Quantitative Proteomics Reveals Changes Induced by TIMP-3 on Cell Membrane Composition and Novel Metalloprotease Substrates
Abstract
1. Introduction
2. Results
2.1. Surfaceome Analysis of TIMP-3 Overexpressing Cells
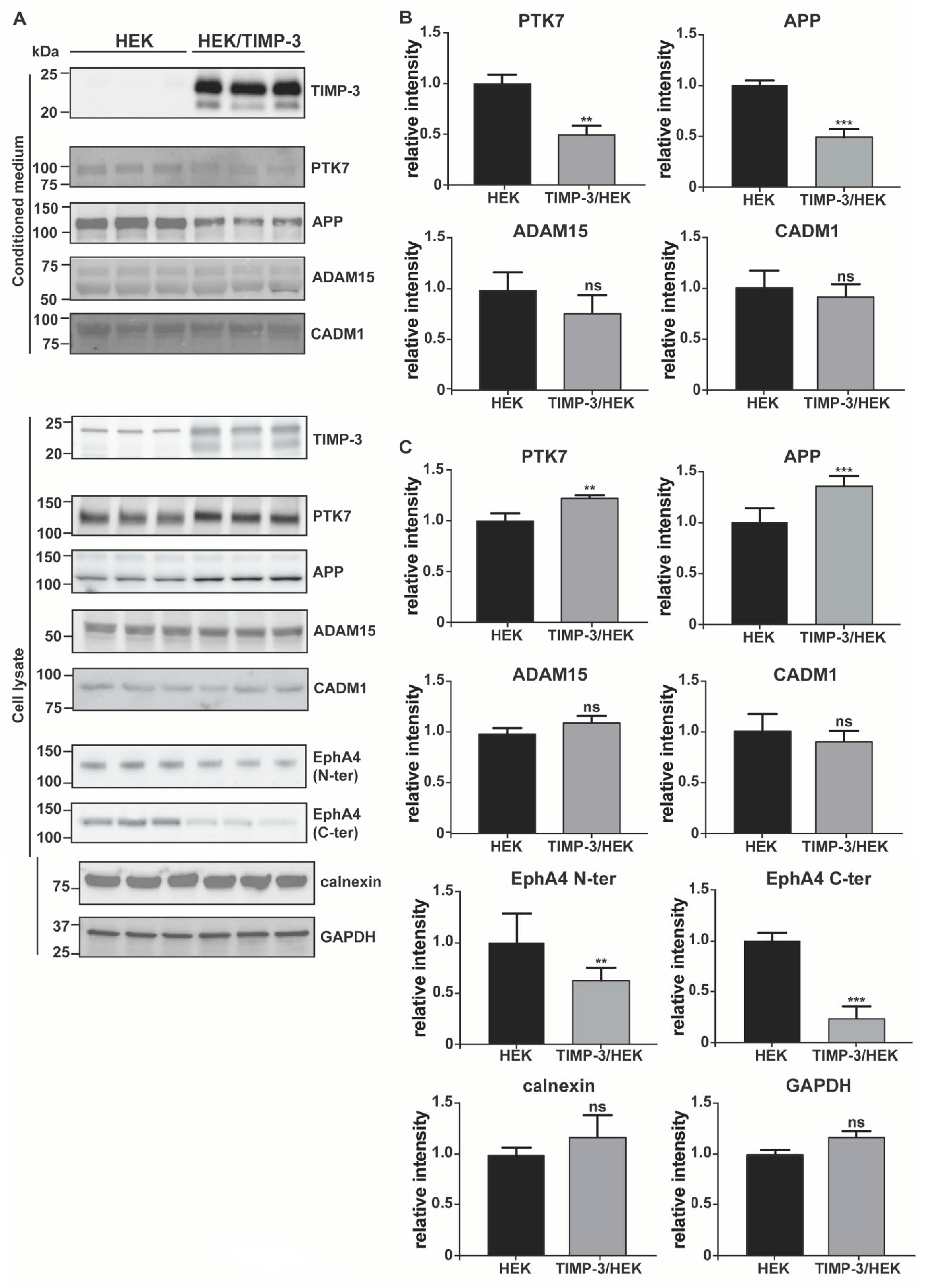
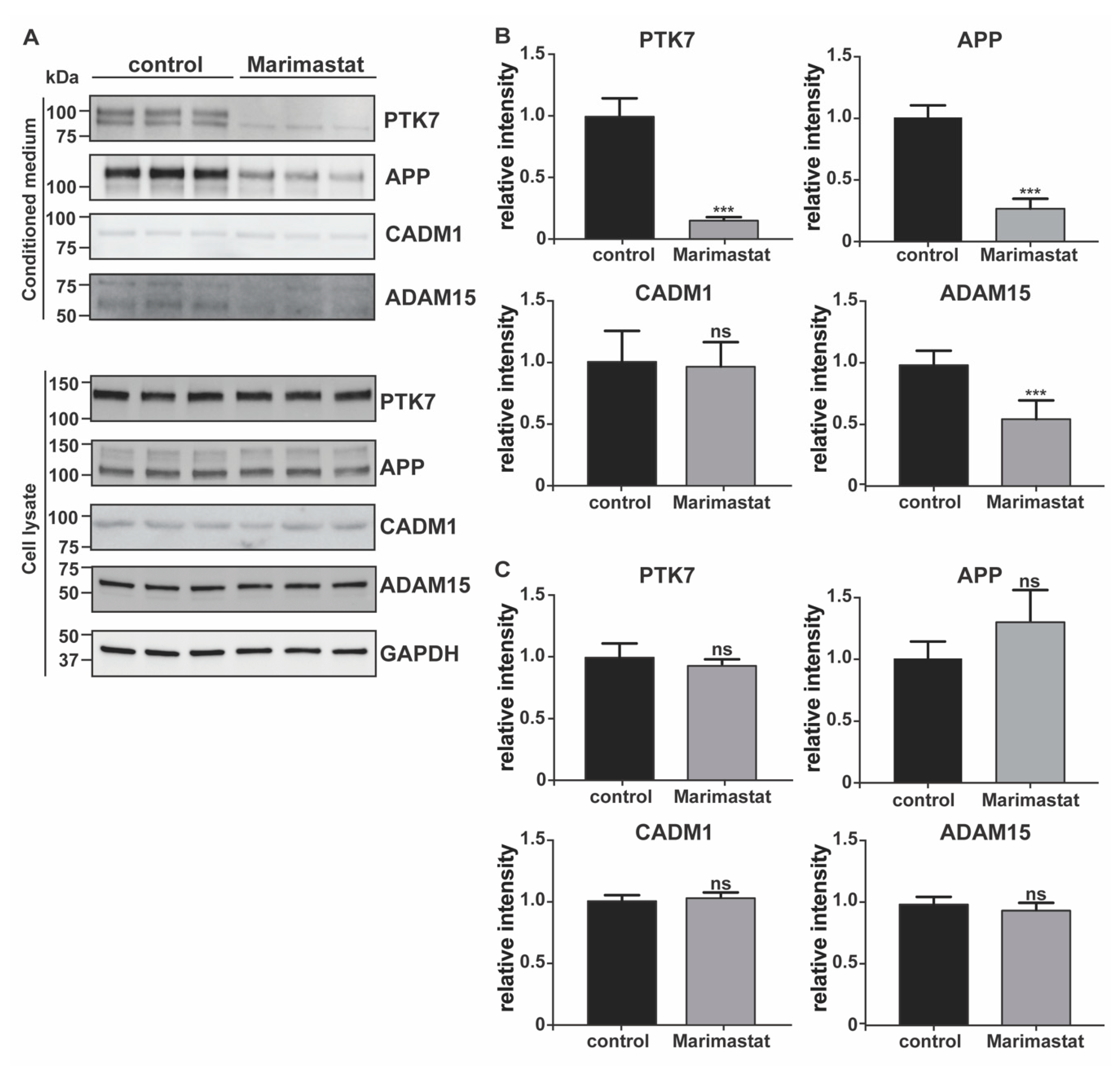
2.2. Validation of the Surfaceome Analysis
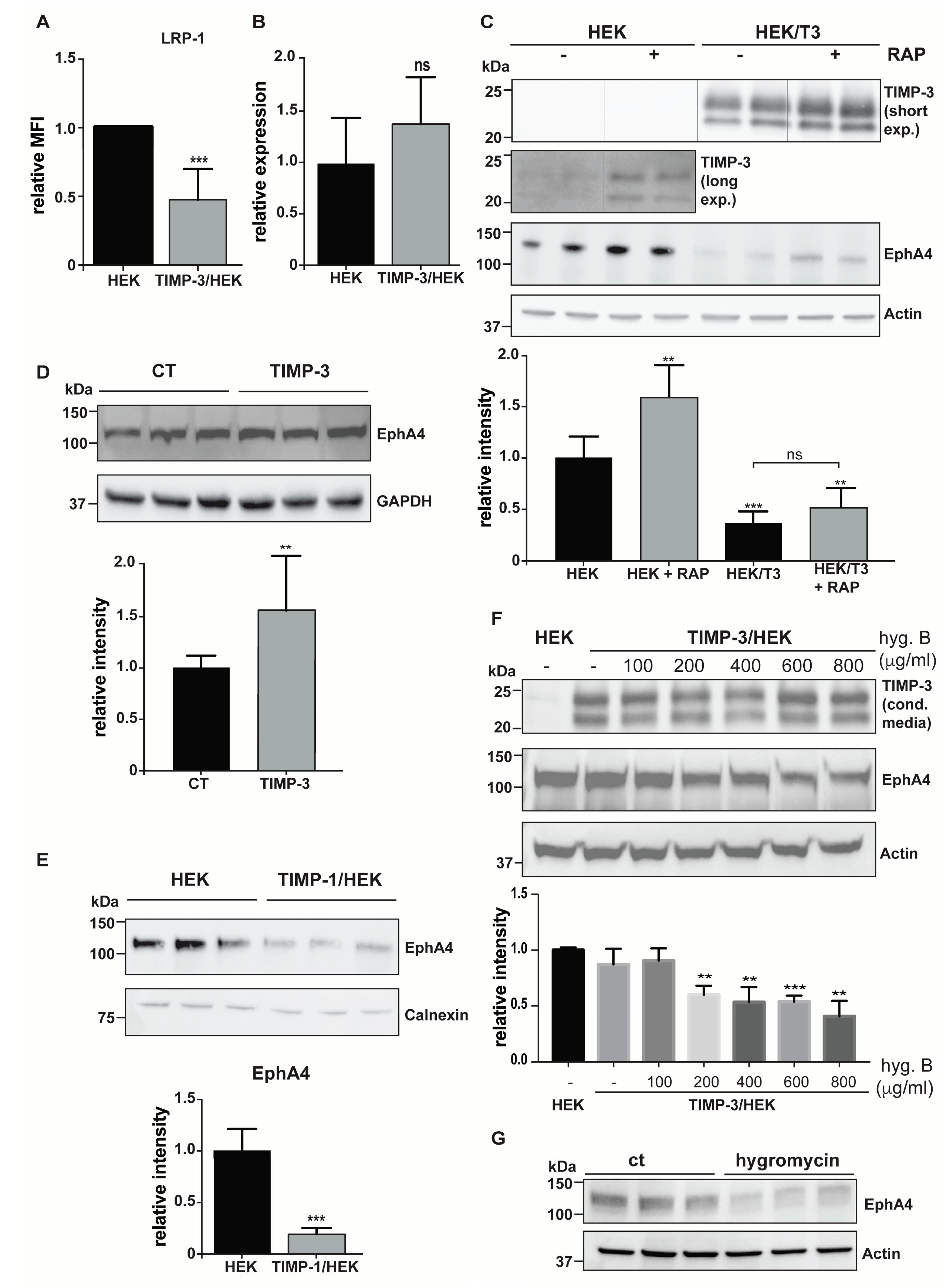
2.3. Hygromycin B Promotes Decrease of EphA4 Levels
3. Discussion
4. Materials and Methods
4.1. Generation of TIMP-3 and TIMP-1 Overexpressing Cells
4.2. Cell Membrane Proteome Analysis of TIMP-3 Overexpressing Cells
4.3. Proteomic Data Analysis
4.4. Methods to Validate Surfaceomics
- Western blotting. Conditioned media were collected and proteins precipitated with 5% v/v trichloroacetic acid (Sigma, Aldrich, St. Louis, MO, USA) before being resuspended in Laemmli sample buffer (Bio-Rad, Hercules, CA, USA). Cells were collected with STET lysis buffer (50 mM Tris, pH 7,5, 150 mM NaCl, 2 mM EDTA, 1% Triton), containing protease inhibitor cocktail (1:500, P-2714, Sigma, Aldrich, St. Louis, MO, USA). Alternatively, cell membranes were labelled by using EZ-Link Sulfo-NHS-LC-Biotin and membrane proteins isolated, as described above (Section 4.2). Either conditioned media, lysate or membrane proteins were loaded onto an acrylamide gel and analyzed using SDS-PAGE electrophoresis, followed by immunoblotting. The following antibodies were used: anti-TIMP-3 (AB6000, Sigma, Aldrich, St. Louis, MO, USA), anti-EphA4 (targeting C-terminal EphA4, 6H7 Sigma, Aldrich, St. Louis, MO, USA; targeting the N-terminal EphA4, 35/EphA4, BD Biosciences, Franklin Lakes, NJ, USA), anti-calnexin (ADI-SPA-860-F, ENZO lifescience, Farmingdale, NY, USA), anti-GAPDH (88845, Cell Signaling, Danvers, MA, USA), anti-PTK7 (AF4499, R&D systems, Minneapolis, MN, USA), anti-APP (clone 22c11, Sigma, Aldrich, St. Louis, MO, USA), CADM1 (TSLC1 H-300, Santa Cruz Biotechnology, Santa Cruz, CA, USA), ADAM15 (HPA011633, Atlas Antibodies, Bromma, Sweden), SDC4 (ab24511, Abcam, Cambridge, UK), and actin (81178, Santa Cruz, Santa Cruz, CA, USA). For each experiment, 3 to 9 biological replicates were analyzed (number and raw quantifications are shown in Supplemental Materials). Bands corresponding to each protein were quantified using Image Lab software (Bio-Rad, Hercules, CA, USA) and normalized to the mean of the original non-normalized control values (HEK 293 cells for TIMP-3/HEK or DMSO-treated HEK 293 cells for marimastat-treated cells). A two-sided Student’s t-test was used to evaluate proteins statistically significantly regulated. A p-value less than 0.05 was set as the significance threshold.
