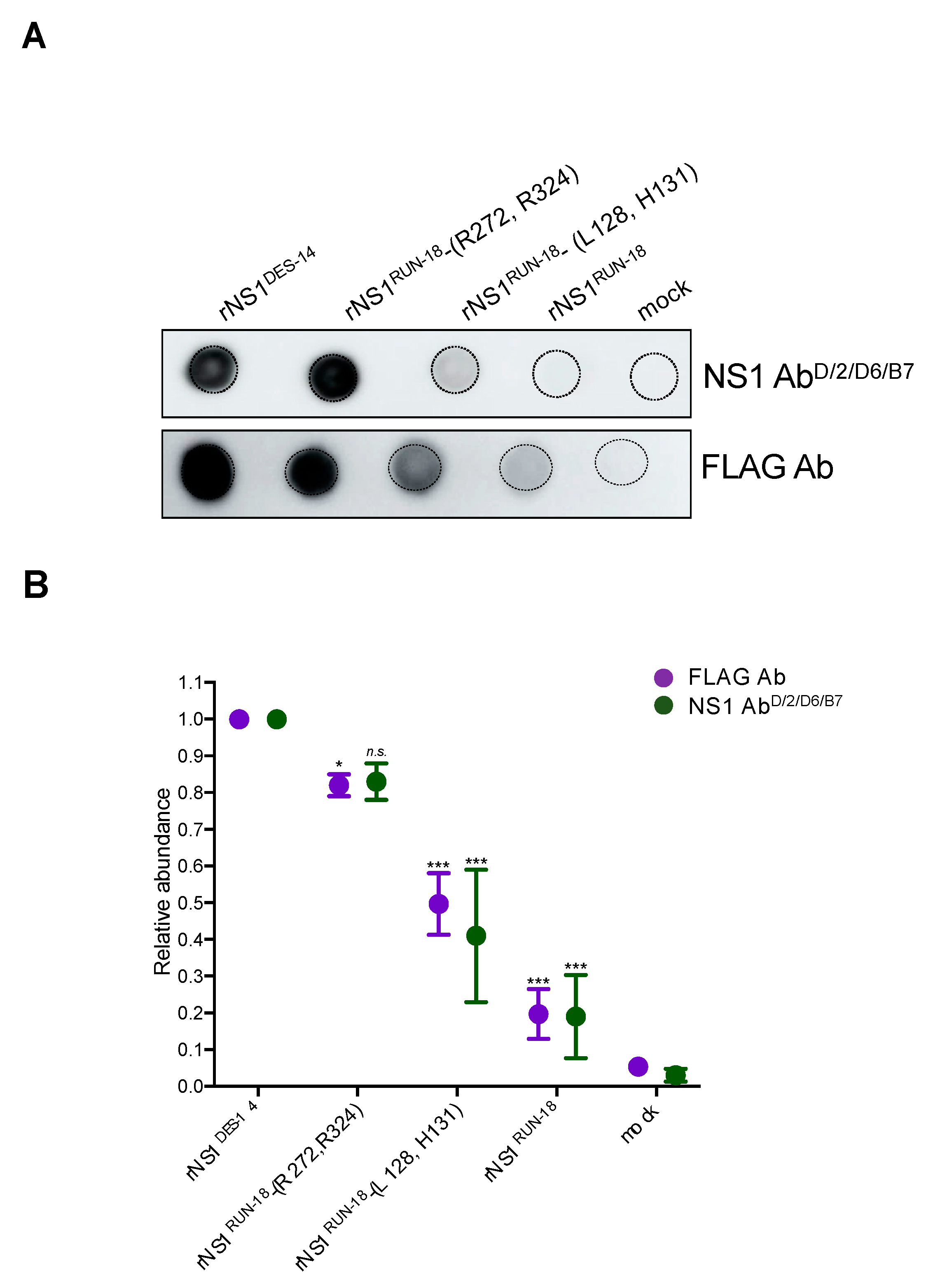
2.2.2. Impact of Mutations on rNS1RUN-18 Expression in Huh7 Cells
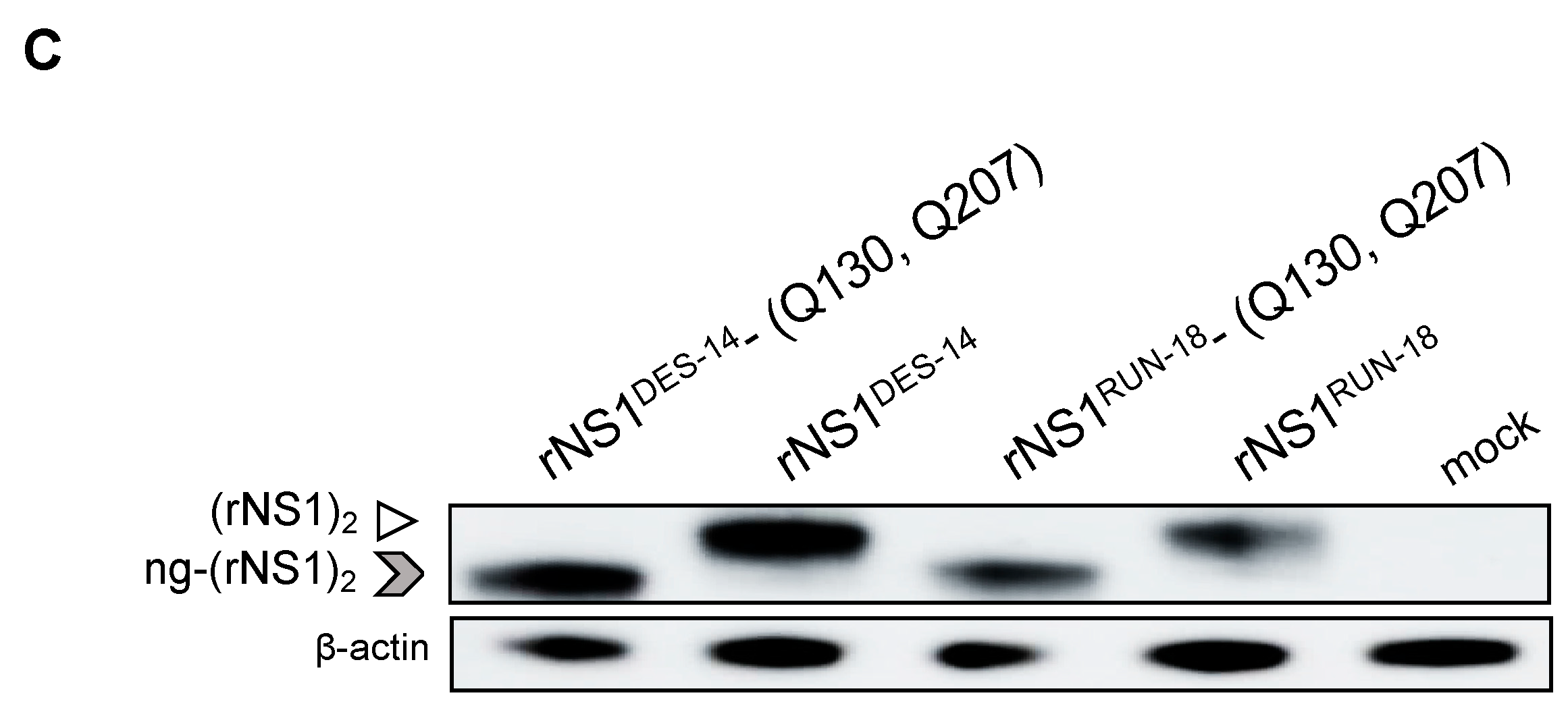
2.2.3. The P128 and Q131 Residues Have No Influence on rNS1RUN-18 Glycosylation
2.2.4. Secretion of rNS1RUN-18 Is Impacted by the NS1-128/131/272/324 Residues
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Antibodies
4.2. Vector Plasmids Expressing Recombinant DENV-2 NS1 Proteins and Their Mutants
4.3. Immunofluorescence Assay
4.4. Immunoblot Assay
4.5. Cytotoxicity Assays
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Ab | Antibody |
| DAPI | 4’,6-diamidino-2-phenylindole |
| DENV | Dengue virus |
| DMEM | Dulbecco’s modified Eagle’s medium |
| DMSO | Dimethyl sulfoxide |
| ECL | Enhanced chemiluminescent |
| ER | Endoplasmic reticulum |
| HEK293Tcells | Human embryonic kidney 293T SV40 cells |
| HRP | Horseradish peroxidase |
| IFI | Immunofluorescence indirect |
| kDa | Kilodalton |
| LDH | Lactate dehydrogenase |
| MAb | Monoclonal antibody |
| MEM | Minimum essential medium |
| MTT | 3-[4,5-Dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide |
| MW | Molecular weight |
| NS1 | Nonstructural protein 1 |
