The Functional Role of Loops and Flanking Sequences of G-Quadruplex Aptamer to the Hemagglutinin of Influenza a Virus
Abstract
:1. Introduction
2. Results and Discussion
2.1. Design of RHA0385 Variants
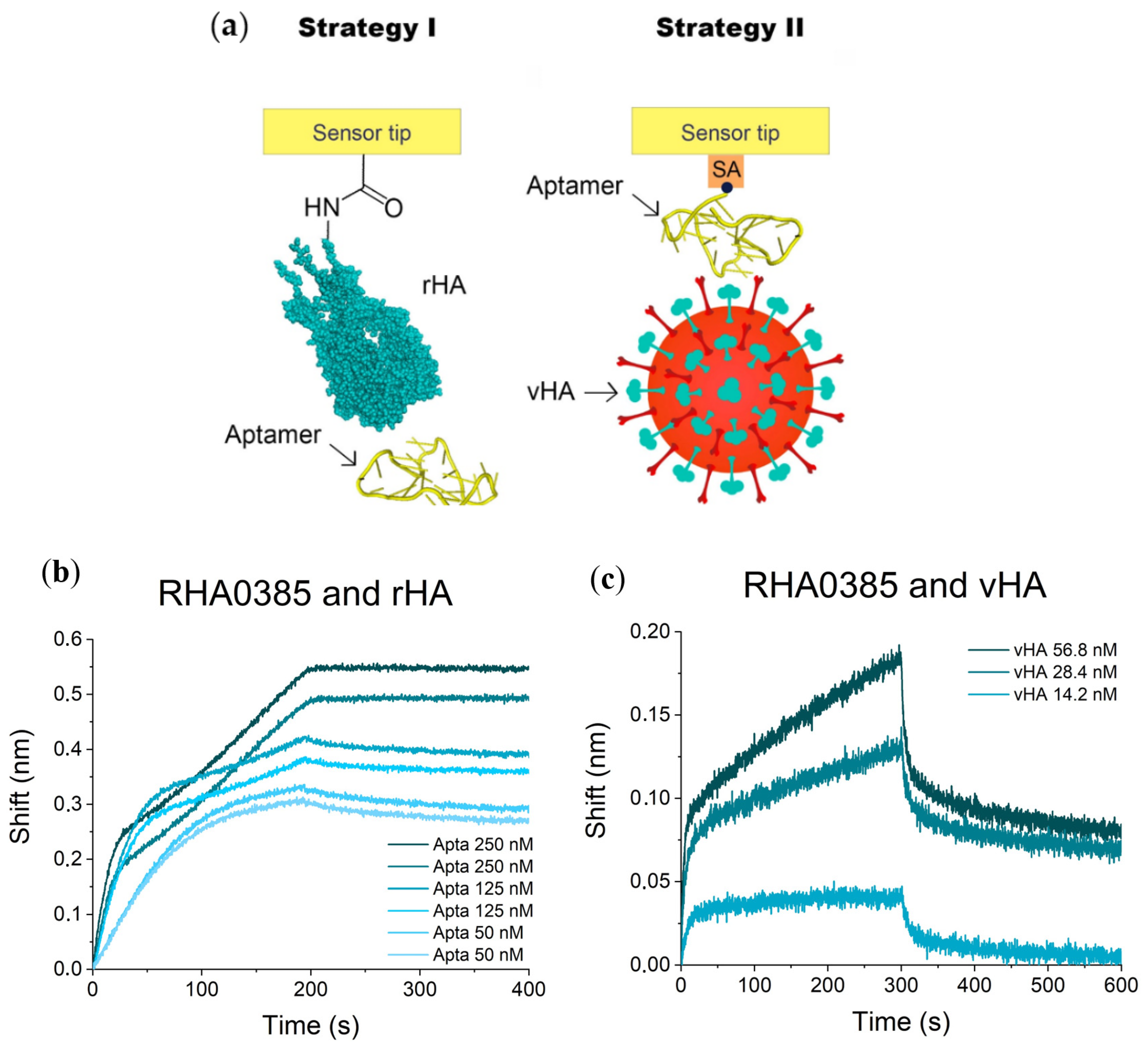
2.2. Affinity of RHA0385 and Its Variants to Viral and Recombinant HA
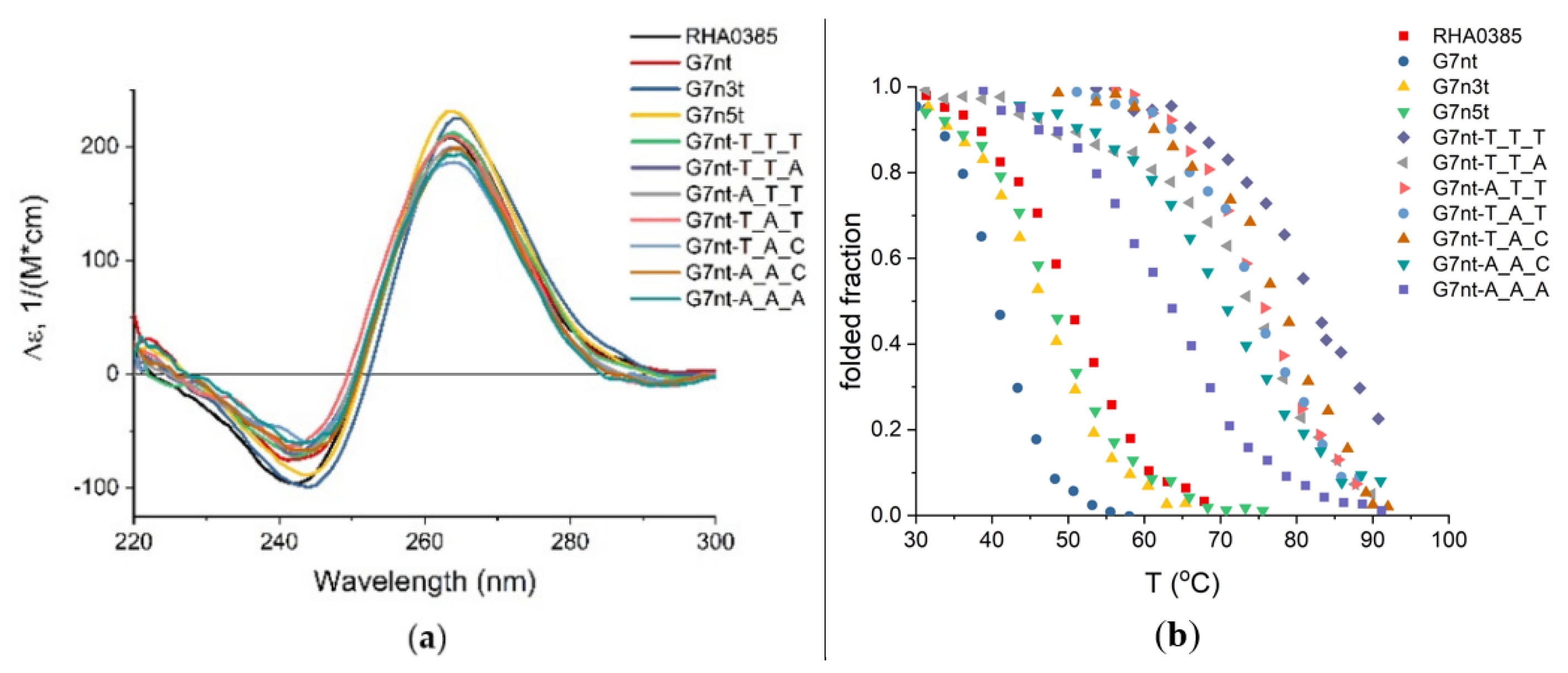
2.3. G-Quadruplex Structure of Aptamers
2.4. Oligomeric Composition of G-Quadruplexes
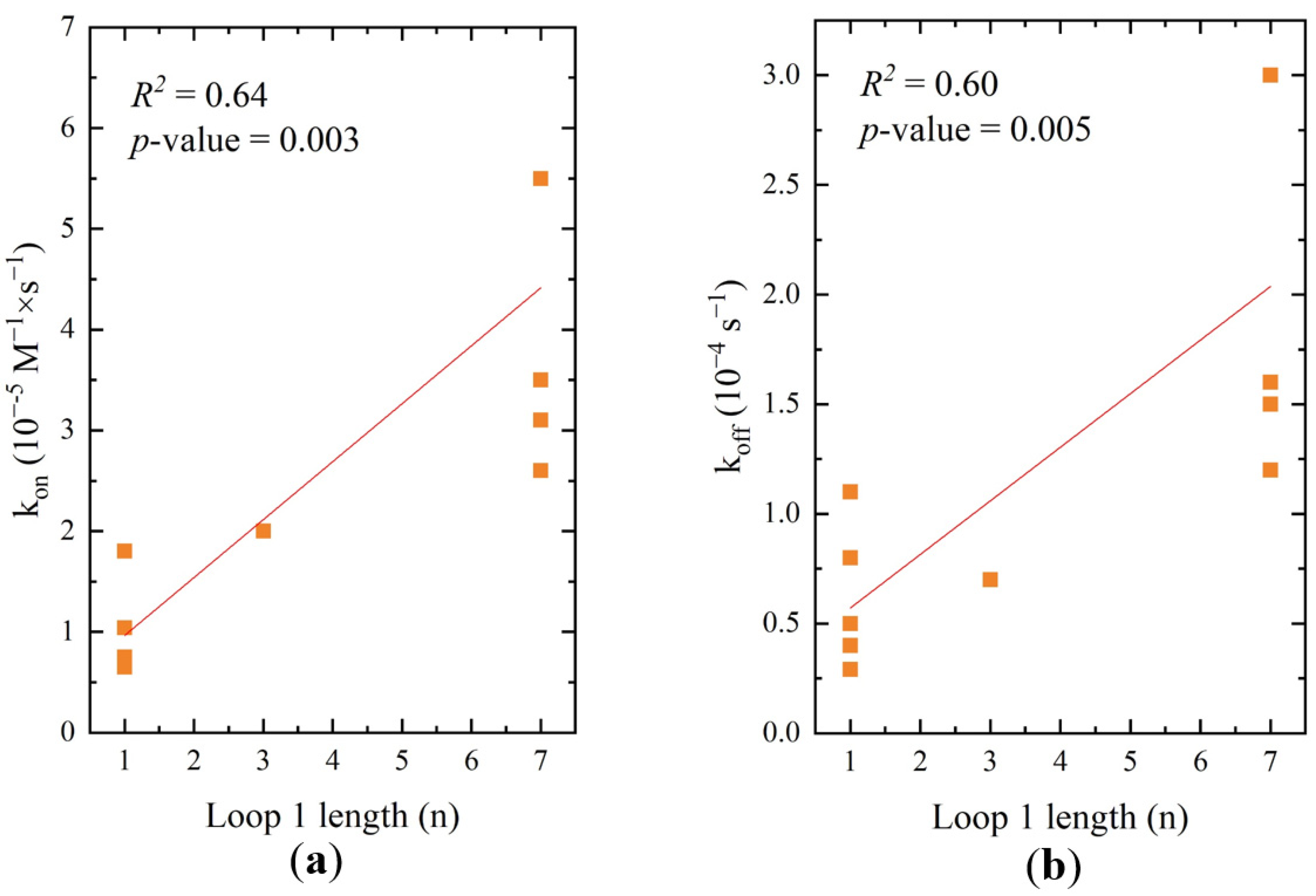
2.5. Correlations between Structure and Affinity of RHA0385 Derivatives
3. Materials and Methods
3.1. Oligonucleotides Preparation
3.2. Influenza A Virus Preparation and Characterization
3.3. BLI Affinity Assay
3.3.1. I Strategy (rHA Immobilized + Aptamer in Solution)
3.3.2. II Strategy (vHA in Solution + Aptamer Immobilized)
3.4. HAI Test
3.5. Circular Dichroism (CD) Spectroscopic Study
3.6. Size-Exclusion High Performance Liquid Chromatography (SE-HPLC)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| BLI | Biolayer interferometry |
| HA | Hemagglutinin |
| HAI | Hemagglutination inhibition assay |
| HPLC | High-performance liquid chromatography |
| rHA | Recombinant hemagglutinin |
| vHA | Viral hemagglutinin |
| SA | Streptavidin |
| SAR | Structure−affinity relationship |
| SE-HPLC | Size-exclusion high-performance liquid chromatography |
| SPR | Surface plasmon resonance |
References
- Rohloff, J.C.; Gelinas, A.D.; Jarvis, T.C.; Ochsner, U.A.; Schneider, D.J.; Gold, L.; Janjic, N. Nucleic acid ligands with protein-like side chains: Modified aptamers and their use as diagnostic and therapeutic agents. Mol. Ther. Nucleic Acids 2014, 3, e201. [Google Scholar] [CrossRef] [PubMed]
- Gelinas, A.D.; Davies, D.R.; Janjic, N. Embracing proteins: Structural themes in aptamer–protein complexes. Curr. Opin. Struct. Biol. 2016, 36, 122–132. [Google Scholar] [CrossRef] [Green Version]
- Bjerregaard, N.; Andreasen, P.A.; Dupont, D.M. Expected and unexpected features of protein-binding RNA aptamers. Wiley Interdiscip. Rev. RNA 2016, 7, 744–757. [Google Scholar] [CrossRef]
