Pseudomonas Flagella: Generalities and Specificities
Abstract
:1. Introduction
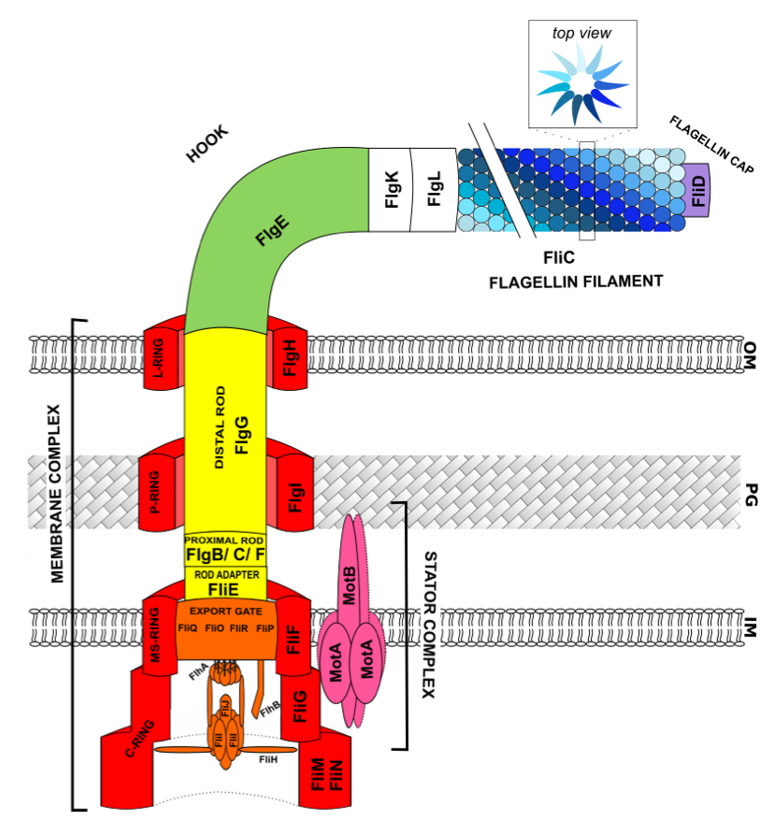
2. Flagellar Structure
3. Dynamics of Flagellar Assembly
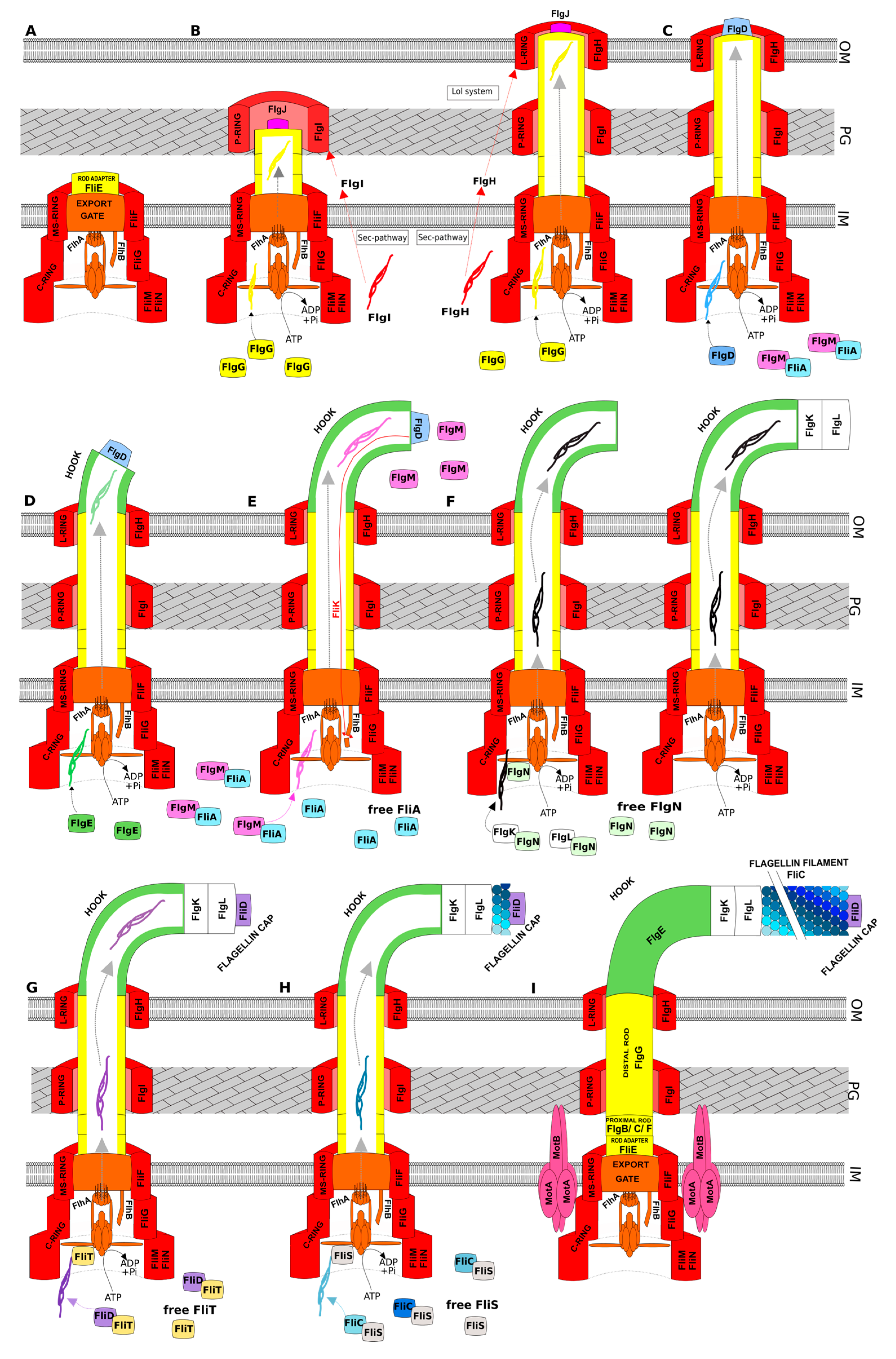
3.1. General Scenario of Assembly
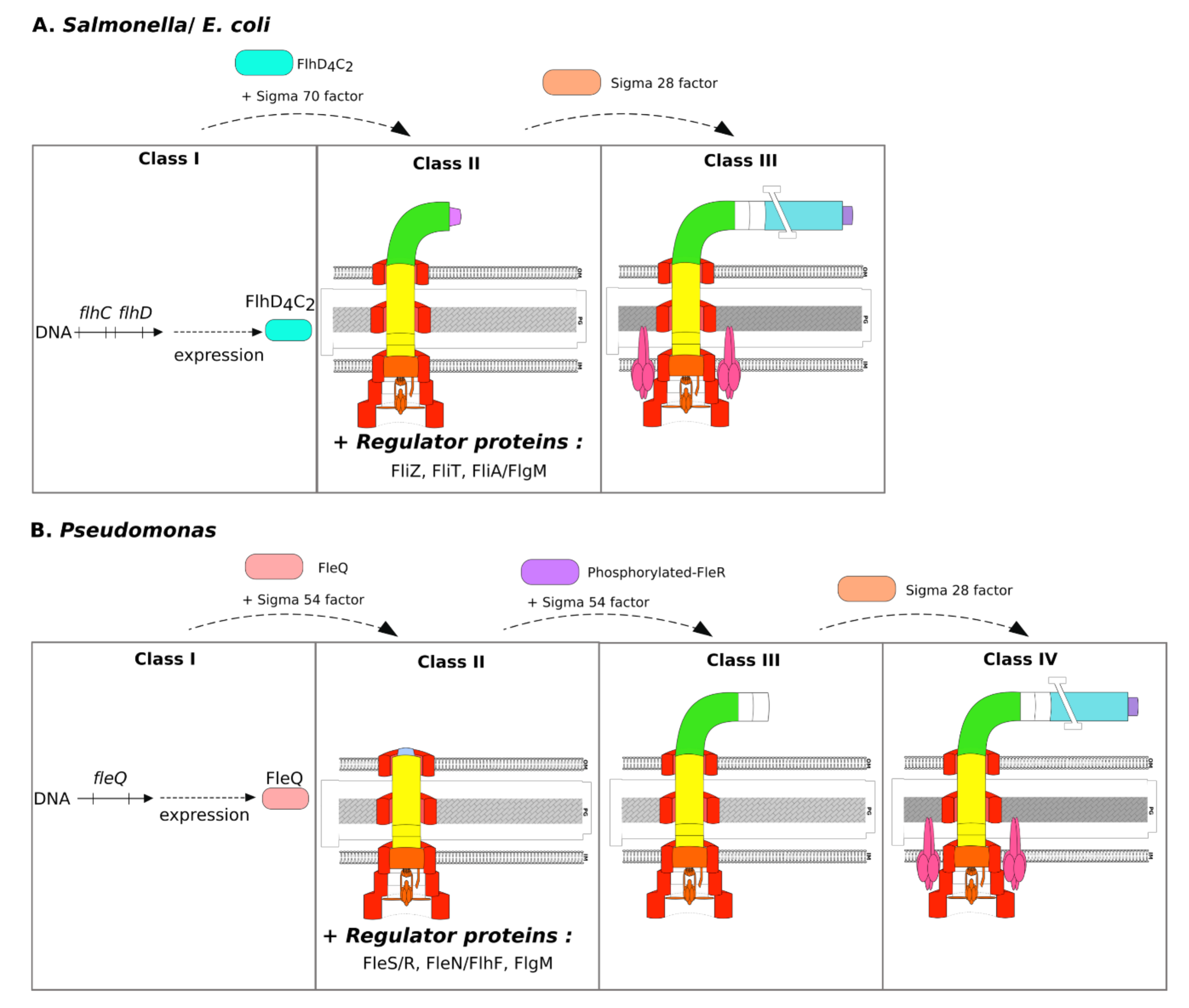
3.2. Specificities of Salmonella and E. coli
3.3. Specificities of Pseudomonas
4. Fuelling of the Flagellar Machinery
4.1. Export of Flagellar Proteins
4.2. Generation of Flagellar Torque
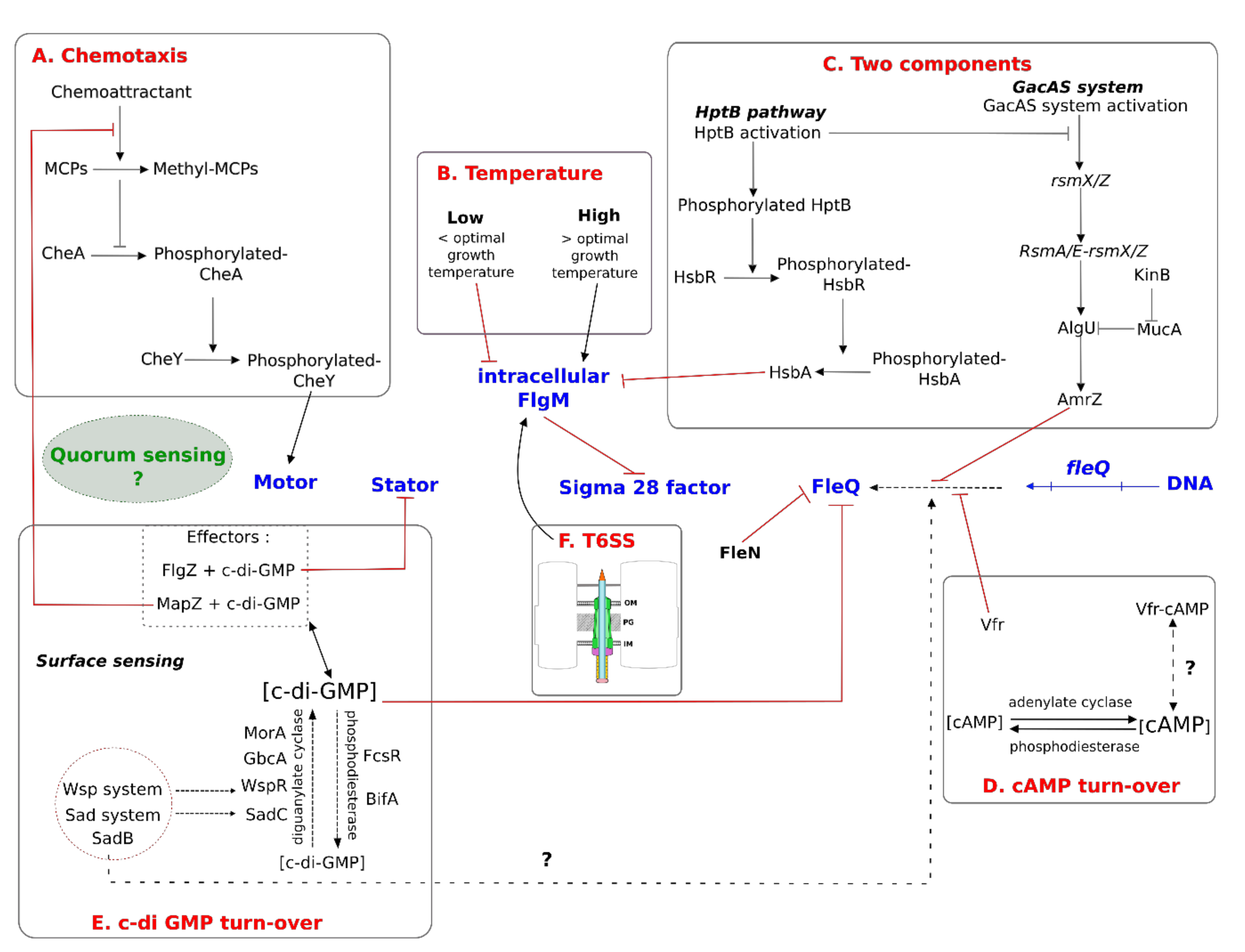
4.2.1. Chemotaxis
4.2.2. Energy Generation
5. Regulation of Flagella
5.1. Extrinsic Factors and General Mechanisms
5.1.1. Biochemical Molecules
5.1.2. Temperature
5.1.3. DNA Topology
5.1.4. Membrane Stress
5.2. Regulation by Two-Component Systems
5.3. Regulation by Second Messengers
5.3.1. Adenosine 3′-5′ Cyclic Monophosphate (cAMP)
5.3.2. Bis-(3′-5′)-Cyclic Dimeric Guanosine Monophosphate (c-di-GMP)
- First, c-di-GMP is able to act at the transcriptional level. For example, in V. cholerae, overexpression of CdgF, a DGC, induces a decrease in transcript levels of many class III and class IV genes and thus a decrease in “swimming” motility [21].
- Second, c-di-GMP regulates flagella at the post-transcriptional level. In the polar flagellated species Caulobacter crescentus, TipF protein appears to function as a PDE. Deletion of the tipF gene in this bacterial species affects motility but does not impair fliC or flgE transcription. Only the hook and flagellar filament are missing. In E. coli, MifA and MifB are two DGCs that promote c-di-GMP production. Both enzymes have been reported to act at a post-transcriptional level to decrease flagella production.