| RIPA | Radioimmunoprecipitation assay |
| rNS1 | Recombinant nonstructural protein 1 |
| RNA | Ribonucleic acid |
| SDS–PAGE | Sodium dodecyl sulfate-polyacrylamide gel electrophoresis |
| SP | Signal peptide |
| SWIO | South West Indian Ocean |
| TMD | Transmembrane domain |
| Ub | Ubiquitin |
References
- Pascalis, H.; Turpin, J.; Roche, M.; Krejbich, P.; Gadea, G.; Nten, C.A.; Desprès, P.; Mavingui, P.; Krejbich-Trotot, P. The epidemic of Dengue virus type-2 Cosmopolitan genotype on Reunion Island relates to its active circulation in the Southwestern Indian Ocean neighboring islands. Heliyon 2019, 5, e01455. [Google Scholar] [CrossRef]
- Bhatt, S.; Gething, P.W.; Brady, O.J.; Messina, J.P.; Farlow, A.W.; Moyes, C.L.; Drake, J.M.; Brownstein, J.S.; Hoen, A.G.; Sankoh, O.; et al. The global distribution and burden of dengue. Nature 2013, 496, 504–507. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Special Programme for Research and Training in Tropical Diseases. In Dengue Guidelines for Diagnosis, Treatment, Prevention and Control; World Health Organization: Geneva, Switzerland, 2009; 147 p. [Google Scholar]
