Non-TZF Protein AtC3H59/ZFWD3 Is Involved in Seed Germination, Seedling Development, and Seed Development, Interacting with PPPDE Family Protein Desi1 in Arabidopsis
Abstract
1. Introduction
2. Results
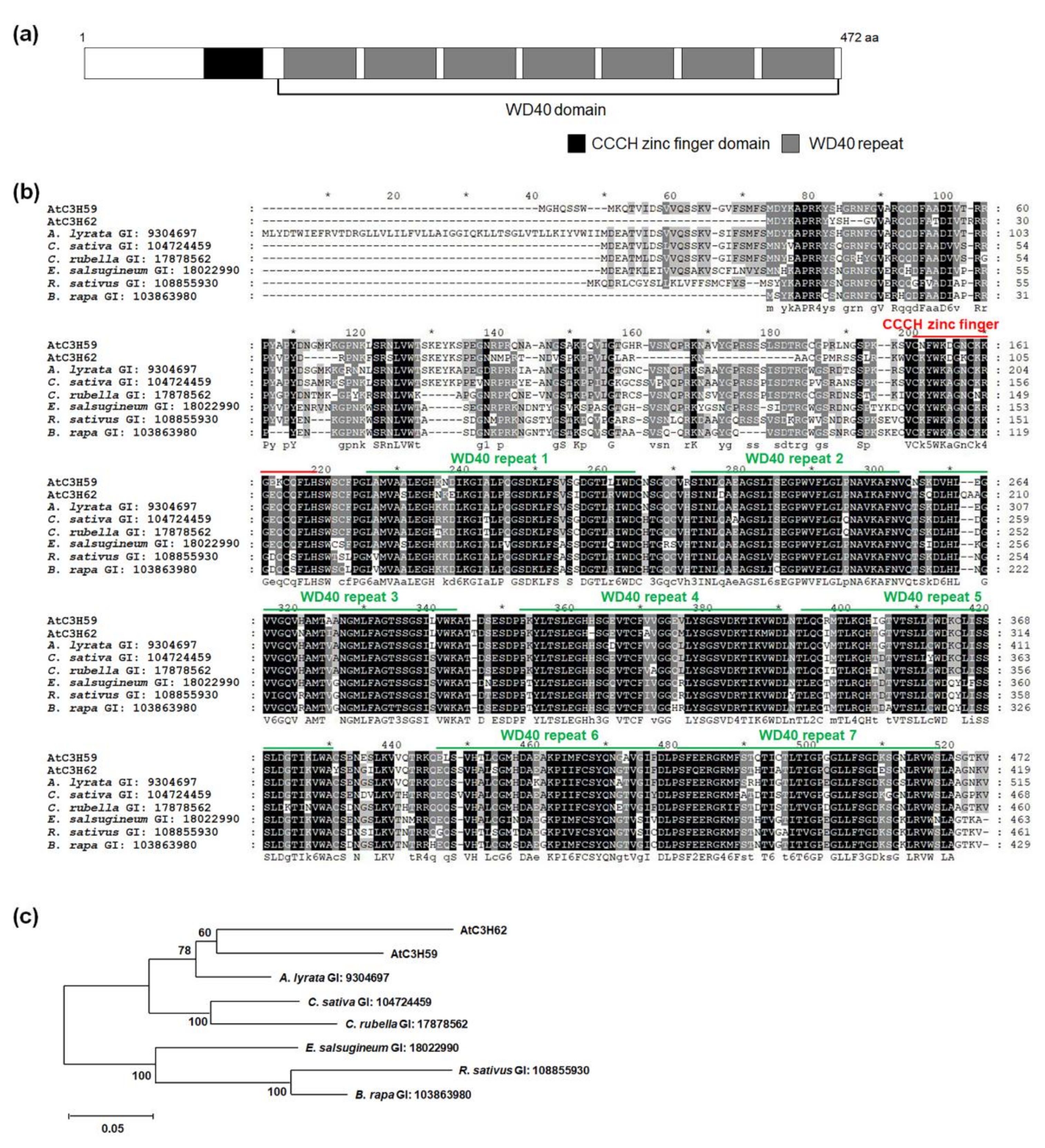
2.1. AtC3H59 Has One CCCH Zinc Finger Domain and One WD40 Domain
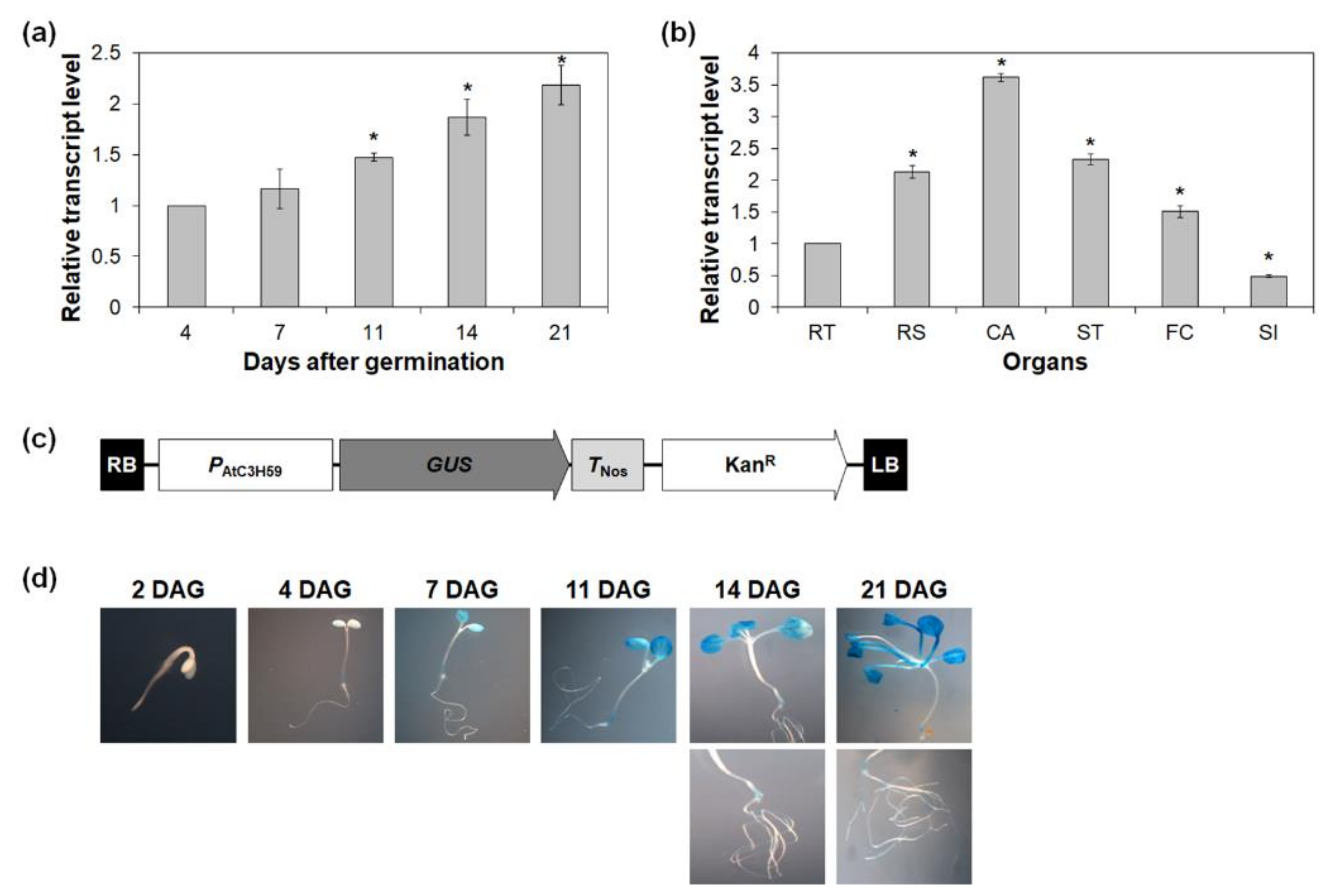
2.2. Expression of AtC3H59 during Development and in Organs of Arabidopsis
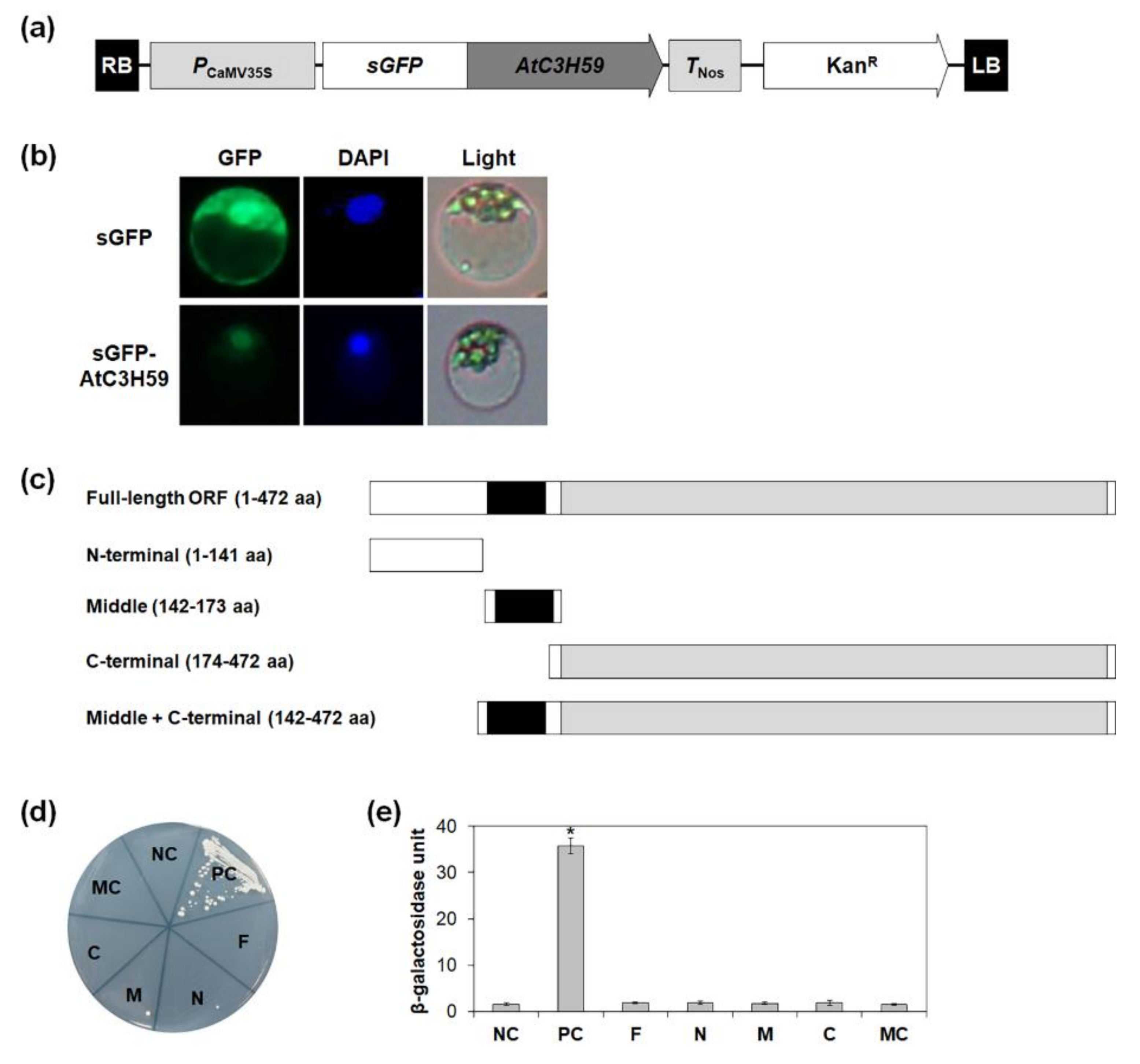
2.3. AtC3H59 Protein Is Subcellularly Localized in the Nucleus and Does Not Have Transactivation Activity
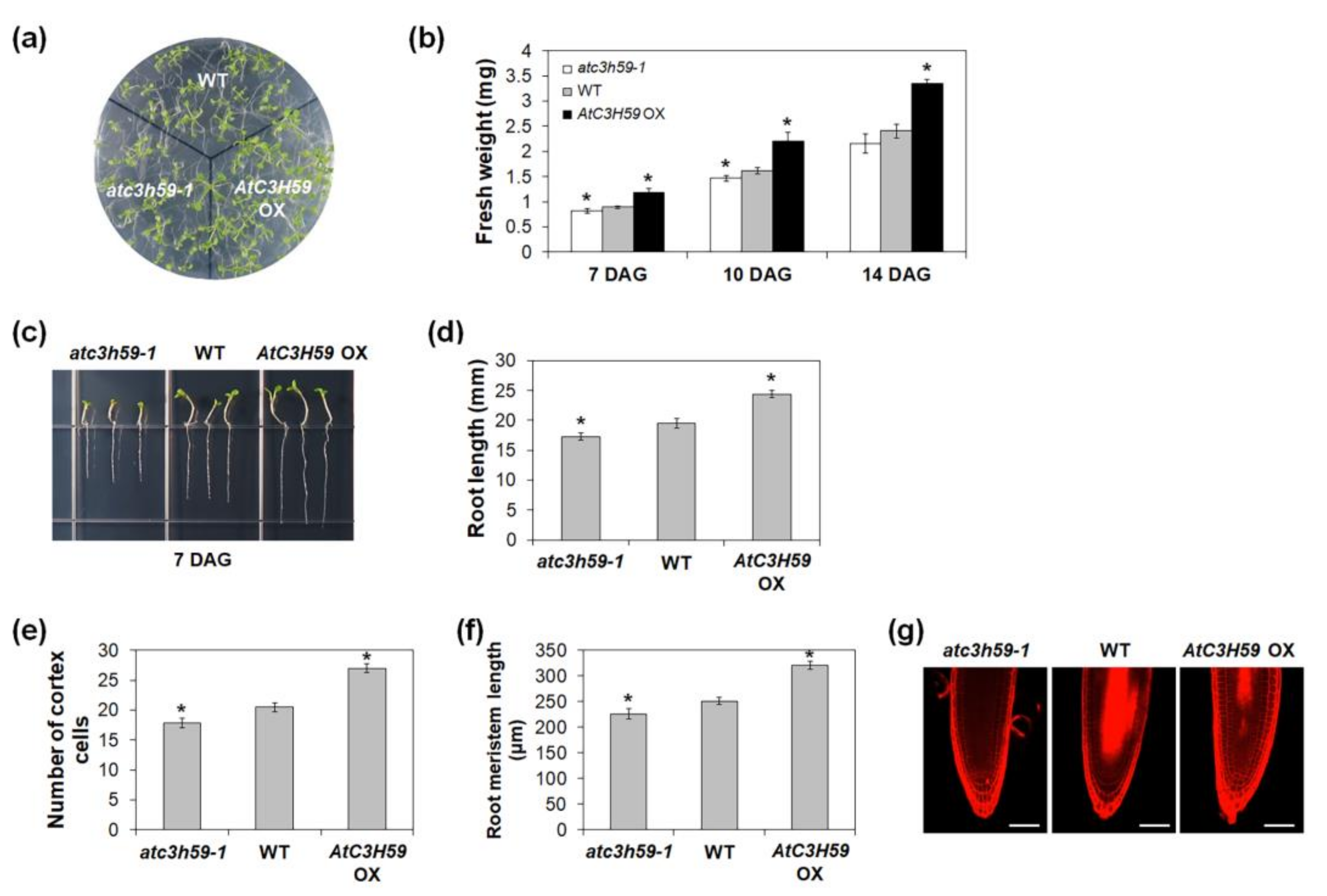
2.4. AtC3H59 Is Involved in Regulation of Seed Germination and Seedling Development
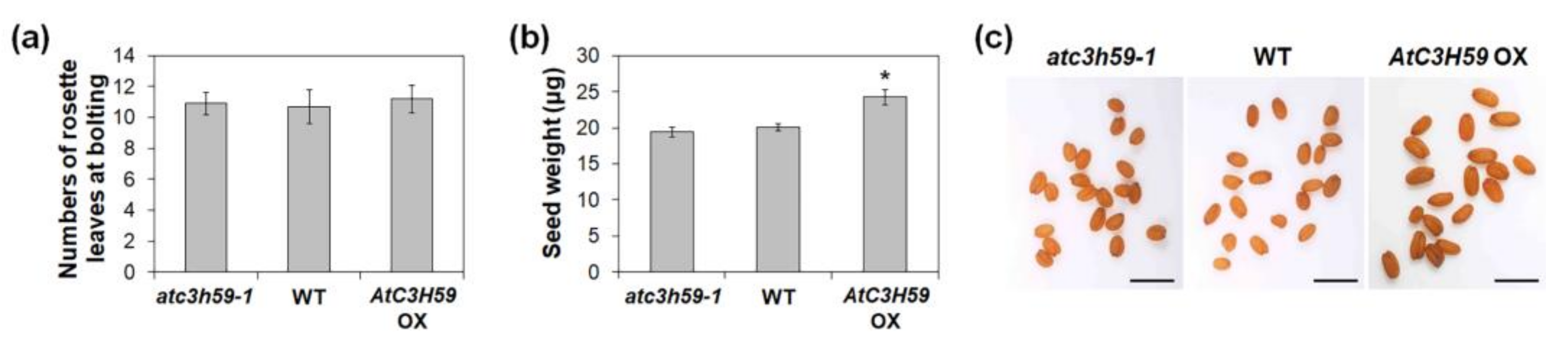
2.5. AtC3H59 Is Also involved in Regulation of Seed Development
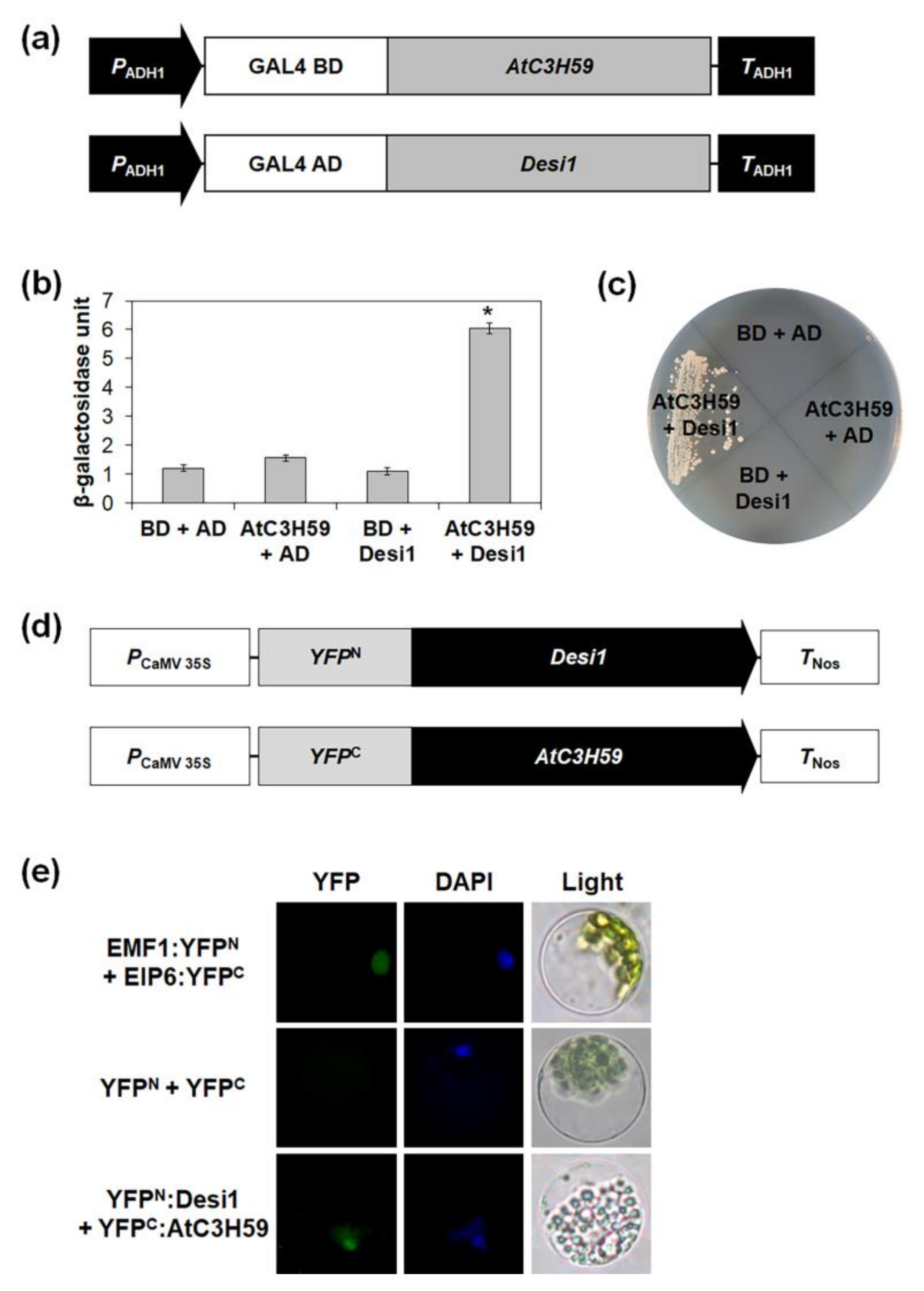
2.6. AtC3H59 Interacts with Desi1 in the Arabidopsis Nucleus
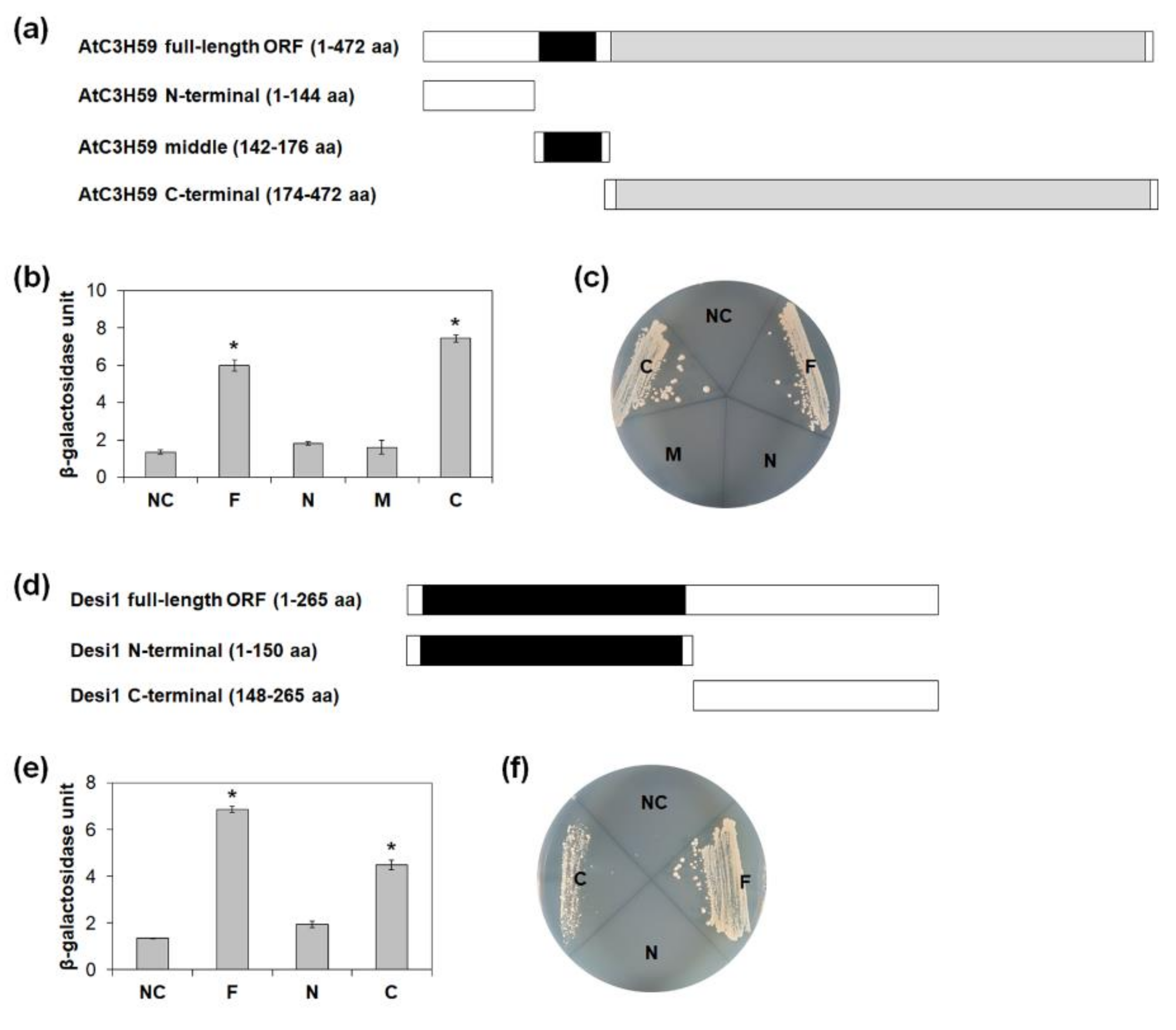
2.7. AtC3H59 Interacts with Desi1 via the WD40 Domain
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Plasmid Construction
4.3. Plant Transformation and Selection of Transgenic Plants