- Cells were collected with ice-cold staining buffer (PBS 1X, FBS 2%, 2 mM EDTA), washed, and resuspended in 100 μL of staining buffer and incubated with appropriate antibodies for 30 min at 4 °C—LRP-1 (CD91-PE, Clone REA709, 130-111-412, Miltenyi Biotec, Bergisch Gladbach, Germany). After washing in PBS, cells were analyzed with a FACSCanto II flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA).
4.5. Analysis of EphA4 mRNA Levels by qPCR
4.6. Exogenous TIMP-3 Induces Accumulation of EphA4
4.7. Dose-Dependent Effects of Hygromycin B on EphA4 Levels
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| TIMPs | Tissue inhibitor of metalloproteases |
| ADAMs | A disintegrin and metalloproteinases |
| MT-MMP | Membrane-type metalloproteases |
| TNF | Tumor necrosis factor α |
| HEK | Human embryonic kidney |
| LC-MS/MS | Liquid chromatography-tandem mass spectrometry |
| EphA4 | Ephrin type-A receptor 4 |
| LRP-1 | Low-density lipoprotein receptor-related protein 1 |
References
- Lichtenthaler, S.F.; Lemberg, M.K.; Fluhrer, R. Proteolytic ectodomain shedding of membrane proteins in mammals—hardware, concepts, and recent developments. EMBO J. 2018, 37, e99456. [Google Scholar] [CrossRef] [PubMed]
- Edwards, D.R.; Handsley, M.M.; Pennington, C.J. The ADAM metalloproteinases. Mol. Asp. Med. 2008, 29, 258–289. [Google Scholar] [CrossRef]
- Itoh, Y. Membrane-type matrix metalloproteinases: Their functions and regulations. Matrix Biol. 2015, 44-46, 207–223. [Google Scholar] [CrossRef]
- Brew, K.; Nagase, H. The tissue inhibitors of metalloproteinases (TIMPs): An ancient family with structural and functional diversity. Biochim. Biophys. Acta (BBA)-Bioenerg. 2010, 1803, 55–71. [Google Scholar] [CrossRef]
- Federici, M.; Hribal, M.L.; Menghini, R.; Kanno, H.; Marchetti, V.; Porzio, O.; Sunnarborg, S.W.; Rizza, S.; Serino, M.; Cunsolo, V.; et al. Timp3 deficiency in insulin receptor-haploinsufficient mice promotes diabetes and vascular inflammation via increased TNF-. J. Clin. Investig. 2005, 115, 3494–3505. [Google Scholar] [CrossRef]
- Mohammed, F.F.; Smookler, D.S.; Taylor, S.E.M.; Fingleton, B.; Kassiri, Z.; Sanchez, O.H.; English, J.L.; Matrisian, L.M.; Au, B.; Yeh, W.-C.; et al. Abnormal TNF activity in Timp3−/− mice leads to chronic hepatic inflammation and failure of liver regeneration. Nat. Genet. 2004, 36, 969–977. [Google Scholar] [CrossRef] [PubMed]
- Fedak, P.W.M.; Smookler, D.S.; Kassiri, Z.; Ohno, N.; Leco, K.J.; Verma, S.; Mickle, D.A.G.; Watson, K.L.; Hojilla, C.V.; Cruz, W.; et al. TIMP-3 Deficiency Leads to Dilated Cardiomyopathy. Circulaltion 2004, 110, 2401–2409. [Google Scholar] [CrossRef] [PubMed]
- Fujii, T.; Duarte, S.; Lee, E.; Ke, B.; Busuttil, R.W.; Coito, A.J. Tissue Inhibitor of Metalloproteinase 3 Deficiency Disrupts the Hepatocyte E-Cadherin/β-Catenin Complex and Induces Cell Death in Liver Ischemia/Reperfusion Injury. Liver Transplant. 2020, 26, 113–126. [Google Scholar] [CrossRef]