- Hung, N.T.; Lei, H.; Lan, N.T.; Lin, Y.; Huang, K.; Lien, L.B.; Lin, C.; Yeh, T.; Ha, D.Q.; Huong, V.T.Q.; et al. Dengue Hemorrhagic Fever in Infants: A Study of Clinical and Cytokine Profiles. J. Infect. Dis. 2004, 189, 221–232. [Google Scholar] [CrossRef] [PubMed]
- Flores-Mendoza, L.K.; Estrada-Jiménez, T.; Sedeño-Monge, V.; Moreno, M.; Manjarrez, M.D.C.; González-Ochoa, G.; Peña, L.M.-P.; Reyes-Leyva, J. IL-10 and socs3 Are Predictive Biomarkers of Dengue Hemorrhagic Fever. Mediat. Inflamm. 2017, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Soo, K.-M.; Khalid, B.; Ching, S.-M.; Chee, H.-Y. Meta-Analysis of Dengue Severity during Infection by Different Dengue Virus Serotypes in Primary and Secondary Infections. PLoS ONE 2016, 11, e0154760. [Google Scholar] [CrossRef]
- Chareonsirisuthigul, T.; Kalayanarooj, S.; Ubol, S. Dengue virus (DENV) antibody-dependent enhancement of infection upregulates the production of anti-inflammatory cytokines, but suppresses anti-DENV free radical and pro-inflammatory cytokine production, in THP-1 cells. J. Gen. Virol. 2007, 88, 365–375. [Google Scholar] [CrossRef] [PubMed]
- Santé Publique France. Dengue à La Réunion: Inter épidémie 2020. Available online: www.santepubliquefrance.fr (accessed on 3 September 2020).