- Finally, c-di-GMP acts directly by altering flagellar function. The PilZ-like protein, YcgR, in E. coli or S. enterica, binds to c-di-GMP and consequently interferes with the association of Mot protein with FliG, which impairs flagellar rotation [21,22]. In E. coli, YhjH is a PDE and DgcE a DGC. These two proteins are essential for controlling c-di-GMP levels, and mediate flagellar activity [107]. This second messenger is recognized by two diguanylate receptors (DgrA/DgrB) that impair flagella function.
5.4. Regulation by Quorum Sensing
5.5. Crosstalk between Flagella and the T6SS
6. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Soutourina, O.A.; Bertin, P.N. Regulation Cascade of Flagellar Expression in Gram-Negative Bacteria. FEMS Microbiol. Rev. 2003, 27, 505–523. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Burrage, A.M.; Postel, S.; Clark, R.E.; Orlova, A.; Sundberg, E.J.; Kearns, D.B.; Egelman, E.H. A Structural Model of Flagellar Filament Switching across Multiple Bacterial Species. Nat. Commun. 2017, 8, 960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hockett, K.L.; Burch, A.Y.; Lindow, S.E. Thermo-Regulation of Genes Mediating Motility and Plant Interactions in Pseudomonas Syringae. PLoS ONE 2013, 8, e59850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haiko, J.; Westerlund-Wikström, B. The Role of the Bacterial Flagellum in Adhesion and Virulence. Biology 2013, 2, 1242–1267. [Google Scholar] [CrossRef] [Green Version]
- Toutain, C.M.; Caizza, N.C.; Zegans, M.E.; O’Toole, G.A. Roles for Flagellar Stators in Biofilm Formation by Pseudomonas Aeruginosa. Res. Microbiol. 2007, 158, 471–477. [Google Scholar] [CrossRef]
- Diepold, A.; Armitage, J.P. Type III Secretion Systems: The Bacterial Flagellum and the Injectisome. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20150020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belas, R. Biofilms, Flagella, and Mechanosensing of Surfaces by Bacteria. Trends Microbiol. 2014, 22, 517–527. [Google Scholar] [CrossRef]
- Minamino, T.; Imada, K. The Bacterial Flagellar Motor and Its Structural Diversity. Trends Microbiol. 2015, 23, 267–274. [Google Scholar] [CrossRef]
- Gewirtz, A.T.; Navas, T.A.; Lyons, S.; Godowski, P.J.; Madara, J.L. Cutting Edge: Bacterial Flagellin Activates Basolaterally Expressed TLR5 to Induce Epithelial Proinflammatory Gene Expression. J. Immunol. 2001, 167, 1882–1885. [Google Scholar] [CrossRef] [Green Version]
- Stecher, B.; Hapfelmeier, S.; Müller, C.; Kremer, M.; Stallmach, T.; Hardt, W.-D. Flagella and Chemotaxis Are Required for Efficient Induction of Salmonella Enterica Serovar Typhimurium Colitis in Streptomycin-Pretreated Mice. Infect. Immun. 2004, 72, 4138–4150. [Google Scholar] [CrossRef] [Green Version]
- Fleiszig, S.M.J.; Arora, S.K.; Van, R.; Ramphal, R. FlhA, a Component of the Flagellum Assembly Apparatus of Pseudomonas Aeruginosa, Plays a Role in Internalization by Corneal Epithelial Cells. Infect. Immun. 2001, 69, 4931–4937. [Google Scholar] [CrossRef] [Green Version]
- Taguchi, F.; Shimizu, R.; Inagaki, Y.; Toyoda, K.; Shiraishi, T.; Ichinose, Y. Post-Translational Modification of Flagellin Determines the Specificity of HR Induction. Plant Cell Physiol. 2003, 44, 342–349. [Google Scholar] [CrossRef] [Green Version]
- Osterman, I.A.; Dikhtyar, Y.Y.; Bogdanov, A.A.; Dontsova, O.A.; Sergiev, P.V. Regulation of Flagellar Gene Expression in Bacteria. Biochem. Mosc. 2015, 80, 1447–1456. [Google Scholar] [CrossRef]
- Kojima, S.; Terashima, H.; Homma, M. Regulation of the Single Polar Flagellar Biogenesis. Biomolecules 2020, 10, 533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rossignol, G.; Sperandio, D.; Guerillon, J.; Duclairoir Poc, C.; Soum-Soutera, E.; Orange, N.; Feuilloley, M.G.J.; Merieau, A. Phenotypic Variation in the Pseudomonas Fluorescens Clinical Strain MFN1032. Res. Microbiol. 2009, 160, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, N.; Ramphal, R. Interaction of the Antiactivator FleN with the Transcriptional Activator FleQ Regulates Flagellar Number InPseudomonas Aeruginosa. J. Bacteriol. 2001, 183, 6636–6644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bergeau, D.; Mazurier, S.; Barbey, C.; Merieau, A.; Chane, A.; Goux, D.; Bernard, S.; Driouich, A.; Lemanceau, P.; Vicré, M.; et al. Unusual Extracellular Appendages Deployed by the Model Strain Pseudomonas Fluorescens C7R12. PLoS ONE 2019, 14, e0221025. [Google Scholar] [CrossRef]
- Chevance, F.F.V.; Hughes, K.T. Coordinating Assembly of a Bacterial Macromolecular Machine. Nat. Rev. Microbiol. 2008, 6, 455–465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herlihey, F.A.; Clarke, A.J. Controlling Autolysis During Flagella Insertion in Gram-Negative Bacteria. In Protein Reviews. Advances in Experimental Medicine and Biology; Atassi, M.Z., Ed.; Springer Singapore: Singapore, 2016; Volume 925, pp. 41–56. ISBN 978-981-10-3709-2. [Google Scholar]
- Dasgupta, N.; Wolfgang, M.C.; Goodman, A.L.; Arora, S.K.; Jyot, J.; Lory, S.; Ramphal, R. A Four-Tiered Transcriptional Regulatory Circuit Controls Flagellar Biogenesis in Pseudomonas Aeruginosa. Mol. Microbiol. 2003, 50, 809–824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolfe, A.J.; Visick, K.L. Get the Message Out: Cyclic-Di-GMP Regulates Multiple Levels of Flagellum-Based Motility. J. Bacteriol. 2008, 190, 463–475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baker, A.E.; O’Toole, G.A. Bacteria, Rev Your Engines: Stator Dynamics Regulate Flagellar Motility. J. Bacteriol. 2017, 199, e00088-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kitao, A.; Hata, H. Molecular Dynamics Simulation of Bacterial Flagella. Biophys. Rev. 2018, 10, 617–629. [Google Scholar] [CrossRef] [Green Version]
- Minamino, T.; Inoue, Y.; Kinoshita, M.; Namba, K. FliK-Driven Conformational Rearrangements of FlhA and FlhB Are Required for Export Switching of the Flagellar Protein Export Apparatus. J. Bacteriol. 2019, 202, e00637-19. [Google Scholar] [CrossRef]
- Evans, L.D.B.; Hughes, C.; Fraser, G.M. Building a Flagellum Outside the Bacterial Cell. Trends Microbiol. 2014, 22, 566–572. [Google Scholar] [CrossRef] [Green Version]
- Terashima, H.; Kawamoto, A.; Tatsumi, C.; Namba, K.; Minamino, T.; Imada, K. In Vitro Reconstitution of Functional Type III Protein Export and Insights into Flagellar Assembly. mBio 2018, 9, e00988-18. [Google Scholar] [CrossRef] [Green Version]