4.4. RNA Isolation, cDNA Synthesis, Semi-Quantitative RT-PCR, and Quantitative RT-PCR
4.5. GUS Activity Analysis
4.6. Protoplast Transformation
4.7. cDNA Library Generation and Yeast Two-hybrid Screening
4.8. Yeast Transformation and Assay
4.9. Multiple Alignment Analysis
4.10. Phylogenetic Tree
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ciftci-Yilmaz, S.; Mittler, R. The zinc finger network of plants. Cell. Mol. Life Sci. 2008, 65, 1150–1160. [Google Scholar] [CrossRef] [PubMed]
- Pomeranz, M.; Finer, J.; Jang, J.C. Putative molecular mechanisms underlying tandem CCCH zinc finger protein mediated plant growth, stress, and gene expression responses. Plant Signal. Behav. 2011, 6, 647–651. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Guo, Y.; Wu, C.; Yang, G.; Li, Y.; Zheng, C. Genome-wide analysis of CCCH zinc finger family in Arabidopsis and rice. BMC Genom. 2008, 9, 44. [Google Scholar] [CrossRef] [PubMed]
- Laity, J.H.; Lee, B.M.; Wright, P.E. Zinc finger proteins: New insights into structural and functional diversity. Curr. Opin. Struct. Biol. 2001, 11, 39–46. [Google Scholar] [CrossRef]