- Anand-Apte, B.; Bao, L.; Smith, R.; Zetter, B.; Iwata, K.; Olsen, B.R.; Apte, S.S. A review of tissue inhibitor of metalloproteinases-3 (TIMP-3) and experimental analysis of its effect on primary tumor growth. Biochem. Cell Biol. 1996, 74, 853–862. [Google Scholar] [CrossRef]
- Mahmoodi, M.; Sahebjam, S.; Smookler, D.; Khokha, R.; Mort, J.S. Lack of Tissue Inhibitor of Metalloproteinases-3 Results in an Enhanced Inflammatory Response in Antigen-Induced Arthritis. Am. J. Pathol. 2005, 166, 1733–1740. [Google Scholar] [CrossRef]
- Casagrande, V.; Menghini, R.; Menini, S.; Marino, A.; Marchetti, V.; Cavalera, M.; Fabrizi, M.; Hribal, M.L.; Pugliese, G.; Gentileschi, P.; et al. Overexpression of Tissue Inhibitor of Metalloproteinase 3 in Macrophages Reduces Atherosclerosis in Low-Density Lipoprotein Receptor Knockout Mice. Arter. Thromb. Vasc. Biol. 2012, 32, 74–81. [Google Scholar] [CrossRef]
- Menghini, R.; Casagrande, V.; Menini, S.; Marino, A.; Marzano, V.; Hribal, M.L.; Gentileschi, P.; Lauro, D.; Schillaci, O.; Pugliese, G.; et al. TIMP3 Overexpression in Macrophages Protects From Insulin Resistance, Adipose Inflammation, and Nonalcoholic Fatty Liver Disease in Mice. Diabetes 2012, 61, 454–462. [Google Scholar] [CrossRef] [PubMed]
- Anania, M.C.; Sensi, M.; Radaelli, E.; Miranda, C.; Vizioli, M.G.; Pagliardini, S.; Favini, E.; Cleris, L.; Supino, R.; Formelli, F.; et al. TIMP3 regulates migration, invasion and in vivo tumorigenicity of thyroid tumor cells. Oncogene 2011, 30, 3011–3023. [Google Scholar] [CrossRef]
- Lammich, S.; Kojro, E.; Postina, R.; Gilbert, S.; Pfeiffer, R.; Jasionowski, M.; Haass, C.; Fahrenholz, F. Constitutive and regulated -secretase cleavage of Alzheimer’s amyloid precursor protein by a disintegrin metalloprotease. Proc. Natl. Acad. Sci. USA 1999, 96, 3922–3927. [Google Scholar] [CrossRef]
- Conn, E.M.; Madsen, M.A.; Cravatt, B.F.; Ruf, W.; Deryugina, E.I.; Quigley, J.P. Cell Surface Proteomics Identifies Molecules Functionally Linked to Tumor Cell Intravasation. J. Biol. Chem. 2008, 283, 26518–26527. [Google Scholar] [CrossRef] [PubMed]
- Roesli, C.; Neri, D.; Rybak, J.-N. In vivo protein biotinylation and sample preparation for the proteomic identification of organ- and disease-specific antigens accessible from the vasculature. Nat. Protoc. 2006, 1, 192–199. [Google Scholar] [CrossRef] [PubMed]
- Schiapparelli, L.M.; McClatchy, D.B.; Liu, H.-H.; Sharma, P.; Yates, J.R.; Cline, H.T. Direct Detection of Biotinylated Proteins by Mass Spectrometry. J. Proteome Res. 2014, 13, 3966–3978. [Google Scholar] [CrossRef]
- Kuhn, P.-H.; Colombo, A.V.; Schusser, B.; Dreymueller, D.; Wetzel, S.; Schepers, U.; Herber, J.; Ludwig, A.; Kremmer, E.; Montag, D.; et al. Systematic substrate identification indicates a central role for the metalloprotease ADAM10 in axon targeting and synapse function. eLife 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Kamekura, R.; Nava, P.; Feng, M.; Quiros, M.; Nishio, H.; Weber, D.A.; Parkos, C.A.; Nusrat, A. Inflammation-induced desmoglein-2 ectodomain shedding compromises the mucosal barrier. Mol. Biol. Cell 2015, 26, 3165–3177. [Google Scholar] [CrossRef]