- Kucera, J.; Terentjev, E.M. FliI6-FliJ Molecular Motor Assists with Unfolding in the Type III Secretion Export Apparatus. Sci. Rep. 2020, 10, 7127. [Google Scholar] [CrossRef]
- Bai, F.; Morimoto, Y.V.; Yoshimura, S.D.J.; Hara, N.; Kami-ike, N.; Namba, K.; Minamino, T. Assembly Dynamics and the Roles of FliI ATPase of the Bacterial Flagellar Export Apparatus. Sci. Rep. 2015, 4, 6528. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, S.; Minamino, T. Flagella-Driven Motility of Bacteria. Biomolecules 2019, 9, 279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujii, T.; Matsunami, H.; Inoue, Y.; Namba, K. Evidence for the Hook Supercoiling Mechanism of the Bacterial Flagellum. Biophys. Physicobiology 2018, 15, 28–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samatey, F.A.; Imada, K.; Nagashima, S.; Vonderviszt, F.; Kumasaka, T.; Yamamoto, M.; Namba, K. Structure of the Bacterial Flagellar Protofilament and Implications for a Switch for Supercoiling. Nature 2001, 410, 331–337. [Google Scholar] [CrossRef] [PubMed]
- Bandara, D.Y.M.N.; Tang, J.; Saharia, J.; Rogowski, L.W.; Ahn, C.W.; Kim, M.J. Characterization of Flagellar Filaments and Flagellin through Optical Microscopy and Label-Free Nanopore Responsiveness. Anal. Chem. 2019, 91, 13665–13674. [Google Scholar] [CrossRef]
- Van Gerven, N.; Waksman, G.; Remaut, H. Pili and Flagella. In Progress in Molecular Biology and Translational Science; Elsevier: Amsterdam, The Netherlands, 2011; Volume 103, pp. 21–72. ISBN 978-0-12-415906-8. [Google Scholar]
- Forstnerič, V.; Ivičak-Kocjan, K.; Plaper, T.; Jerala, R.; Benčina, M. The Role of the C-Terminal D0 Domain of Flagellin in Activation of Toll like Receptor 5. PLoS Pathog. 2017, 13, e1006574. [Google Scholar] [CrossRef]
- Spangenberg, C.; Heuer, T.; Bürger, C.; Tümmler, B. Genetic Diversity of Flagellins of Pseudomonas aeruginosa. FEBS Lett. 1996, 396, 213–217. [Google Scholar] [CrossRef] [Green Version]
- Arora, S.K.; Bangera, M.; Lory, S.; Ramphal, R. A Genomic Island in Pseudomonas Aeruginosa Carries the Determinants of Flagellin Glycosylation. Proc. Natl. Acad. Sci. USA 2001, 98, 9342–9347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horstmann, J.A.; Lunelli, M.; Cazzola, H.; Heidemann, J.; Kühne, C.; Steffen, P.; Szefs, S.; Rossi, C.; Lokareddy, R.K.; Wang, C.; et al. Methylation of Salmonella Typhimurium Flagella Promotes Bacterial Adhesion and Host Cell Invasion. Nat. Commun. 2020, 11, 2013. [Google Scholar] [CrossRef] [Green Version]
- Takeuchi, K.; Taguchi, F.; Inagaki, Y.; Toyoda, K.; Shiraishi, T.; Ichinose, Y. Flagellin Glycosylation Island in Pseudomonas Syringae Pv. Glycinea and Its Role in Host Specificity. J. Bacteriol. 2003, 185, 6658–6665. [Google Scholar] [CrossRef] [Green Version]
- Arora, S.K.; Neely, A.N.; Blair, B.; Lory, S.; Ramphal, R. Role of Motility and Flagellin Glycosylation in the Pathogenesis of Pseudomonas Aeruginosa Burn Wound Infections. Infect. Immun. 2005, 73, 4395–4398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merino, S.; Tomás, J.M. Gram-Negative Flagella Glycosylation. Int. J. Mol. Sci. 2014, 15, 2840–2857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taguchi, F.; Shibata, S.; Suzuki, T.; Ogawa, Y.; Aizawa, S.-I.; Takeuchi, K.; Ichinose, Y. Effects of Glycosylation on Swimming Ability and Flagellar Polymorphic Transformation in Pseudomonas Syringae Pv. Tabaci 6605. J. Bacteriol. 2008, 190, 764–768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalir, S. Ordering Genes in a Flagella Pathway by Analysis of Expression Kinetics from Living Bacteria. Science 2001, 292, 2080–2083. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCarter, L.L. Regulation of Flagella. Curr. Opin. Microbiol. 2006, 9, 180–186. [Google Scholar] [CrossRef] [PubMed]
- Matsunami, H.; Yoon, Y.-H.; Meshcheryakov, V.A.; Namba, K.; Samatey, F.A. Structural Flexibility of the Periplasmic Protein, FlgA, Regulates Flagellar P-Ring Assembly in Salmonella Enterica. Sci. Rep. 2016, 6, 27399. [Google Scholar] [CrossRef]
- Narita, S.; Tokuda, H. Sorting of Bacterial Lipoproteins to the Outer Membrane by the Lol System. In Protein Secretion; Economou, A., Ed.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2010; Volume 619, pp. 117–129. ISBN 978-1-60327-167-7. [Google Scholar]
- Lee, H.J.; Hughes, K.T. Posttranscriptional Control of the Salmonella Enterica Flagellar Hook Protein FlgE. J. Bacteriol. 2006, 188, 3308–3316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wood, T.K.; González Barrios, A.F.; Herzberg, M.; Lee, J. Motility Influences Biofilm Architecture in Escherichia coli. Appl. Microbiol. Biotechnol. 2006, 72, 361–367. [Google Scholar] [CrossRef]
- Das, C.; Mokashi, C.; Mande, S.S.; Saini, S. Dynamics and Control of Flagella Assembly in Salmonella Typhimurium. Front. Cell. Infect. Microbiol. 2018, 8, 36. [Google Scholar] [CrossRef] [Green Version]
- Kinoshita, M.; Hara, N.; Imada, K.; Namba, K.; Minamino, T. Interactions of Bacterial Flagellar Chaperone-Substrate Complexes with FlhA Contribute to Co-Ordinating Assembly of the Flagellar Filament: Interactions of Flagellar Chaperones with FlhA. Mol. Microbiol. 2013, 90, 1249–1261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galeva, A.; Moroz, N.; Yoon, Y.-H.; Hughes, K.T.; Samatey, F.A.; Kostyukova, A.S. Bacterial Flagellin-Specific Chaperone FliS Interacts with Anti-Sigma Factor FlgM. J. Bacteriol. 2014, 196, 1215–1221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rudenko, I.; Ni, B.; Glatter, T.; Sourjik, V. Inefficient Secretion of Anti-Sigma Factor FlgM Inhibits Bacterial Motility at High Temperature. iScience 2019, 16, 145–154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saini, S.; Brown, J.D.; Aldridge, P.D.; Rao, C.V. FliZ Is a Posttranslational Activator of FlhD4C2-Dependent Flagellar Gene Expression. J. Bacteriol. 2008, 190, 4979–4988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Partridge, J.D.; Nieto, V.; Harshey, R.M. A New Player at the Flagellar Motor: FliL Controls Both Motor Output and Bias. mBio 2015, 6, e02367-14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferris, H.U.; Minamino, T. Flipping the Switch: Bringing Order to Flagellar Assembly. Trends Microbiol. 2006, 14, 519–526. [Google Scholar] [CrossRef] [PubMed]