- Seok, H.Y.; Woo, D.H.; Park, H.Y.; Lee, S.Y.; Tran, H.T.; Lee, E.H.; Nguyen, L.V.; Moon, Y.H. AtC3H17, a non-tandem CCCH zinc finger protein, functions as a nuclear transcriptional activator and has pleiotropic effects on vegetative development, flowering and seed development in Arabidopsis. Plant Cell Physiol. 2016, 57, 603–615. [Google Scholar] [CrossRef]
- Lee, S.J.; Jung, H.J.; Kang, H.; Kim, S.Y. Arabidopsis zinc finger proteins AtC3H49/AtTZF3 and AtC3H20/AtTZF2 are involved in ABA and JA responses. Plant Cell Physiol. 2012, 53, 673–686. [Google Scholar] [CrossRef]
- Li, Z.; Thomas, T.L. PEI1, an embryo-specific zinc finger protein gene required for heart-stage embryo formation in Arabidopsis. Plant Cell 1998, 10, 383–398. [Google Scholar] [CrossRef]
- Chai, G.; Kong, Y.; Zhu, M.; Yu, L.; Qi, G.; Tang, X.; Wang, Z.; Cao, Y.; Yu, C.; Zhou, G. Arabidopsis C3H14 and C3H15 have overlapping roles in the regulation of secondary wall thickening and anther development. J. Exp. Bot. 2015, 66, 2595–2609. [Google Scholar] [CrossRef]