- Novoseltseva, A.; Zavyalova, E.; Golovin, A.; Kopylov, A. An insight into aptamer–protein complexes. Aptamers 2018, 2, 1–19. [Google Scholar]
- Zavyalova, E.; Kopylov, A. Energy Transfer as A Driving Force in Nucleic Acid⁻Protein Interactions. Molecules 2019, 24, 1443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zavyalova, E.G.; Legatova, V.A.; Alieva, R.S.; Zalevsky, A.O.; Tashlitsky, V.N.; Arutyunyan, A.M.; Kopylov, A.M. Putative mechanisms underlying high inhibitory activities of bimodular DNA aptamers to thrombin. Biomolecules 2019, 9, 41. [Google Scholar] [CrossRef] [Green Version]
- Harris, A.; Cardone, G.; Winkler, D.C.; Heymann, J.B.; Brecher, M.; White, J.M.; Steven, A.C. Influenza virus pleiomorphy characterized by cryoelectron tomography. Proc. Natl. Acad. Sci. USA 2006, 103, 19123–19127. [Google Scholar] [CrossRef] [Green Version]
- Zavyalova, E.; Kopylov, A. Aptamers to Hemagglutinin: A Novel Tool for Influenza Virus Recognition and Neutralization. Curr. Pharm. Des. 2016, 22, 4835–4853. [Google Scholar] [CrossRef]
- Shiratori, I.; Akitomi, J.; Boltz, D.A.; Horii, K.; Furuichi, M.; Waga, I. Selection of DNA aptamers that bind to influenza A viruses with high affinity and broad subtype specificity. Biochem. Biophys. Res. Commun. 2014, 443, 37–41. [Google Scholar] [CrossRef] [Green Version]
- Woo, H.M.; Lee, J.M.; Yim, S.; Jeong, Y.J. Isolation of single-stranded DNA aptamers that distinguish influenza virus hemagglutinin subtype H1 from H5. PLoS ONE 2015, 10, e0125060. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Yu, Z.; Jiang, F.; Fu, P.; Shen, J.; Wu, W.; Li, J. Two DNA aptamers against avian influenza H9N2 virus prevent viral infection in cells. PLoS ONE 2015, 10, e0123060. [Google Scholar] [CrossRef] [PubMed]
- Burge, S.; Parkinson, G.N.; Hazel, P.; Todd, A.K.; Neidle, S. Quadruplex DNA: Sequence, topology and structure. Nucleic Acids Res. 2006, 34, 5402–5415. [Google Scholar] [CrossRef] [Green Version]
- Platella, C.; Riccardi, C.; Montesarchio, D.; Roviello, G.N.; Musumeci, D. G-quadruplex-based aptamers against protein targets in therapy and diagnostics. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 1429–1447. [Google Scholar] [CrossRef] [PubMed]