- Bos, S.; Gadea, G.; Desprès, P. Dengue: A growing threat requiring vaccine development for disease prevention. Pathog. Glob. Health 2018. [Google Scholar] [CrossRef]
- Guy, B.; Jackson, N. Dengue vaccine: Hypotheses to understand CYD-TDV-induced protection. Nat. Rev. Genet. 2015, 14, 45–54. [Google Scholar] [CrossRef]
- Pascalis, H.; Biscornet, L.; Toty, C.; Hafsia, S.; Roche, M.; Desprès, P.; Nten, C.A.; Bibi, J.; Louange, M.; Gedeon, J.; et al. Complete Genome Sequences of Dengue Virus Type 2 Epidemic Strains from Reunion Island and the Seychelles. Microbiol. Resour. Announc. 2020, 9. [Google Scholar] [CrossRef]
- Vairo, F.; Mboera, L.E.; De Nardo, P.; Oriyo, N.M.; Meschi, S.; Rumisha, S.F.; Colavita, F.; Mhina, A.; Carletti, F.; Mwakapeje, E.; et al. Clinical, Virologic, and Epidemiologic Characteristics of Dengue Outbreak, Dar es Salaam, Tanzania, 2014. Emerg. Infectious Dis. 2016, 22, 895–899. [Google Scholar] [CrossRef]
- Lustig, Y.; Wolf, D.; Halutz, O.; Schwartz, E. An outbreak of dengue virus (DENV) type 2 Cosmopolitan genotype in Israeli travellers returning from the Seychelles, April 2017. Eurosurveillance 2017, 22. [Google Scholar] [CrossRef]
- Welsch, S.; Miller, S.; Romero-Brey, I.; Merz, A.; Bleck, C.K.; Walther, P.; Fuller, S.D.; Antony, C.; Krijnse-Locker, J.; Bartenschlager, R. Composition and Three-Dimensional Architecture of the Dengue Virus Replication and Assembly Sites. Cell Host Microbe 2009, 5, 365–375. [Google Scholar] [CrossRef]
- Khromykh, A.A.; Sedlak, P.L.; Westaway, E.G. Cis- and trans-Acting Elements in Flavivirus RNA Replication. J. Virol. 2000, 74, 3253–3263. [Google Scholar] [CrossRef] [PubMed]
- Lindenbach, B.D.; Rice, C.M. Molecular biology of flaviviruses. In Advances in Virus Research; Elsevier: Amsterdam, The Netherlands, 2003; Volume 59, pp. 23–61. ISBN 978-0-12-039859-1. [Google Scholar]
- Randolph, B.; Cleaves, R.; Ryan, E.; Stollar, V. Evidence that the Mature Form of the Flavivirus Nonstructural Protein NSl Is a Dimer. Virology 1988, 162, 187–196. [Google Scholar]
- Mackenzie, J.M.; Jones, M.K.; Young, P.R. Immunolocalization of the Dengue Virus Nonstructural Glycoprotein NS1 Suggests a Role in Viral RNA Replication. Virology 1996, 220, 232–240. [Google Scholar] [CrossRef] [PubMed]
- Avirutnan, P.; Zhang, L.; Punyadee, N.; Manuyakorn, A.; Puttikhunt, C.; Kasinrerk, W.; Malasit, P.; Atkinson, J.P.; Diamond, M.S. Secreted NS1 of Dengue Virus Attaches to the Surface of Cells via Interactions with Heparan Sulfate and Chondroitin Sulfate E. PLOS Pathog. 2007, 3, e183. [Google Scholar] [CrossRef] [PubMed]
- Gutsche, I.; Coulibaly, F.; Voss, J.E.; Salmon, J.; D’Alayer, J.; Ermonval, M.; Larquet, E.; Charneau, P.; Krey, T.; Mégret, F.; et al. Secreted dengue virus nonstructural protein NS1 is an atypical barrel-shaped high-density lipoprotein. Proc. Natl. Acad. Sci. USA 2011, 108, 8003–8008. [Google Scholar] [CrossRef] [PubMed]