- Liu, Q.; Zhang, J.; Tran, H.; Verbeek, M.M.; Reiss, K.; Estus, S.; Bu, G. LRP1 shedding in human brain: Roles of ADAM10 and ADAM17. Mol. Neurodegener. 2009, 4, 17. [Google Scholar] [CrossRef]
- Scilabra, S.D.; Pigoni, M.; Pravatá, V.; Schätzl, T.; Müller, S.A.; Troeberg, L.; Lichtenthaler, S.F. Increased TIMP-3 expression alters the cellular secretome through dual inhibition of the metalloprotease ADAM10 and ligand-binding of the LRP-1 receptor. Sci. Rep. 2018, 8, 14697. [Google Scholar] [CrossRef] [PubMed]
- Seki, M.; Watanabe, A.; Enomoto, S.; Kawamura, T.; Ito, H.; Kodama, T.; Hamakubo, T.; Aburatani, H. Human ROBO1 is cleaved by metalloproteinases and γ-secretase and migrates to the nucleus in cancer cells. FEBS Lett. 2010, 584, 2909–2915. [Google Scholar] [CrossRef] [PubMed]
- Atapattu, L.; Saha, N.; Chheang, C.; Eissman, M.F.; Xu, K.; Vail, M.E.; Hii, L.; Llerena, C.; Liu, Z.; Horvay, K.; et al. An activated form of ADAM10 is tumor selective and regulates cancer stem-like cells and tumor growth. J. Exp. Med. 2016, 213, 1741–1757. [Google Scholar] [CrossRef] [PubMed]
- Schlecker, E.; Fiegler, N.; Arnold, A.; Altevogt, P.; Rose-John, S.; Moldenhauer, G.; Sucker, A.; Paschen, A.; Von Strandmann, E.P.; Textor, S.; et al. Metalloprotease-Mediated Tumor Cell Shedding of B7-H6, the Ligand of the Natural Killer Cell–Activating Receptor NKp30. Cancer Res. 2014, 74, 3429–3440. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.C.; Liu, X.; Li, Y.; Covington, M.; Wynn, R.; Huber, R.; Hillman, M.; Yang, G.; Ellis, D.; Marando, C.; et al. Identification of ADAM10 as a major source of HER2 ectodomain sheddase activity in HER2 overexpressing breast cancer cells. Cancer Biol. Ther. 2006, 5, 657–664. [Google Scholar] [CrossRef]
- Golubkov, V.S.; Prigozhina, N.L.; Zhang, Y.; Stoletov, K.; Lewis, J.D.; Schwartz, P.E.; Hoffman, R.M.; Strongin, A.Y. Protein-tyrosine Pseudokinase 7 (PTK7) Directs Cancer Cell Motility and Metastasis. J. Biol. Chem. 2014, 289, 24238–24249. [Google Scholar] [CrossRef]
- Zhang, G.; Hou, J.; Shi, J.; Yu, G.; Lu, B.; Zhang, X. Soluble CD276 (B7-H3) is released from monocytes, dendritic cells and activated T cells and is detectable in normal human serum. Immunology 2008, 123, 538–546. [Google Scholar] [CrossRef] [PubMed]
- Asher, R.A.; Morgenstern, D.A.; Properzi, F.; Nishiyama, A.; Levine, J.M.; Fawcett, J.W. Two separate metalloproteinase activities are responsible for the shedding and processing of the NG2 proteoglycan in vitro. Mol. Cell. Neurosci. 2005, 29, 82–96. [Google Scholar] [CrossRef]