- Kwok, C.K.; Merrick, C.J. G-Quadruplexes: Prediction, Characterization, and Biological Application. Trends Biotechnol. 2017, 35, 997–1013. [Google Scholar] [CrossRef]
- Roxo, C.; Kotkowiak, W.; Pasternak, A. G-Quadruplex-Forming Aptamers-Characteristics, Applications, and Perspectives. Molecules 2019, 24, 3781. [Google Scholar] [CrossRef] [Green Version]
- Carvalho, J.; Mergny, J.L.; Salgado, G.F.; Queiroz, J.A.; Cruz, C. G-quadruplex, Friend or Foe: The Role of the G-quartet in Anticancer Strategies. Trends Mol. Med. 2020, 26, 848–861. [Google Scholar] [CrossRef]
- Kukushkin, V.I.; Ivanov, N.M.; Novoseltseva, A.A.; Gambaryan, A.S.; Yaminsky, I.V.; Kopylov, A.M.; Zavyalova, E.G. Highly sensitive detection of influenza virus with SERS aptasensor. PLoS ONE 2019, 14, e0216247. [Google Scholar] [CrossRef] [Green Version]
- Pang, Y.; Rong, Z.; Wang, J.; Xiao, R.; Wang, S. A fluorescent aptasensor for H5N1 influenza virus detection based-on the core-shell nanoparticles metal-enhanced fluorescence (MEF). Biosens. Bioelectron. 2015, 66, 527–532. [Google Scholar] [CrossRef]
- Diba, F.S.; Kim, S.; Lee, H.J. Amperometric bioaffinity sensing platform for avian influenza virus proteins with aptamer modified gold nanoparticles on carbon chips. Biosens. Bioelectron. 2015, 72, 355–361. [Google Scholar] [CrossRef]
- Novoseltseva, A.A.; Ivanov, N.M.; Novikov, R.A.; Tkachev, Y.V.; Bunin, D.A.; Gambaryan, A.S.; Tashlitsky, V.N.; Arutyunyan, A.M.; Kopylov, A.M.; Zavyalova, E.G. Structural and Functional Aspects of G-Quadruplex Aptamers Which Bind a Broad Range of Influenza A Viruses. Biomolecules 2020, 10, 119. [Google Scholar] [CrossRef] [Green Version]
- Lou, X.; Egli, M.; Yang, X. Determining functional aptamer–protein interaction by biolayer interferometry. Curr. Protoc. Nucleic Acid Chem. 2016, 67, 7–25. [Google Scholar] [CrossRef] [PubMed]
- Plach, M.; Schubert, T. Biophysical Characterization of Aptamer–Target Interactions. Adv. Biochem. Eng. Biotechnol. 2019, 174, 1–15. [Google Scholar] [CrossRef]