- Aldridge, P.; Karlinsey, J.E.; Becker, E.; Chevance, F.F.V.; Hughes, K.T. Flk Prevents Premature Secretion of the Anti-Sigma Factor FlgM into the Periplasm. Mol. Microbiol. 2006, 60, 630–643. [Google Scholar] [CrossRef] [Green Version]
- Karlinsey, J.E.; Lonner, J.; Brown, K.L.; Hughes, K.T. Translation/Secretion Coupling by Type III Secretion Systems. Cell 2000, 102, 487–497. [Google Scholar] [CrossRef] [Green Version]
- Ping, L.; Birkenbeil, J.; Monajembashi, S. Swimming Behavior of the Monotrichous Bacterium Pseudomonas Fluorescens SBW25. FEMS Microbiol. Ecol. 2013, 86, 36–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouteiller, M.; Gallique, M.; Bourigault, Y.; Kosta, A.; Hardouin, J.; Massier, S.; Konto-Ghiorghi, Y.; Barbey, C.; Latour, X.; Chane, A.; et al. Crosstalk between the Type VI Secretion System and the Expression of Class IV Flagellar Genes in the Pseudomonas Fluorescens MFE01 Strain. Microorganisms 2020, 8, 622. [Google Scholar] [CrossRef]
- Baraquet, C.; Harwood, C.S. Cyclic Diguanosine Monophosphate Represses Bacterial Flagella Synthesis by Interacting with the Walker A Motif of the Enhancer-Binding Protein FleQ. Proc. Natl. Acad. Sci. USA 2013, 110, 18478–18483. [Google Scholar] [CrossRef] [Green Version]
- Capdevila, S.; Martínez-Granero, F.M.; Sánchez-Contreras, M.; Rivilla, R.; Martín, M. Analysis of Pseudomonas Fluorescens F113 Genes Implicated in Flagellar Filament Synthesis and Their Role in Competitive Root Colonization. Microbiology 2004, 150, 3889–3897. [Google Scholar] [CrossRef] [Green Version]
- Murray, T.S.; Kazmierczak, B.I. FlhF Is Required for Swimming and Swarming in Pseudomonas Aeruginosa. J. Bacteriol. 2006, 188, 6995–7004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandza, S.; Baetens, M.; Park, C.H.; Au, T.; Keyhan, M.; Matin, A. The G-Protein FlhF Has a Role in Polar Flagellar Placement and General Stress Response Induction in Pseudomonas Putida. Mol. Microbiol. 2000, 36, 414–423. [Google Scholar] [CrossRef]
- Echazarreta, M.A.; Klose, K.E. Vibrio Flagellar Synthesis. Front. Cell. Infect. Microbiol. 2019, 9, 131. [Google Scholar] [CrossRef] [PubMed]
- Kondo, S.; Imura, Y.; Mizuno, A.; Homma, M.; Kojima, S. Biochemical Analysis of GTPase FlhF Which Controls the Number and Position of Flagellar Formation in Marine Vibrio. Sci. Rep. 2018, 8, 12115. [Google Scholar] [CrossRef]
- Schniederberend, M.; Abdurachim, K.; Murray, T.S.; Kazmierczak, B.I. The GTPase Activity of FlhF Is Dispensable for Flagellar Localization, but Not Motility, in Pseudomonas Aeruginosa. J. Bacteriol. 2013, 195, 1051–1060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schniederberend, M.; Williams, J.F.; Shine, E.; Shen, C.; Jain, R.; Emonet, T.; Kazmierczak, B.I. Modulation of Flagellar Rotation in Surface-Attached Bacteria: A Pathway for Rapid Surface-Sensing after Flagellar Attachment. PLoS Pathog. 2019, 15, e1008149. [Google Scholar] [CrossRef]
- Burnham, P.M.; Kolar, W.P.; Hendrixson, D.R. A Polar Flagellar Transcriptional Program Mediated by Diverse Two-Component Signal Transduction Systems and Basal Flagellar Proteins Is Broadly Conserved in Polar Flagellates. mBio 2020, 11, e03107-19. [Google Scholar] [CrossRef] [Green Version]
- Frisk, A.; Jyot, J.; Arora, S.K.; Ramphal, R. Identification and Functional Characterization of FlgM, a Gene Encoding the Anti-Sigma 28 Factor in Pseudomonas Aeruginosa. J. Bacteriol. 2002, 184, 1514–1521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez-Herva, J.J.; Duque, E.; Molina-Henares, M.A.; Navarro-Avilés, G.; Van Dillewijn, P.; De La Torre, J.; Molina-Henares, A.J.; La Campa, A.S.; Ran, F.A.; Segura, A.; et al. Physiological and Transcriptomic Characterization of a FliA Mutant of Pseudomonas Putida KT2440: Physiological and Transcriptiomic Characterization of KT2440. Environ. Microbiol. Rep. 2009, 2, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Barahona, E.; Navazo, A.; Garrido-Sanz, D.; Muriel, C.; Martínez-Granero, F.; Redondo-Nieto, M.; Martín, M.; Rivilla, R. Pseudomonas Fluorescens F113 Can Produce a Second Flagellar Apparatus, Which Is Important for Plant Root Colonization. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Galán, J.E. Energizing Type III Secretion Machines: What Is the Fuel? Nat. Struct. Mol. Biol. 2008, 15, 127–128. [Google Scholar] [CrossRef]
- De Keyzer, J.; van der Does, C.; Driessen, A.J.M. The Bacterial Translocase: A Dynamic Protein Channel Complex. Cell. Mol. Life Sci. CMLS 2003, 60, 2034–2052. [Google Scholar] [CrossRef] [PubMed]
- Paul, K.; Erhardt, M.; Hirano, T.; Blair, D.F.; Hughes, K.T. Energy Source of Flagellar Type III Secretion. Nature 2008, 451, 489–492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minamino, T.; Namba, K. Distinct Roles of the FliI ATPase and Proton Motive Force in Bacterial Flagellar Protein Export. Nature 2008, 451, 485–488. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.-C.; Rietsch, A. Fueling Type III Secretion. Trends Microbiol. 2015, 23, 296–300. [Google Scholar] [CrossRef] [Green Version]
- Stern, A.S.; Berg, H.C. Single-File Diffusion of Flagellin in Flagellar Filaments. Biophys. J. 2013, 105, 182–184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Weert, S.; Vermeiren, H.; Mulders, I.H.M.; Kuiper, I.; Hendrickx, N.; Bloemberg, G.V.; Vanderleyden, J.; De Mot, R.; Lugtenberg, B.J.J. Flagella-Driven Chemotaxis Towards Exudate Components Is an Important Trait for Tomato Root Colonization by Pseudomonas fluorescens. Mol. Plant. Microbe Interact. 2002, 15, 1173–1180. [Google Scholar] [CrossRef] [Green Version]
- Oku, S.; Komatsu, A.; Tajima, T.; Nakashimada, Y.; Kato, J. Identification of Chemotaxis Sensory Proteins for Amino Acids in Pseudomonas Fluorescens Pf0-1 and Their Involvement in Chemotaxis to Tomato Root Exudate and Root Colonization. Microbes Environ. 2012, 27, 462–469. [Google Scholar] [CrossRef] [Green Version]
- Oku, S.; Komatsu, A.; Nakashimada, Y.; Tajima, T.; Kato, J. Identification of Pseudomonas Fluorescens Chemotaxis Sensory Proteins for Malate, Succinate, and Fumarate, and Their Involvement in Root Colonization. Microbes Environ. 2014, 29, 413–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sampedro, I.; Parales, R.E.; Krell, T.; Hill, J.E. Pseudomonas Chemotaxis. FEMS Microbiol. Rev. 2015, 39, 17–46. [Google Scholar] [CrossRef] [Green Version]