- Jan, A.; Maruyama, K.; Todaka, D.; Kidokoro, S.; Abo, M.; Yoshimura, E.; Shinozaki, K.; Nakashima, K.; Yamaguchi-Shinozaki, K. OsTZF1, a CCCH-tandem zinc finger protein, confers delayed senescence and stress tolerance in rice by regulating stress-related genes. Plant Physiol. 2013, 161, 1202–1216. [Google Scholar] [CrossRef]
- Seong, S.Y.; Shim, J.S.; Bang, S.W.; Kim, J.K. Overexpression of OsC3H10, a CCCH-zinc finger, improves drought tolerance in rice by regulating stress-related genes. Plants 2020, 9, 1298. [Google Scholar] [CrossRef]
- Li, J.; Jia, D.; Chen, X. HUA1, a regulator of stamen and carpel identities in Arabidopsis, codes for a nuclear RNA binding protein. Plant Cell 2001, 13, 2269–2281. [Google Scholar] [CrossRef]
- Seok, H.Y.; Nguyen, L.V.; Park, H.Y.; Tarte, V.N.; Ha, J.; Lee, S.Y.; Moon, Y.H. Arabidopsis non-TZF gene AtC3H17 functions as a positive regulator in salt stress response. Biochem. Biophys. Res. Commun. 2018, 498, 954–959. [Google Scholar] [CrossRef]
- Xu, L.; Liu, T.; Xiong, X.; Liu, W.; Yu, Y.; Cao, J. Overexpression of two CCCH-type zinc-finger protein genes leads to pollen abortion in Brassica campestris ssp. chinensis. Genes 2020, 11, 1287. [Google Scholar] [CrossRef]