- Zavyalova, E.; Turashev, A.; Novoseltseva, A.; Legatova, V.; Antipova, O.; Savchenko, E.; Balk, S.; Golovin, A.; Pavlova, G.; Kopylov, A. Pyrene-Modified DNA Aptamers with High Affinity to Wild-Type EGFR and EGFRvIII. Nucleic Acid Ther. 2020, 30, 175–187. [Google Scholar] [CrossRef]
- KissCC0. Available online: https://www.kisscc0.com/clipart/avian-influenza-influenza-a-virus-subtype-h7n9-com-85jpxh/ (accessed on 27 January 2020).
- Dolot, R.; Lam, C.H.; Sierant, M.; Zhao, Q.; Liu, F.W.; Nawrot, B.; Egli, M.; Yang, X. Crystal structures of thrombin in complex with chemically modified thrombin DNA aptamers reveal the origins of enhanced affinity. Nucleic Acids Res. 2018, 46, 4819–4830. [Google Scholar] [CrossRef] [PubMed]
- Vorlickova, M.; Kejnovska, I.; Sagi, J.; Renciuk, D.; Bednarova, K.; Motlova, J.; Kypr, J. Circular dichroism and guanine quadruplexes. Methods 2012, 57, 64–75. [Google Scholar] [CrossRef] [PubMed]
- Kypr, J.; Kejnovská, I.; Renciuk, D.; Vorlícková, M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009, 37, 1713–1725. [Google Scholar] [CrossRef] [Green Version]
- Karsisiotis, A.I.; Hessari, N.M.; Novellino, E.; Spada, G.P.; Randazzo, A.; Webba da Silva, M. Topological characterization of nucleic acid G-quadruplexes by UV absorption and circular dichroism. Angew. Chem. Int. Ed. Engl. 2011, 50, 10645–10648. [Google Scholar] [CrossRef]
- Del Villar-Guerra, R.; Trent, J.O.; Chaires, J.B. G-Quadruplex Secondary Structure Obtained from Circular Dichroism Spectroscopy. Angew. Chem. Int. Ed. Engl. 2018, 57, 7171–7175. [Google Scholar] [CrossRef]
- Hatzakis, E.; Okamoto, K.; Yang, D. Thermodynamic stability and folding kinetics of the major G-quadruplex and its loop isomers formed in the nuclease hypersensitive element in the human c-Myc promoter: Effect of loops and flanking segments on the stability of parallel-stranded intramolecular G-quadruplexes. Biochemistry 2010, 49, 9152–9160. [Google Scholar] [CrossRef] [Green Version]