- Güvener, Z.T.; Tifrea, D.F.; Harwood, C.S. Two Different Pseudomonas aeruginosa Chemosensory Signal Transduction Complexes Localize to Cell Poles and Form and Remould in Stationary Phase: Localization of Pseudomonas Chemotaxis Proteins. Mol. Microbiol. 2006, 61, 106–118. [Google Scholar] [CrossRef]
- Cai, Q.; Li, Z.; Ouyang, Q.; Luo, C.; Gordon, V.D. Singly Flagellated Pseudomonas aeruginosa Chemotaxes Efficiently by Unbiased Motor Regulation. mBio 2016, 7, e00013-16. [Google Scholar] [CrossRef] [Green Version]
- Mandadapu, K.K.; Nirody, J.A.; Berry, R.M.; Oster, G. Mechanics of Torque Generation in the Bacterial Flagellar Motor. Proc. Natl. Acad. Sci. USA 2015, 112, E4381–E4389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Che, Y.-S.; Nakamura, S.; Morimoto, Y.V.; Kami-ike, N.; Namba, K.; Minamino, T. Load-Sensitive Coupling of Proton Translocation and Torque Generation in the Bacterial Flagellar Motor: Effect of the MotB(D33E) Mutation on the Speed Stability of the Flagellar Motor. Mol. Microbiol. 2014, 91, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Toutain, C.M.; Zegans, M.E.; O’Toole, G.A. Evidence for Two Flagellar Stators and Their Role in the Motility of Pseudomonas Aeruginosa. J. Bacteriol. 2005, 187, 771–777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doyle, T.B.; Hawkins, A.C.; McCarter, L.L. The Complex Flagellar Torque Generator of Pseudomonas Aeruginosa. J. Bacteriol. 2004, 186, 6341–6350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harrison, J.J.; Almblad, H.; Irie, Y.; Wolter, D.J.; Eggleston, H.C.; Randall, T.E.; Kitzman, J.O.; Stackhouse, B.; Emerson, J.C.; Mcnamara, S.; et al. Elevated Exopolysaccharide Levels in Pseudomonas Aeruginosa Flagellar Mutants Have Implications for Biofilm Growth and Chronic Infections. PLoS Genet. 2020, 16, e1008848. [Google Scholar] [CrossRef] [PubMed]
- Minamino, T.; Imae, Y.; Oosawa, F.; Kobayashi, Y.; Oosawa, K. Effect of Intracellular PH on Rotational Speed of Bacterial Flagellar Motors. J. Bacteriol. 2003, 185, 1190–1194. [Google Scholar] [CrossRef] [Green Version]
- Muriel, C.; Jalvo, B.; Redondo-Nieto, M.; Rivilla, R.; Martín, M. Chemotactic Motility of Pseudomonas Fluorescens F113 under Aerobic and Denitrification Conditions. PLoS ONE 2015, 10, e0132242. [Google Scholar] [CrossRef] [Green Version]
- Soutourina, O.; Kolb, A.; Krin, E.; Laurent-Winter, C.; Rimsky, S.; Danchin, A.; Bertin, P. Multiple Control of Flagellum Biosynthesis in Escherichia coli: Role of H-NS Protein and the Cyclic AMP-Catabolite Activator Protein Complex in Transcription of the FlhDC Master Operon. J. Bacteriol. 1999, 181, 7500–7508. [Google Scholar] [CrossRef] [Green Version]
- Kim, E.A.; Blair, D.F. Function of the Histone-Like Protein H-NS in Motility of Escherichia coli: Multiple Regulatory Roles Rather than Direct Action at the Flagellar Motor. J. Bacteriol. 2015, 197, 3110–3120. [Google Scholar] [CrossRef] [Green Version]
- Spöring, I.; Felgner, S.; Preuße, M.; Eckweiler, D.; Rohde, M.; Häussler, S.; Weiss, S.; Erhardt, M. Regulation of Flagellum Biosynthesis in Response to Cell Envelope Stress in Salmonella enterica Serovar Typhimurium. mBio 2018, 9, e00736-17. [Google Scholar] [CrossRef] [Green Version]
- Hews, C.L.; Cho, T.; Rowley, G.; Raivio, T.L. Maintaining Integrity Under Stress: Envelope Stress Response Regulation of Pathogenesis in Gram-Negative Bacteria. Front. Cell. Infect. Microbiol. 2019, 9, 313. [Google Scholar] [CrossRef] [Green Version]
- Latour, X. The Evanescent GacS Signal. Microorganisms 2020, 8, 1746. [Google Scholar] [CrossRef] [PubMed]
- Navazo, A.; Barahona, E.; Redondo-Nieto, M.; Martínez-Granero, F.; Rivilla, R.; Martín, M. Three Independent Signalling Pathways Repress Motility in Pseudomonas fluorescens F113. Microb. Biotechnol. 2009, 2, 489–498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez-Granero, F.; Navazo, A.; Barahona, E.; Redondo-Nieto, M.; Rivilla, R.; Martín, M. The Gac-Rsm and SadB Signal Transduction Pathways Converge on AlgU to Downregulate Motility in Pseudomonas Fluorescens. PLoS ONE 2012, 7, e31765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez-Granero, F.; Navazo, A.; Barahona, E.; Redondo-Nieto, M.; González de Heredia, E.; Baena, I.; Martín-Martín, I.; Rivilla, R.; Martín, M. Identification of FlgZ as a Flagellar Gene Encoding a PilZ Domain Protein That Regulates Swimming Motility and Biofilm Formation in Pseudomonas. PLoS ONE 2014, 9, e87608. [Google Scholar] [CrossRef] [Green Version]
- Damron, F.H.; Qiu, D.; Yu, H.D. The Pseudomonas Aeruginosa Sensor Kinase KinB Negatively Controls Alginate Production through AlgW-Dependent MucA Proteolysis. J. Bacteriol. 2009, 191, 2285–2295. [Google Scholar] [CrossRef] [Green Version]
- Chand, N.S.; Lee, J.S.-W.; Clatworthy, A.E.; Golas, A.J.; Smith, R.S.; Hung, D.T. The Sensor Kinase KinB Regulates Virulence in Acute Pseudomonas Aeruginosa Infection. J. Bacteriol. 2011, 193, 2989–2999. [Google Scholar] [CrossRef] [Green Version]
- Valentini, M.; Filloux, A. Biofilms and Cyclic Di-GMP (c-Di-GMP) Signaling: Lessons from Pseudomonas aeruginosa and Other Bacteria. J. Biol. Chem. 2016, 291, 12547–12555. [Google Scholar] [CrossRef] [Green Version]
- Bouillet, S.; Ba, M.; Houot, L.; Iobbi-Nivol, C.; Bordi, C. Connected Partner-Switches Control the Life Style of Pseudomonas aeruginosa through RpoS Regulation. Sci. Rep. 2019, 9, 6496. [Google Scholar] [CrossRef]
- Hengge, R. Principles of C-Di-GMP Signalling in Bacteria. Nat. Rev. Microbiol. 2009, 7, 263–273. [Google Scholar] [CrossRef]
- Dasgupta, N.; Ferrell, E.P.; Kanack, K.J.; West, S.E.H.; Ramphal, R. FleQ, the Gene Encoding the Major Flagellar Regulator of Pseudomonas Aeruginosa, Is Σ70 Dependent and Is Downregulated by Vfr, a Homolog of Escherichia Coli Cyclic AMP Receptor Protein. J. Bacteriol. 2002, 184, 5240–5250. [Google Scholar] [CrossRef] [Green Version]