- Addepalli, B.; Hunt, A.G. Ribonuclease activity is a common property of Arabidopsis CCCH-containing zinc-finger proteins. FEBS Lett. 2008, 582, 2577–2582. [Google Scholar] [CrossRef]
- Xie, Z.; Lin, W.; Yu, G.; Cheng, Q.; Xu, B.; Huang, B. Improved cold tolerance in switchgrass by a novel CCCH-type zinc finger transcription factor gene, PvC3H72, associated with ICE1—CBF—COR regulon and ABA-responsive genes. Biotechnol. Biofuels 2019, 12, 224. [Google Scholar] [CrossRef]
- Zhang, H.; Gao, X.; Zhi, Y.; Li, X.; Zhang, Q.; Niu, J.; Wang, J.; Zhai, H.; Zhao, N.; Li, J.; et al. A non-tandem CCCH-type zinc-finger protein, IbC3H18, functions as a nuclear transcriptional activator and enhances abiotic stress tolerance in sweet potato. New Phytol. 2019, 223, 1918–1936. [Google Scholar] [CrossRef]
- Zhuang, Y.; Wang, C.; Zhang, Y.; Chen, S.; Wang, D.; Liu, Q.; Zhou, G.; Chai, G. Overexpression of PdC3H17 confers tolerance to drought stress depending on its CCCH domain in Populus. Front. Plant Sci. 2020, 10, 1748. [Google Scholar] [CrossRef]
- Fong, H.K.; Hurley, J.B.; Hopkins, R.S.; Miake-Lye, R.; Johnson, M.S.; Doolittle, R.F.; Simon, M.I. Repetitive segmental structure of the transducin beta subunit: Homology with the CDC4 gene and identification of related mRNAs. Proc. Natl. Acad. Sci. USA 1986, 83, 2162–2166. [Google Scholar] [CrossRef]