- Bugaut, A.; Balasubramanian, S. A sequence-independent study of the influence of short loop lengths on the stability and topology of intramolecular DNA G-quadruplexes. Biochemistry 2008, 47, 689–697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guédin, A.; Gros, J.; Alberti, P.; Mergny, J.L. How long is too long? Effects of loop size on G-quadruplex stability. Nucleic Acids Res. 2010, 38, 7858–7868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelley, S.; Boroda, S.; Musier-Forsyth, K.; Kankia, B.I. HIV-integrase aptamer folds into a parallel quadruplex: A thermodynamic study. Biophys. Chem. 2011, 155, 82–88. [Google Scholar] [CrossRef]
- Largy, E.; Mergny, J.L. Shape matters: Size-exclusion HPLC for the study of nucleic acid structural polymorphism. Nucleic Acids Res. 2014, 42, e149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dailey, M.M.; Miller, C.M.; Bates, P.J.; Lane, A.N.; Trent, J.O. Resolution and characterization of the structural polymorphism of a single quadruplex-forming sequence. Nucleic Acids Res. 2010, 38, 4877–4888. [Google Scholar] [CrossRef]
- Riccardi, C.; Musumeci, D.; Russo Krauss, I.; Piccolo, M.; Irace, C.; Paduano, L.; Montesarchio, D. Exploring the conformational behaviour and aggregation properties of lipid-conjugated AS1411 aptamers. Int. J. Biol. Macromol. 2018, 118, 1384–1399. [Google Scholar] [CrossRef] [PubMed]
- Alieva, R.R.; Zavyalova, E.G.; Tashlitsky, V.N.; Kopylov, A.M. Quantitative characterization of oligomeric state of G-quadruplex antithrombin aptamers by size exclusion HPLC. Mendeleev Commun. 2019, 29, 424–425. [Google Scholar] [CrossRef]
- Aliyeva, R.; Tashlitsky, V.; Kopylov, A.; Zavyalova, E. Bimodular thrombin aptamers with two types of non-covalent locks. Bioconjugate Chem. under review.
- Di Bucchianico, A. Coefficient of determination (R 2). In Encyclopedia of Statistics in Quality and Reliability; John Wiley & Sons, Ltd.: New York, NY, USA, 2008; p. 1. [Google Scholar] [CrossRef]
- Benjamin, D.J.; Berger, J.O. Three recommendations for improving the use of p-values. Am. Stat. 2019, 73, 186–191. [Google Scholar] [CrossRef] [Green Version]