- Muller, D.A.; Landsberg, M.J.; Bletchly, C.; Rothnagel, R.; Waddington, L.; Hankamer, B.; Young, P.R. Structure of the dengue virus glycoprotein non-structural protein 1 by electron microscopy and single-particle analysis. J. Gen. Virol. 2012, 93, 771–779. [Google Scholar] [CrossRef]
- Avirutnan, P.; Fuchs, A.; Hauhart, R.E.; Somnuke, P.; Youn, S.; Diamond, M.S.; Atkinson, J.P. Antagonism of the complement component C4 by flavivirus nonstructural protein NS1. J. Exp. Med. 2010, 207, 793–806. [Google Scholar] [CrossRef]
- Beatty, P.R.; Puerta-Guardo, H.; Killingbeck, S.S.; Glasner, D.R.; Hopkins, K.; Harris, E. Dengue virus NS1 triggers endothelial permeability and vascular leak that is prevented by NS1 vaccination. Sci. Transl. Med. 2015, 7, 304ra141. [Google Scholar] [CrossRef]
- Alayli, F.; Scholle, F. Dengue virus NS1 enhances viral replication and pro-inflammatory cytokine production in human dendritic cells. Virology 2016, 496, 227–236. [Google Scholar] [CrossRef]
- Modhiran, N.; Watterson, D.; Muller, D.A.; Panetta, A.K.; Sester, D.P.; Liu, L.; Hume, D.A.; Stacey, K.J.; Young, P.R. Dengue virus NS1 protein activates cells via Toll-like receptor 4 and disrupts endothelial cell monolayer integrity. Sci. Transl. Med. 2015, 7, 304ra142. [Google Scholar] [CrossRef]
- Puerta-Guardo, H.; Glasner, D.R.; Harris, E. Dengue Virus NS1 Disrupts the Endothelial Glycocalyx, Leading to Hyperpermeability. PLOS Pathog. 2016, 12, e1005738. [Google Scholar] [CrossRef] [PubMed]
- Libraty, D.H.; Young, P.R.; Pickering, D.; Endy, T.P.; Kalayanarooj, S.; Green, S.; Vaughn, D.W.; Nisalak, A.; Ennis, F.A.; Rothman, A.L. High Circulating Levels of the Dengue Virus Nonstructural Protein NS1 Early in Dengue Illness Correlate with the Development of Dengue Hemorrhagic Fever. J. Infect. Dis. 2002, 186, 1165–1168. [Google Scholar] [CrossRef] [PubMed]
- Paranavitane, S.A.; Gomes, L.; Kamaladasa, A.; Adikari, T.N.; Wickramasinghe, N.; Jeewandara, C.; Shyamali, N.L.A.; Ogg, G.S.; Malavige, G.N. Dengue NS1 antigen as a marker of severe clinical disease. BMC Infect. Dis. 2014, 14, 570. [Google Scholar] [CrossRef] [PubMed]
- Bessoff, K.; DeLorey, M.J.; Sun, W.; Hunsperger, E. Comparison of Two Commercially Available Dengue Virus (DENV) NS1 Capture Enzyme-Linked Immunosorbent Assays Using a Single Clinical Sample for Diagnosis of Acute DENV Infection. Clin. Vaccine Immunol. 2008, 15, 1513–1518. [Google Scholar] [CrossRef]
- Hunsperger, E.A.; Yoksan, S.; Buchy, P.; Nguyen, V.C.; Sekaran, S.D.; Enria, D.A.; Vazquez, S.; Cartozian, E.; Pelegrino, J.L.; Artsob, H.; et al. Evaluation of Commercially Available Diagnostic Tests for the Detection of Dengue Virus NS1 Antigen and Anti-Dengue Virus IgM Antibody. PLoS Negl. Trop. Dis. 2014, 8, e3171. [Google Scholar] [CrossRef]