- Hoa, N.; Tsui, S.; Afifiyan, N.F.; Hikim, A.S.; Li, B.; Douglas, R.S.; Smith, T.J. Nuclear Targeting of IGF-1 Receptor in Orbital Fibroblasts from Graves’ Disease: Apparent Role of ADAM17. PLoS ONE 2012, 7, e34173. [Google Scholar] [CrossRef] [PubMed]
- Albrechtsen, R.; Albrechtsen, N.J.W.; Gnosa, S.; Schwarz, J.; Dyrskjøt, L.; Kveiborg, M. Identification of ADAM12 as a Novel Basigin Sheddase. Int. J. Mol. Sci. 2019, 20, 1957. [Google Scholar] [CrossRef] [PubMed]
- Kryczka, J.; Stasiak, M.; Dziki, L.; Mik, M.; Dziki, A.; Cierniewski, C.S. Matrix Metalloproteinase-2 Cleavage of the β1 Integrin Ectodomain Facilitates Colon Cancer Cell Motility. J. Biol. Chem. 2012, 287, 36556–36566. [Google Scholar] [CrossRef] [PubMed]
- Hsia, H.-E.; Tüshaus, J.; Brummer, T.; Zheng, Y.; Scilabra, S.D.; Lichtenthaler, S.F. Functions of ‘A disintegrin and metalloproteases (ADAMs)’ in the mammalian nervous system. Cell. Mol. Life Sci. 2019, 76, 3055–3081. [Google Scholar] [CrossRef] [PubMed]
- Nagase, H.; Visse, R.; Murphy, G. Structure and function of matrix metalloproteinases and TIMPs. Cardiovasc. Res. 2006, 69, 562–573. [Google Scholar] [CrossRef]
- Nagara, Y.; Hagiyama, M.; Hatano, N.; Futai, E.; Suo, S.; Takaoka, Y.; Murakami, Y.; Ito, A.; Ishiura, S. Tumor suppressor cell adhesion molecule 1 (CADM1) is cleaved by a disintegrin and metalloprotease 10 (ADAM10) and subsequently cleaved by γ-secretase complex. Biochem. Biophys. Res. Commun. 2012, 417, 462–467. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, M.L.; Wang, Z.; Park, P.W.; Murphy, G.A.; Bernfield, M. Shedding of Syndecan-1 and -4 Ectodomains Is Regulated by Multiple Signaling Pathways and Mediated by a Timp-3–Sensitive Metalloproteinase. J. Cell Biol. 2000, 148, 811–824. [Google Scholar] [CrossRef]
- Carreca, A.P.; Pravatà, V.M.; Markham, M.; Bonelli, S.; Murphy, G.; Nagase, H.; Troeberg, L.; Scilabra, S.D. TIMP-3 facilitates binding of target metalloproteinases to the endocytic receptor LRP-1 and promotes scavenging of MMP-1. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef]
- Gopal, U.; Bohonowych, J.E.; Lema-Tome, C.; Liu, A.; Garrett-Mayer, E.; Wang, B.; Isaacs, J.S. A Novel Extracellular Hsp90 Mediated Co-Receptor Function for LRP1 Regulates EphA2 Dependent Glioblastoma Cell Invasion. PLoS ONE 2011, 6, e17649. [Google Scholar] [CrossRef]
- Lillis, A.P.; Van Duyn, L.B.; Murphy-Ullrich, J.E.; Strickland, D.K. LDL Receptor-Related Protein 1: Unique Tissue-Specific Functions Revealed by Selective Gene Knockout Studies. Physiol. Rev. 2008, 88, 887–918. [Google Scholar] [CrossRef]
- Amour, A.; Slocombe, P.M.; Webster, A.; Butler, M.; Knight, C.; Smith, B.J.; Stephens, P.E.; Shelley, C.; Hutton, M.; Knäuper, V.; et al. TNF-α converting enzyme (TACE) is inhibited by TIMP-3. FEBS Lett. 1998, 435, 39–44. [Google Scholar] [CrossRef]
- Weber, S.; Saftig, P. Ectodomain shedding and ADAMs in development. Development 2012, 139, 3693–3709. [Google Scholar] [CrossRef]