- Ustinov, N.B.; Zavyalova, E.G.; Smirnova, I.G.; Kopylov, A.M. The power and limitations of influenza virus hemagglutinin assays. Biochem. Mosc. 2017, 84, 1234–1248. [Google Scholar] [CrossRef]
- Ambartsumyan, O.; Gribanyov, D.; Kukushkin, V.; Kopylov, A.; Zavyalova, E. SERS-Based Biosensors for Virus Determination with Oligonucleotides as Recognition Elements. Int. J. Mol. Sci. 2020, 21, 3373. [Google Scholar] [CrossRef]
- Magbanua, E.; Zivkovic, T.; Hansen, B.; Beschorner, N.; Meyer, C.; Lorenzen, I.; Grötzinger, J.; Hauber, J.; Torda, A.E.; Mayer, G.; et al. d(GGGT) 4 and r(GGGU) 4 are both HIV-1 inhibitors and interleukin-6 receptor aptamers. RNA Biol. 2013, 10, 216–227. [Google Scholar] [CrossRef] [Green Version]
- Jing, N.; De Clercq, E.; Rando, R.F.; Pallansch, L.; Lackman-Smith, C.; Lee, S.; Hogan, M.E. Stability-activity relationships of a family of G-tetrad forming oligonucleotides as potent HIV inhibitors. A basis for anti-HIV drug design. J. Biol. Chem. 2000, 275, 3421–3430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jing, N.; Li, Y.; Xu, X.; Sha, W.; Li, P.; Feng, L.; Tweardy, D.J. Targeting Stat3 with G-quartet oligodeoxynucleotides in human cancer cells. DNA Cell Biol. 2003, 22, 685–696. [Google Scholar] [CrossRef] [PubMed]
- Kramberger, P.; Ciringer, M.; Strancar, A.; Peterka, M. Evaluation of nanoparticle tracking analysis for total virus particle determination. Virol. J. 2012, 9, 265. [Google Scholar] [CrossRef] [PubMed]
- O’Shannessy, D.J.; Brigham-Burke, M.; Soneson, K.K.; Hensley, P.; Brooks, I. Determination of rate and equilibrium binding constants for macromolecular interactions using surface plasmon resonance: Use of nonlinear least squares analysis methods. Anal. Biochem. 1993, 212, 457–468. [Google Scholar] [CrossRef]


| ID | Sequence, 5′-3′ | Length, nt | ɛ, cm−1 × M−1 | Mw, Da |
|---|---|---|---|---|
| RHA0385 | TTGGGGTTATTTTGGGAGGGCGGGGGTT | 28 | 268,100 | 8834 |
| G7nt | GGGTTATTTTGGGAGGGCGGG | 21 | 205,600 | 6629 |
| G7n3t | TTGGGGTTATTTTGGGAGGGCGGG | 24 | 231,300 | 7567 |