- Hickman, J.W.; Harwood, C.S. Identification of FleQ from Pseudomonas aeruginosa as a c-Di-GMP-Responsive Transcription Factor. Mol. Microbiol. 2008, 69, 376–389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryjenkov, D.A.; Simm, R.; Römling, U.; Gomelsky, M. The PilZ Domain Is a Receptor for the Second Messenger C-Di-GMP: The PilZ Domain Protein YcgR Controls Motility in Enterobacteria. J. Biol. Chem. 2006, 281, 30310–30314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choy, W.-K.; Zhou, L.; Syn, C.K.-C.; Zhang, L.-H.; Swarup, S. MorA Defines a New Class of Regulators Affecting Flagellar Development and Biofilm Formation in Diverse Pseudomonas Species. J. Bacteriol. 2004, 186, 7221–7228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suchanek, V.M.; Esteban-López, M.; Colin, R.; Besharova, O.; Fritz, K.; Sourjik, V. Chemotaxis and Cyclic-di-GMP Signalling Control Surface Attachment of Escherichia coli. Mol. Microbiol. 2020, 113, 728–739. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Xin, L.; Zeng, Y.; Yam, J.K.H.; Ding, Y.; Venkataramani, P.; Cheang, Q.W.; Yang, X.; Tang, X.; Zhang, L.-H.; et al. A Cyclic Di-GMP-Binding Adaptor Protein Interacts with a Chemotaxis Methyltransferase to Control Flagellar Motor Switching. Sci. Signal. 2016, 9, ra102. [Google Scholar] [CrossRef]
- Jiménez-Fernández, A.; López-Sánchez, A.; Jiménez-Díaz, L.; Navarrete, B.; Calero, P.; Platero, A.I.; Govantes, F. Complex Interplay between FleQ, Cyclic Diguanylate and Multiple σ Factors Coordinately Regulates Flagellar Motility and Biofilm Development in Pseudomonas Putida. PLoS ONE 2016, 11, e0163142. [Google Scholar] [CrossRef]
- Navarrete, B.; Leal-Morales, A.; Serrano-Ron, L.; Sarrió, M.; Jiménez-Fernández, A.; Jiménez-Díaz, L.; López-Sánchez, A.; Govantes, F. Transcriptional Organization, Regulation and Functional Analysis of FlhF and FleN in Pseudomonas Putida. PLoS ONE 2019, 14, e0214166. [Google Scholar] [CrossRef]
- Xiao, Y.; Nie, H.; Liu, H.; Chen, W.; Huang, Q. Expression of the Diguanylate Cyclase GcbA Is Regulated by FleQ in Response to Cyclic Di-GMP in Pseudomonas putida KT2440: FleQ Regulates GcbA in Response to c-Di-GMP. Environ. Microbiol. Rep. 2016, 8, 993–1002. [Google Scholar] [CrossRef]
- Merritt, J.H.; Brothers, K.M.; Kuchma, S.L.; O’Toole, G.A. SadC Reciprocally Influences Biofilm Formation and Swarming Motility via Modulation of Exopolysaccharide Production and Flagellar Function. J. Bacteriol. 2007, 189, 8154–8164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewis, K.A.; Baker, A.E.; Chen, A.I.; Harty, C.E.; Kuchma, S.L.; O’Toole, G.A.; Hogan, D.A. Ethanol Decreases Pseudomonas Aeruginosa Flagellar Motility through the Regulation of Flagellar Stators. J. Bacteriol. 2019, 201, e00285-19. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, J.R.; Kuwada, N.J.; Huangyutitham, V.; Wiggins, P.A.; Harwood, C.S. Surface Sensing and Lateral Subcellular Localization of WspA, the Receptor in a Chemosensory-like System Leading to c-Di-GMP Production: WspA Functional Domains. Mol. Microbiol. 2012, 86, 720–729. [Google Scholar] [CrossRef] [Green Version]
- Huangyutitham, V.; Güvener, Z.T.; Harwood, C.S. Subcellular Clustering of the Phosphorylated WspR Response Regulator Protein Stimulates Its Diguanylate Cyclase Activity. mBio 2013, 4, e00242-13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rossello, J.; Lima, A.; Gil, M.; Rodríguez Duarte, J.; Correa, A.; Carvalho, P.C.; Kierbel, A.; Durán, R. The EAL-Domain Protein FcsR Regulates Flagella, Chemotaxis and Type III Secretion System in Pseudomonas Aeruginosa by a Phosphodiesterase Independent Mechanism. Sci. Rep. 2017, 7, 10281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jang, M.S.; Goo, E.; An, J.H.; Kim, J.; Hwang, I. Quorum Sensing Controls Flagellar Morphogenesis in Burkholderia Glumae. PLoS ONE 2014, 9, e84831. [Google Scholar] [CrossRef]
- Sperandio, V.; Torres, A.G.; Kaper, J.B. Quorum Sensing Escherichia coli Regulators B and C (QseBC): A Novel Two-Component Regulatory System Involved in the Regulation of Flagella and Motility by Quorum Sensing in E. coli: QseBC Regulates Flagella and Motility in E. coli. Mol. Microbiol. 2002, 43, 809–821. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kang, Y.; Choi, O.; Jeong, Y.; Jeong, J.-E.; Lim, J.Y.; Kim, M.; Moon, J.S.; Suga, H.; Hwang, I. Regulation of Polar Flagellum Genes Is Mediated by Quorum Sensing and FlhDC in Burkholderia Glumae: Regulation of Flagellum Genes in Burkholderia Glumae. Mol. Microbiol. 2007, 64, 165–179. [Google Scholar] [CrossRef]
- Cheng, F.; Ma, A.; Zhuang, G.; Fray, R.G. Exogenous N -Acyl-Homoserine Lactones Enhance the Expression of Flagella of Pseudomonas syringae and Activate Defence Responses in Plants: Interactions between Plants and Phytopathogens. Mol. Plant Pathol. 2018, 19, 104–115. [Google Scholar] [CrossRef]
- Cha, C.; Gao, P.; Chen, Y.-C.; Shaw, P.D.; Farrand, S.K. Production of Acyl-Homoserine Lactone Quorum-Sensing Signals by Gram-Negative Plant-Associated Bacteria. Mol. Plant Microbe Interact. 1998, 11, 1119–1129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martins, M.L.; Uelinton, M.P.; Riedel, K.; Vanetti, M.C.D.; Mantovani, H.C.; Araújo, E.F. De Lack of AHL-Based Quorum Sensing in Pseudomonas Fluorescens Isolated from Milk. Braz. J. Microbiol. 2014, 45, 1039–1046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tashiro, Y.; Yawata, Y.; Toyofuku, M.; Uchiyama, H.; Nomura, N. Interspecies Interaction between Pseudomonas Aeruginosa and Other Microorganisms. Microbes Environ. 2013, 28, 13–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallique, M.; Decoin, V.; Barbey, C.; Rosay, T.; Feuilloley, M.G.J.; Orange, N.; Merieau, A. Contribution of the Pseudomonas Fluorescens MFE01 Type VI Secretion System to Biofilm Formation. PLoS ONE 2017, 12. [Google Scholar] [CrossRef] [Green Version]
- Gallique, M.; Bouteiller, M.; Merieau, A. The Type VI Secretion System: A Dynamic System for Bacterial Communication? Front. Microbiol. 2017, 8, 1454. [Google Scholar] [CrossRef] [PubMed]
- Pukatzki, S.; Ma, A.T.; Sturtevant, D.; Krastins, B.; Sarracino, D.; Nelson, W.C.; Heidelberg, J.F.; Mekalanos, J.J. Identification of a Conserved Bacterial Protein Secretion System in Vibrio Cholerae Using the Dictyostelium Host Model System. Proc. Natl. Acad. Sci. USA 2006, 103, 1528–1533. [Google Scholar] [CrossRef] [Green Version]
- Cascales, E.; Cambillau, C. Structural Biology of Type VI Secretion Systems. Philos. Trans. R. Soc. B Biol. Sci. 2012, 367, 1102–1111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ho, B.T.; Dong, T.G.; Mekalanos, J.J. A View to a Kill: The Bacterial Type VI Secretion System. Cell Host Microbe 2014, 15, 9–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Planamente, S.; Salih, O.; Manoli, E.; Albesa-Jové, D.; Freemont, P.S.; Filloux, A. TssA Forms a Gp6-like Ring Attached to the Type VI Secretion Sheath. EMBO J. 2016, 35, 1613–1627. [Google Scholar] [CrossRef] [PubMed]