- Sahebjam, S.; Khokha, R.; Mort, J.S. Increased collagen and aggrecan degradation with age in the joints ofTimp3−/− mice. Arthritis Rheum. 2007, 56, 905–909. [Google Scholar] [CrossRef]
- Black, R.A.; Castner, B.; Slack, J.; Tocker, J.; Eisenman, J.; Jacobson, E.; Delaney, J.; Winters, D.; Hecht, R.; Bendele, A. Injected TIMP-3 protects cartilage in a rat meniscal tear model. Osteoarthr. Cartil. 2006. [Google Scholar] [CrossRef][Green Version]
- Cardellini, M.; Menghini, R.; Martelli, E.; Casagrande, V.; Marino, A.; Rizza, S.; Porzio, O.; Mauriello, A.; Solini, A.; Ippoliti, A.; et al. TIMP3 Is Reduced in Atherosclerotic Plaques From Subjects With Type 2 Diabetes and Increased by SirT1. Diabetes 2009, 58, 2396–2401. [Google Scholar] [CrossRef] [PubMed]
- Müller, S.A.; Scilabra, S.D.; Lichtenthaler, S.F. Proteomic Substrate Identification for Membrane Proteases in the Brain. Front. Mol. Neurosci. 2016, 9, 96. [Google Scholar] [CrossRef] [PubMed]
- Brummer, T.; Müller, S.A.; Pan-Montojo, F.; Yoshida, F.; Fellgiebel, A.; Tomita, T.; Endres, K.; Lichtenthaler, S.F. Nr CAM is a marker for substrate-selective activation of ADAM 10 in Alzheimer’s disease. EMBO Mol. Med. 2019, 11, e9695. [Google Scholar] [CrossRef]
- Matthews, A.L.; Noy, P.J.; Reyat, J.S.; Tomlinson, M.G. Regulation of A disintegrin and metalloproteinase (ADAM) family sheddases ADAM10 and ADAM17: The emerging role of tetraspanins and rhomboids. Platelets 2017, 28, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Dulloo, I.; Muliyil, S.; Freeman, M. The molecular, cellular and pathophysiological roles of iRhom pseudoproteases. Open Biol. 2019, 9, 190003. [Google Scholar] [CrossRef]
- Maretzky, T.; McIlwain, D.R.; Issuree, P.D.A.; Li, X.; Malapeira, J.; Amin, S.; Lang, P.A.; Mak, T.W.; Blobel, C.P. iRhom2 controls the substrate selectivity of stimulated ADAM17-dependent ectodomain shedding. Proc. Natl. Acad. Sci. USA 2013, 110, 11433–11438. [Google Scholar] [CrossRef] [PubMed]
- Maretzky, T.; Yang, G.; Ouerfelli, O.; Overall, C.M.; Worpenberg, S.; Hassiepen, U.; Eder, J.; Blobel, C.P. Characterization of the catalytic activity of the membrane-anchored metalloproteinase ADAM15 in cell-based assays. Biochem. J. 2009, 420, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Böhm, B.B.; Aigner, T.; Roy, B.; Brodie, T.A.; Blobel, C.P.; Burkhardt, H. Homeostatic effects of the metalloproteinase disintegrin ADAM15 in degenerative cartilage remodeling. Arthritis Rheum. 2005, 52, 1100–1109. [Google Scholar] [CrossRef] [PubMed]
- Aigner, T.; Blobel, C.P.; Kalden, J.R.; Burkhardt, H. Highly enhanced expression of the disintegrin metalloproteinase MDC15 (metargidin) in rheumatoid synovial tissue. Arthritis Rheum. 2001, 44, 2046–2054. [Google Scholar] [CrossRef]