| G7n5t | GGGTTATTTTGGGAGGGCGGGGGTT | 25 | 242,400 | 7896 |
| G7nt-T_T_T | GGGTGGGTGGGTGGG | 15 | 148,700 | 4801 |
| G7nt-A_T_T | GGGAGGGTGGGTGGG | 15 | 153,200 | 4810 |
| G7nt-T_A_T | GGGTGGGAGGGTGGG | 15 | 153,200 | 4810 |
| G7nt-T_T_A | GGGTGGGTGGGAGGG | 15 | 153,200 | 4810 |
| G7nt-T_A_C | GGGTGGGAGGGCGGG | 15 | 151,100 | 4795 |
| G7nt-A_A_C | GGGAGGGAGGGCGGG | 15 | 155,600 | 4804 |
| G7nt-A_A_A | GGGAGGGAGGGAGGG | 15 | 162,200 | 4828 |
| G7-TTATTAA_A_C | TTGGGGTTATTAAGGGAGGGCGGGGGTT | 28 | 277,900 | 8851 |
| G7-TTA_A_C | TTGGGGTTAGGGAGGGCGGGGGTT | 24 | 235,700 | 7617 |
| I Strategy, rHA | II Strategy, vHA | |||||
|---|---|---|---|---|---|---|
| kon, 1 × 105 M−1 × s−1 | koff, 1 × 10−4 s−1 | aKD, nM | kon, 1 × 105 M−1 × s−1 | koff, 1 × 10−4 s−1 | aKD, nM | |
| RHA0385 | 2.6 ± 0.2 | 1.5 ± 0.7 | 0.6 ± 0.3 | 41 ± 6 | 2.0 ± 0.6 | 0.05 ± 0.02 |
| G7nt | 3.5 ± 0.2 | 3 ± 1 | 0.7 ± 0.3 | 19 ± 5 | 2.6 ± 0.6 | 0.13 ± 0.07 |
| G7n3t | 7.4 ± 0.6 | 3 ± 1 | 0.4 ± 0.2 | 28 ± 5 | 4 ± 1 | 0.16 ± 0.07 |
| G7n5t | 5.5 ± 0.2 | 1.6 ± 0. 8 | 0.3 ± 0.2 | 33 ± 6 | 1.5 ± 0.5 | 0.04 ± 0.02 |
| G7nt-T_T_T | 1.04 ± 0.04 | 0.8 ± 0.2 | 0.7 ± 0.2 | 8.0 ± 0.8 | 5 ± 2 | 0.6 ± 0.3 |
| G7nt-A_T_T | 0.65 ± 0.08 | 0.29 ± 0.09 | 0.4 ± 0.2 | 21 ± 4 | 4 ± 1 | 0.2 ± 0.1 |
| G7nt-T_A_T | 1.80 ± 0.05 | 1.1 ± 0.4 | 0.6 ± 0.2 | 37 ± 9 | 2.6 ± 0.7 | 0.07 ± 0.04 |
| G7nt-T_T_A | 0.65 ± 0.01 | 0.5 ± 0.1 | 0.8 ± 0.2 | 10 ± 2 | 7 ± 2 | 0.7 ± 0.3 |
| G7nt-T_A_C | - | - | - | 16 ± 4 | 5 ± 1 | 0.3 ± 0.1 |
| G7nt-A_A_C | - | - | - | 15 ± 3 | 4 ± 1 | 0.3 ± 0.1 |
| G7nt-A_A_A | 0.75 ± 0.09 | 0.4 ± 0.2 | 0.6 ± 0.4 | 22 ± 3 | 5 ± 2 | 0.2 ± 0.1 |
| G7-TTATTAA_A_C | 3.10 ± 0.09 | 1.2 ± 0.6 | 0.4 ± 0.2 | 33 ± 6 | 3 ± 1 | 0.09 ± 0.05 |
| G7-TTA_A_C | 2.0 ± 0.2 | 0.7 ± 0.4 | 0.4 ± 0.2 | 54 ± 7 | 2.0 ± 0.3 | 0.04 ± 0.01 |
| ID | Monomer content, % | Tm, °C |
|---|---|---|
| RHA0385 | 81 [15] | 50.1 ± 0.2 [15] |
| G7nt | 74 | 40.6 ± 0.1 |
| G7n3t | 100 | 46.5 ± 0.1 |
| G7n5t | 74 | 47.8 ± 0.2 |
| G7nt-T_T_T | 60 | >80 |
| G7nt-A_T_T | 69 | 75.4 ± 0.3 |
| G7nt-T_A_T | 43 | 74.8 ± 0.5 |
| G7nt-T_T_A | 66 | 78 ± 2 |
| G7nt-T_A_C | 60 | 82 ± 2 |