- Duong, V.; Ly, S.; Try, P.L.; Tuiskunen, A.; Ong, S.; Chroeung, N.; Lundkvist, A.; Leparc-Goffart, I.; Deubel, V.; Vong, S.; et al. Clinical and Virological Factors Influencing the Performance of a NS1 Antigen-Capture Assay and Potential Use as a Marker of Dengue Disease Severity. PLoS Negl. Trop. Dis. 2011, 5, e1244. [Google Scholar] [CrossRef]
- Edeling, M.A.; Diamond, M.S.; Fremont, D.H. Structural basis of Flavivirus NS1 assembly and antibody recognition. Proc. Natl. Acad. Sci. USA 2014, 111, 4285–4290. [Google Scholar] [CrossRef]
- Post, P.R.; Carvalho, R.; Galler, R. Glycosylation and secretion of yellow fever virus non-structural protein NS1. Virus Res. 1991, 18, 291–302. [Google Scholar] [CrossRef]
- Polak, K.; Greze, N.; Lachat, M.; Merle, D.; Chiumento, S.; Bertrand-Gaday, C.; Trentin, B.; Mamoun, R.Z. Extracellular vesicle-based vaccine platform displaying native viral envelope proteins elicits a robust anti-SARS-CoV-2 response. BioRxiv 2020. [Google Scholar] [CrossRef]
- Jacobs, S.C.; Stephenson, J.R.; Wilkinson, G.W. High-level expression of the tick-borne encephalitis virus NS1 protein by using an adenovirus-based vector: Protection elicited in a murine model. J. Virol. 1992, 66, 2086–2095. [Google Scholar] [CrossRef]
- Pryor, M.J.; Wright, P.J. Glycosylation Mutants of Dengue Virus NS1 Protein. J. Gen. Virol. 1994, 75, 1183–1187. [Google Scholar] [CrossRef]
- Ujwal, S.; Sabeena, S.; Bhaskar, R.; D’Souza, G.; Santhosha, D.; Auti, A.; Kumar, R.; Ramachandran, S.; Hindol, M.; Aithal, A.; et al. Circulation of Asian-I and Cosmopolitan genotypes of Dengue-2 virus in northeast India, 2016–2017. J. Vector Borne Dis. 2019, 56, 231–236. [Google Scholar] [PubMed]
- Phadungsombat, J.; Lin, M.Y.; Srimark, N.; Yamanaka, A.; Nakayama, E.E.; Moolasart, V.; Suttha, P.; Shioda, T.; Uttayamakul, S. Emergence of genotype Cosmopolitan of dengue virus type 2 and genotype III of dengue virus type 3 in Thailand. PLoS ONE 2018, 13, e0207220. [Google Scholar] [CrossRef]
- Akram, M.; Fatima, Z.; Purdy, M.A.; Sue, A.; Saleem, S.; Amin, I.; Shahid, M.; Idrees, M.; Nawaz, R. Introduction and evolution of dengue virus type 2 in Pakistan: A phylogeographic analysis. J. Virol. 2015, 12, 148. [Google Scholar] [CrossRef]
- Zhao, H.; Deng, Y.Q.; Hong, W.X.; Yu, X.D.; Jiang, T.; Yu, M.; Hu, F.Y.; Zhu, S.Y.; Li, X.F.; Song, K.Y.; et al. Complete Genome Sequence of Dengue Virus Serotype 2 Cosmopolitan Genotype Strain in Guangdong, China. J. Virol. 2012, 86, 13808. [Google Scholar] [CrossRef][Green Version]
- Yu, H.; Kong, Q.; Wang, J.; Qiu, X.; Wen, Y.; Yu, X.; Liu, M.; Wang, H.; Pan, J.; Sun, Z. Multiple Lineages of Dengue Virus Serotype 2 Cosmopolitan Genotype Caused a Local Dengue Outbreak in Hangzhou, Zhejiang Province, China, in 2017. Sci. Rep. 2019, 9, 7345. [Google Scholar] [CrossRef] [PubMed]