- Decoin, V.; Barbey, C.; Bergeau, D.; Latour, X.; Feuilloley, M.G.J.; Orange, N.; Merieau, A. A Type VI Secretion System Is Involved in Pseudomonas Fluorescens Bacterial Competition. PLoS ONE 2014, 9, e89411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tao, Z.; Zhou, T.; Zhou, S.; Wang, G. Temperature-Regulated Expression of Type VI Secretion Systems in Fish Pathogen Pseudomonas plecoglossicida Revealed by Comparative Secretome Analysis. FEMS Microbiol. Lett. 2016, 363, fnw261. [Google Scholar] [CrossRef] [Green Version]
- Leung, K.Y.; Siame, B.A.; Snowball, H.; Mok, Y.-K. Type VI Secretion Regulation: Crosstalk and Intracellular Communication. Curr. Opin. Microbiol. 2011, 14, 9–15. [Google Scholar] [CrossRef]
- Allsopp, L.P.; Wood, T.E.; Howard, S.A.; Maggiorelli, F.; Nolan, L.M.; Wettstadt, S.; Filloux, A. RsmA and AmrZ Orchestrate the Assembly of All Three Type VI Secretion Systems in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2017, 114, 7707–7712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyata, S.T.; Unterweger, D.; Rudko, S.P.; Pukatzki, S. Dual Expression Profile of Type VI Secretion System Immunity Genes Protects Pandemic Vibrio Cholerae. PLoS Pathog. 2013, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.H.; Galán, J.E. Salmonella Type III Secretion-Associated Chaperones Confer Secretion-Pathway Specificity: Type III Secretion Chaperones. Mol. Microbiol. 2004, 51, 483–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Pace, F.; Boldrin de Paiva, J.; Nakazato, G.; Lancellotti, M.; Sircili, M.P.; Guedes Stehling, E.; Dias da Silveira, W.; Sperandio, V. Characterization of IcmF of the Type VI Secretion System in an Avian Pathogenic Escherichia Coli (APEC) Strain. Microbiology 2011, 157, 2954–2962. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Hao, S.; Lan, R.; Wang, G.; Xiao, D.; Sun, H.; Xu, J. The Type VI Secretion System Modulates Flagellar Gene Expression and Secretion in Citrobacter Freundii and Contributes to Adhesion and Cytotoxicity to Host Cells. Infect. Immun. 2015, 83, 2596–2604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masum, M.; Yang, Y.; Li, B.; Olaitan, O.; Chen, J.; Zhang, Y.; Fang, Y.; Qiu, W.; Wang, Y.; Sun, G. Role of the Genes of Type VI Secretion System in Virulence of Rice Bacterial Brown Stripe Pathogen Acidovorax Avenae Subsp. Avenae Strain RS-2. Int. J. Mol. Sci. 2017, 18, 2024. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Xu, J.; Xu, J.; Zhang, H.; He, L.; Feng, J. TssB Is Essential for Virulence and Required for Type VI Secretion System in Ralstonia Solanacearum. Microb. Pathog. 2014, 74, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Decoin, V.; Gallique, M.; Barbey, C.; Le Mauff, F.; Poc, C.D.; Feuilloley, M.G.; Orange, N.; Merieau, A. A Pseudomonas Fluorescens Type 6 Secretion System Is Related to Mucoidy, Motility and Bacterial Competition. BMC Microbiol. 2015, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Salmonella/E. coli | Pseudomonas | |||
|---|---|---|---|---|
| Gene Name | Transcription Class 1 | Gene Name | Transcription Class 1 | Protein Activity |
| che | III | che | IV | Chemotaxis proteins |
| flgA | II and III | flgA | II | Flagellar basal body P-ring formation protein |
| flgB, C | II and III | flgB, C | III | Proximal rod proteins |
| flgD | II and III | flgD | III | Hook cap protein |
| flgE | II and III | flgE | III | Hook protein |
| flgF | II and III | flgF | III | Proximal rod protein |
| flgG | II and III | flgG | III | Distal rod protein |
| flgH | II and III | flgH | III | L-ring protein |
| flgI | II and III | flgI | III | P-ring protein |
| flgJ | II and III | flgJ | III | Distal rod cap protein |
| flgK, L | II and III | flgK, L | III | Hook/filament junction protein |
| flgM | II and III | flgM | II and IV | Anti-sigma 28 factor |
| flgN | II and III | flgN | II and IV | Hook/filament junction chaperone protein |
| flhA, B | II | flhA, B | II | Export gate proteins |
| flhC, D | I | fleQ (syn. adnA) | I | Master regulator, transcriptional activator |
| fliA, sigma 28 | II and III | fliA, sigma 28 | unknown | Sigma 28 factor |
| fliC, fljB | III | fliC/flaA | IV | Flagellin protein |
| fliD | II and III | fliD | II | Flagellin cap protein |
| fliE | II | fliE | II | Rod adaptor protein |
| fliF | II | fliF | II | MS-ring protein |
| fliG | II | fliG | II | C-ring protein |
| fliH, I, J | II | fliH, I, J | II | ATPase complex |
| fliK | II | fliK | III | Hook length control protein |
| fliL | II | fliL | II | Flagellum associated protein |
| fliM, N | II | fliM, N | II | C-Ring proteins |
| fliO, P, Q, R | II | fliO, P, Q, R | II | Export gate protein |
| fliS | II and III | fliS, fliS’ | II | Flagellin chaperone protein |
| fliT | II and III | fliT, fleP | II and IV | Flagellin cap chaperone protein |
| mot | III | mot | IV | Flagellar motor protein |
| ycgR | III | flgZ | II and IV | c-di-GMP effector |
| fliZ | II and III | FlhD4C2 activator | ||
| yhjH | III | phosphodiesterase | ||
| flaG | IV | Protein involved in filament length control | ||
| fleL | IV | Protein involved in filament length control | ||
| fleR | II | Two component system response regulator | ||
| fleS | II | Two component system sensor protein | ||
| flhF | II | Polar landmark protein | ||
| flhG, fleN | II | FleQ anti-activator protein | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bouteiller, M.; Dupont, C.; Bourigault, Y.; Latour, X.; Barbey, C.; Konto-Ghiorghi, Y.; Merieau, A. Pseudomonas Flagella: Generalities and Specificities. Int. J. Mol. Sci. 2021, 22, 3337. https://doi.org/10.3390/ijms22073337
Bouteiller M, Dupont C, Bourigault Y, Latour X, Barbey C, Konto-Ghiorghi Y, Merieau A. Pseudomonas Flagella: Generalities and Specificities. International Journal of Molecular Sciences. 2021; 22(7):3337. https://doi.org/10.3390/ijms22073337
Chicago/Turabian StyleBouteiller, Mathilde, Charly Dupont, Yvann Bourigault, Xavier Latour, Corinne Barbey, Yoan Konto-Ghiorghi, and Annabelle Merieau. 2021. "Pseudomonas Flagella: Generalities and Specificities" International Journal of Molecular Sciences 22, no. 7: 3337. https://doi.org/10.3390/ijms22073337