- Neer, E.J.; Schmidt, C.J.; Nambudripad, R.; Smith, T.F. The ancient regulatory-protein family of WD-repeat proteins. Nature 1994, 371, 297–300. [Google Scholar] [CrossRef]
- Smith, T.F.; Gaitatzes, C.; Saxena, K.; Neer, E.J. The WD repeat: A common architecture for diverse functions. Trends Biochem. Sci. 1999, 24, 181–185. [Google Scholar] [CrossRef]
- Xu, C.; Min, J. Structure and function of WD40 domain proteins. Protein Cell 2011, 2, 202–214. [Google Scholar] [CrossRef]
- Li, D.; Roberts, R. WD-repeat proteins: Structure characteristics, biological function, and their involvement in human diseases. Cell. Mol. Life Sci. 2001, 58, 2085–2097. [Google Scholar] [CrossRef]
- Hoey, T.; Weinzierl, R.O.; Gill, G.; Chen, J.L.; Dynlacht, B.D.; Tjian, R. Molecular cloning and functional analysis of Drosophila TAF110 reveal properties expected of coactivators. Cell 1993, 72, 247–260. [Google Scholar] [CrossRef]
- Pryer, N.K.; Salama, N.R.; Schekman, R.; Kaiser, C.A. Cytosolic Sec13p complex is required for vesicle formation from the endoplasmic reticulum In Vitro. J. Cell Biol. 1993, 120, 865–875. [Google Scholar] [CrossRef]