| G7nt-A_A_C | 63 | 70.3 ± 0.5 |
| G7nt-A_A_A | 60 | 62.9 ± 0.2 |
| G7-TTATTAA_A_C | 77 [20] | 52.9 ± 0.2 [15] |
| G7-TTA_A_C | 86 [20] | 61.4 ± 0.1 [15] |
| I strategy, rHA | II strategy, vHA | |||||
|---|---|---|---|---|---|---|
| kon, 1 × 105 M−1 × s−1 | koff, 1 × 10−4 s−1 | ΔGb,303, kJ/mol | kon, 1 × 105 M−1 × s−1 | koff, 1 × 10−4 s−1 | ΔGb,303, kJ/mol | |
| Major loop length, nt | 0.64/0.003 | 0.60/0.005 | 0.22/0.2 | 0.18/0.1 | 0.39/0.02 | 0.35/0.03 |
| Monomer content, % | 0.45/0.02 | 0.27/0.1 | 0.23/0.1 | 0.15/0.2 | 0.08/0.3 | 0.11/0.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bizyaeva, A.A.; Bunin, D.A.; Moiseenko, V.L.; Gambaryan, A.S.; Balk, S.; Tashlitsky, V.N.; Arutyunyan, A.M.; Kopylov, A.M.; Zavyalova, E.G. The Functional Role of Loops and Flanking Sequences of G-Quadruplex Aptamer to the Hemagglutinin of Influenza a Virus. Int. J. Mol. Sci. 2021, 22, 2409. https://doi.org/10.3390/ijms22052409
Bizyaeva AA, Bunin DA, Moiseenko VL, Gambaryan AS, Balk S, Tashlitsky VN, Arutyunyan AM, Kopylov AM, Zavyalova EG. The Functional Role of Loops and Flanking Sequences of G-Quadruplex Aptamer to the Hemagglutinin of Influenza a Virus. International Journal of Molecular Sciences. 2021; 22(5):2409. https://doi.org/10.3390/ijms22052409
Chicago/Turabian StyleBizyaeva, Anastasia A., Dmitry A. Bunin, Valeria L. Moiseenko, Alexandra S. Gambaryan, Sonja Balk, Vadim N. Tashlitsky, Alexander M. Arutyunyan, Alexey M. Kopylov, and Elena G. Zavyalova. 2021. "The Functional Role of Loops and Flanking Sequences of G-Quadruplex Aptamer to the Hemagglutinin of Influenza a Virus" International Journal of Molecular Sciences 22, no. 5: 2409. https://doi.org/10.3390/ijms22052409
APA StyleBizyaeva, A. A., Bunin, D. A., Moiseenko, V. L., Gambaryan, A. S., Balk, S., Tashlitsky, V. N., Arutyunyan, A. M., Kopylov, A. M., & Zavyalova, E. G. (2021). The Functional Role of Loops and Flanking Sequences of G-Quadruplex Aptamer to the Hemagglutinin of Influenza a Virus. International Journal of Molecular Sciences, 22(5), 2409. https://doi.org/10.3390/ijms22052409