- Böhm, B.; Hess, S.; Krause, K.; Schirner, A.; Ewald, W.; Aigner, T.; Burkhardt, H. ADAM15 exerts an antiapoptotic effect on osteoarthritic chondrocytes via up-regulation of the X-linked inhibitor of apoptosis. Arthritis Rheum. 2010, 62, 1372–1382. [Google Scholar] [CrossRef]
- Yang, C.; Chanalaris, A.; Bonelli, S.; McClurg, O.; Hiles, G.L.; Cates, A.; Zarebska, J.M.; Vincent, T.; Day, M.; Müller, S.; et al. Interleukin 13 (IL-13)-regulated expression of the chondroprotective metalloproteinase ADAM15 is reduced in aging cartilage. Osteoarthr. Cartil. Open 2020, 2, 100128. [Google Scholar] [CrossRef] [PubMed]
- Scharfenberg, F.; Helbig, A.; Sammel, M.; Benzel, J.; Schlomann, U.; Peters, F.; Wichert, R.; Bettendorff, M.; Schmidt-Arras, D.; Rose-John, S.; et al. Degradome of soluble ADAM10 and ADAM17 metalloproteases. Cell. Mol. Life Sci. 2019, 77, 331–350. [Google Scholar] [CrossRef] [PubMed]
- Pardo, J.M.; Malpartida, F.; Rico, M.; Jiménez, A. Biochemical Basis of Resistance to Hygromycin B in Streptomyces hygroscopicus—The Producing Organism. Microbiology 1985, 131, 1289–1298. [Google Scholar] [CrossRef] [PubMed]
- Aubrecht, J.; Goad, M.E.; Simpson, E.M.; Schiestl, R.H. Expression of hygR in transgenic mice causes resistance to toxic effects of hygromycin B in vivo. J. Pharmacol. Exp. Ther. 1997, 281, 992–997. [Google Scholar]
- Troeberg, L.; Fushimi, K.; Scilabra, S.D.; Nakamura, H.; Dive, V.; Thøgersen, I.B.; Enghild, J.J.; Nagase, H. The C-terminal domains of ADAMTS-4 and ADAMTS-5 promote association with N-TIMP-3. Matrix Biol. 2009, 28, 463–469. [Google Scholar] [CrossRef]
- Shevchenko, A.; Tomas, H.; Havlis, J.; Olsen, J.V.; Mann, M.J. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006, 1, 2856–2860. [Google Scholar] [CrossRef]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate Proteome-wide Label-free Quantification by Delayed Normalization and Maximal Peptide Ratio Extraction, Termed MaxLFQ. Mol. Cell. Proteom. 2014, 13, 2513–2526. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
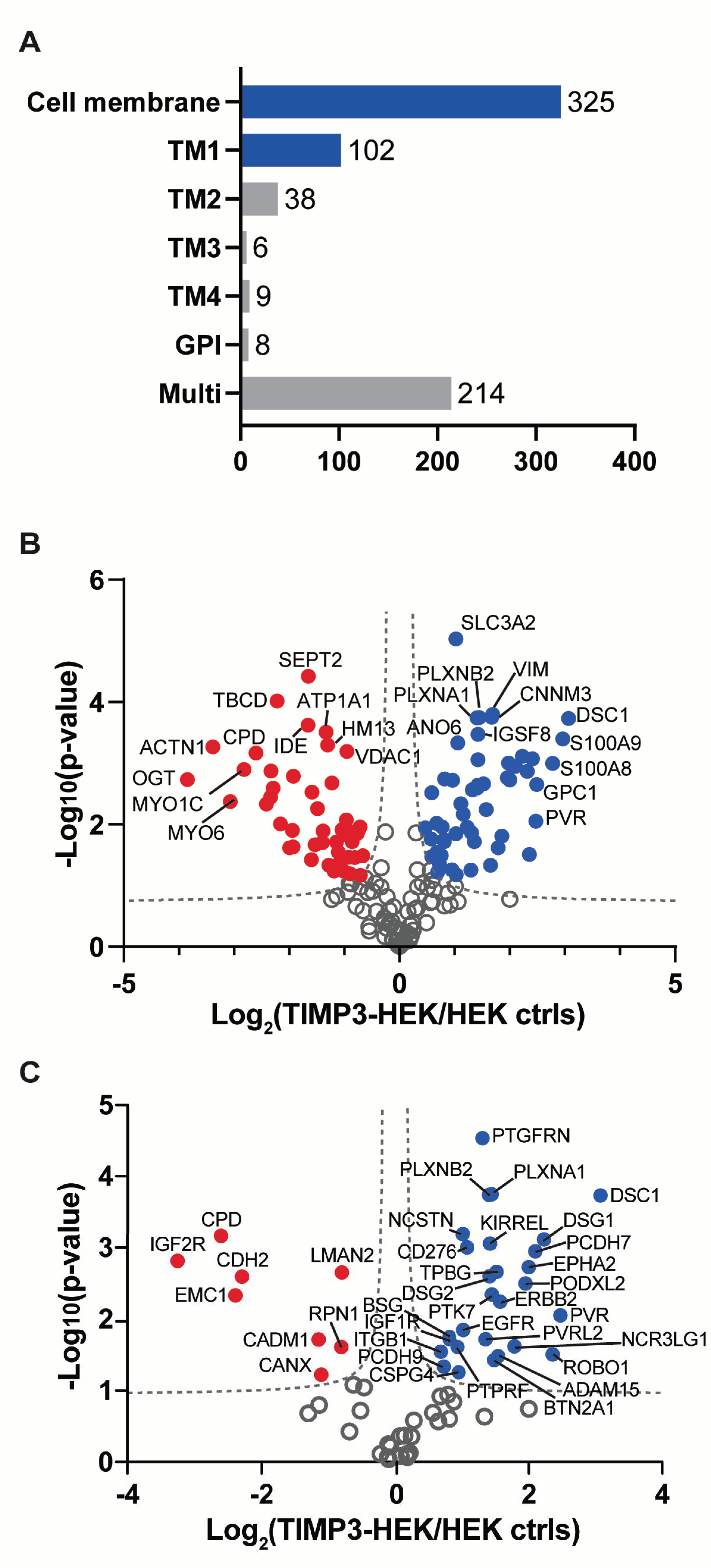

| Protein Name | Protein ID | Gene Name | p-Value | Ratio | MP Substrate |
|---|---|---|---|---|---|
| Desmocollin-1 | Q08554 | DSC1 | 1.86 × 10−4 | 8.40 | unknown |
| Poliovirus receptor | P15151 | PVR | 8.86 × 10−3 | 5.56 | unknown |
| Roundabout homolog 1 | Q9Y6N7 | ROBO1 | 3.11 × 10−2 | 5.13 | [22] |
| Desmoglein-1 | Q02413 | DSG1 | 7.70 × 10−4 | 4.69 | unknown |
| Protocadherin-7 | O60245 | PCDH7 | 1.13 × 10−3 | 4.29 | unknown |
| Ephrin type-A receptor 2 | P29317 | EPHA2 | 1.87 × 10−3 | 4.01 | [23] |
| Podocalyxin-like protein 2 | Q9NZ53 | PODXL2 | 3.19 × 10−3 | 3.87 | [18] |
| Natural cytotoxicity triggering receptor 3 ligand 1 | Q68D85 | NCR3LG1 | 2.40 × 10−2 | 3.46 | [24] |
| Receptor tyrosine-protein kinase erbB-2 | P04626 | ERBB2 | 5.72 × 10−3 | 2.98 | [25] |
| Disintegrin and metalloproteinase domain-containing protein 15 | Q13444 | ADAM15 | 3.32 × 10−2 | 2.92 | unknown |