- Komander, D.; Rape, M. The Ubiquitin Code. Annu. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef] [PubMed]
- Giraldo, M.I.; Vargas-Cuartas, O.; Gallego-Gomez, J.C.; Shi, P.-Y.; Padilla-Sanabria, L.; Castaño-Osorio, J.C.; Rajsbaum, R. K48-linked polyubiquitination of dengue virus NS1 protein inhibits its interaction with the viral partner NS4B. Virus Res. 2018, 246, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Byk, L.A.; Iglesias, N.G.; De Maio, F.A.; Gebhard, L.G.; Rossi, M.; Gamarnik, A.V. Dengue Virus Genome Uncoating Requires Ubiquitination. mBio 2016, 7, e00804-16. [Google Scholar] [CrossRef] [PubMed]
- Kanlaya, R.; Pattanakitsakul, S.-N.; Sinchaikul, S.; Chen, S.-T.; Thongboonkerd, V. The Ubiquitin−Proteasome Pathway Is Important for Dengue Virus Infection in Primary Human Endothelial Cells. J. Proteome Res. 2010, 9, 4960–4971. [Google Scholar] [CrossRef]
- Wohlschlegel, J.A.; Johnson, E.S.; Reed, S.I.; Yates, J.R. Global Analysis of Protein Sumoylation in Saccharomyces cerevisiae. J. Biol. Chem. 2004, 279, 45662–45668. [Google Scholar] [CrossRef]
- Geiss-Friedlander, R.; Melchior, F. Concepts in sumoylation: A decade on. Nat. Rev. Mol. Cell Biol. 2007, 8, 947–956. [Google Scholar] [CrossRef] [PubMed]
- Su, C.-I.; Tseng, C.-H.; Yu, C.-Y.; Lai, M.M.C. SUMO Modification Stabilizes Dengue Virus Nonstructural Protein 5 To Support Virus Replication. J. Virol. 2016, 90, 4308–4319. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ogire, E.; Diaz, O.; Vidalain, P.-O.; Lotteau, V.; Desprès, P.; Roche, M. Instability of the NS1 Glycoprotein from La Reunion 2018 Dengue 2 Virus (Cosmopolitan-1 Genotype) in Huh7 Cells Is Due to Lysine Residues on Positions 272 and 324. Int. J. Mol. Sci. 2021, 22, 1951. https://doi.org/10.3390/ijms22041951
Ogire E, Diaz O, Vidalain P-O, Lotteau V, Desprès P, Roche M. Instability of the NS1 Glycoprotein from La Reunion 2018 Dengue 2 Virus (Cosmopolitan-1 Genotype) in Huh7 Cells Is Due to Lysine Residues on Positions 272 and 324. International Journal of Molecular Sciences. 2021; 22(4):1951. https://doi.org/10.3390/ijms22041951
Chicago/Turabian StyleOgire, Eva, Olivier Diaz, Pierre-Olivier Vidalain, Vincent Lotteau, Philippe Desprès, and Marjolaine Roche. 2021. "Instability of the NS1 Glycoprotein from La Reunion 2018 Dengue 2 Virus (Cosmopolitan-1 Genotype) in Huh7 Cells Is Due to Lysine Residues on Positions 272 and 324" International Journal of Molecular Sciences 22, no. 4: 1951. https://doi.org/10.3390/ijms22041951
APA StyleOgire, E., Diaz, O., Vidalain, P.-O., Lotteau, V., Desprès, P., & Roche, M. (2021). Instability of the NS1 Glycoprotein from La Reunion 2018 Dengue 2 Virus (Cosmopolitan-1 Genotype) in Huh7 Cells Is Due to Lysine Residues on Positions 272 and 324. International Journal of Molecular Sciences, 22(4), 1951. https://doi.org/10.3390/ijms22041951