- Feldman, R.M.; Correll, C.C.; Kaplan, K.B.; Deshaies, R.J. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell 1997, 91, 221–230. [Google Scholar] [CrossRef]
- Groisman, R.; Polanowska, J.; Kuraoka, I.; Sawada, J.; Saijo, M.; Drapkin, R.; Kisselev, A.F.; Tanaka, K.; Nakatani, Y. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell 2003, 113, 357–367. [Google Scholar] [CrossRef]
- Scrima, A.; Konickova, R.; Czyzewski, B.K.; Kawasaki, Y.; Jeffrey, P.D.; Groisman, R.; Nakatani, Y.; Iwai, S.; Pavletich, N.P.; Thoma, N.H. Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex. Cell 2008, 135, 1213–1223. [Google Scholar] [CrossRef]
- Hannah, J.; Zhou, P. Regulation of DNA damage response pathways by the cullin-RING ubiquitin ligases. DNA Repair (Amst) 2009, 8, 536–543. [Google Scholar] [CrossRef]
- Migliori, V.; Mapelli, M.; Guccione, E. On WD40 proteins: Propelling our knowledge of transcriptional control? Epigenetics 2012, 7, 815–822. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Y.; Liang, X.; Zhu, Z.; Sun, H.; He, H.; Min, J.; Liao, S.; Liu, Y. Crystal structure of the WD40 domain of human PRPF19. Biochem. Biophys. Res. Commun. 2017, 493, 1250–1253. [Google Scholar] [CrossRef]