| Trophoblast glycoprotein | Q13641 | TPBG | 2.18 × 10−3 | 2.87 | unknown |
| Butyrophilin subfamily 2 member A1 | Q7KYR7 | BTN2A1 | 3.75 × 10−2 | 2.80 | unknown |
| Inactive tyrosine-protein kinase 7 | Q13308 | PTK7 | 4.54 × 10−3 | 2.73 | [26] |
| Plexin-B2 | O15031 | PLXNB2 | 1.78 × 10−4 | 2.73 | [18] |
| Kin of IRRE-like protein 1 | Q96J84 | KIRREL | 8.80 × 10−4 | 2.69 | unknown |
| Desmoglein-2 | Q14126 | DSG2 | 2.53 × 10−3 | 2.67 | [19] |
| Plexin-A1 | Q9UIW2 | PLXNA1 | 1.82 × 10−4 | 2.65 | unknown |
| Nectin-2 | Q92692 | PVRL2 | 1.91 × 10−2 | 2.56 | unknown |
| Prostaglandin F2 receptor negative regulator | Q9P2B2 | PTGFRN | 2.90 × 10−5 | 2.48 | unknown |
| CD276 antigen | Q5ZPR3 | CD276 | 9.90 × 10−4 | 2.11 | [27] |
| Epidermal growth factor receptor | P00533 | EGFR | 1.43 × 10−2 | 2.03 | unknown |
| Nicastrin | Q92542 | NCSTN | 6.48 × 10−4 | 2.02 | unknown |
| Chondroitin sulfate proteoglycan 4 | Q6UVK1 | CSPG4 | 5.52 × 10−2 | 1.94 | [28] |
| Receptor-type tyrosine-protein phosphatase F | P10586 | PTPRF | 2.45 × 10−2 | 1.91 | [18] |
| Insulin-like growth factor 1 receptor | P08069 | IGF1R | 1.92 × 10−2 | 1.76 | [29] |
| Basigin | P35613 | BSG | 1.79 × 10−2 | 1.76 | [30] |
| Protocadherin-9 | Q9HC56 | PCDH9 | 4.60 × 10−2 | 1.66 | [18] |
| Integrin beta-1 | P05556 | ITGB1 | 2.85 × 10−2 | 1.61 | [31] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carreca, A.P.; Pravatà, V.M.; D’Apolito, D.; Bonelli, S.; Calligaris, M.; Monaca, E.; Müller, S.A.; Lichtenthaler, S.F.; Scilabra, S.D. Quantitative Proteomics Reveals Changes Induced by TIMP-3 on Cell Membrane Composition and Novel Metalloprotease Substrates. Int. J. Mol. Sci. 2021, 22, 2392. https://doi.org/10.3390/ijms22052392
Carreca AP, Pravatà VM, D’Apolito D, Bonelli S, Calligaris M, Monaca E, Müller SA, Lichtenthaler SF, Scilabra SD. Quantitative Proteomics Reveals Changes Induced by TIMP-3 on Cell Membrane Composition and Novel Metalloprotease Substrates. International Journal of Molecular Sciences. 2021; 22(5):2392. https://doi.org/10.3390/ijms22052392
Chicago/Turabian StyleCarreca, Anna Paola, Veronica Maria Pravatà, Danilo D’Apolito, Simone Bonelli, Matteo Calligaris, Elisa Monaca, Stephan A. Müller, Stefan F. Lichtenthaler, and Simone Dario Scilabra. 2021. "Quantitative Proteomics Reveals Changes Induced by TIMP-3 on Cell Membrane Composition and Novel Metalloprotease Substrates" International Journal of Molecular Sciences 22, no. 5: 2392. https://doi.org/10.3390/ijms22052392
APA StyleCarreca, A. P., Pravatà, V. M., D’Apolito, D., Bonelli, S., Calligaris, M., Monaca, E., Müller, S. A., Lichtenthaler, S. F., & Scilabra, S. D. (2021). Quantitative Proteomics Reveals Changes Induced by TIMP-3 on Cell Membrane Composition and Novel Metalloprotease Substrates. International Journal of Molecular Sciences, 22(5), 2392. https://doi.org/10.3390/ijms22052392