- Terol, J.; Bargues, M.; Perez-Alonso, M. ZFWD: A novel subfamily of plant proteins containing a C3H zinc finger and seven WD40 repeats. Gene 2000, 260, 45–53. [Google Scholar] [CrossRef]
- Shin, E.J.; Shin, H.M.; Nam, E.; Kim, W.S.; Kim, J.H.; Oh, B.H.; Yun, Y. DeSUMOylating isopeptidase: A second class of SUMO protease. EMBO Rep. 2012, 13, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Wang, X.; Jiang, D.; Wang, J.; Fei, R.; Cong, X.; Wei, L.; Wang, Y.; Chen, H. PPPDE1 is a novel deubiquitinase belonging to a cysteine isopeptidase family. Biochem. Biophys. Res. Commun. 2017, 488, 291–296. [Google Scholar] [CrossRef] [PubMed]
- Gillies, J.; Hochstrasser, M. A new class of SUMO proteases. EMBO Rep. 2012, 13, 284–285. [Google Scholar] [CrossRef]
- He, Y.; Wang, J.; Gou, L.; Shen, C.; Chen, L.; Yi, C.; Wei, X.; Yang, J. Comprehensive analysis of expression profile reveals the ubiquitous distribution of PPPDE peptidase domain 1, a Golgi apparatus component, and its implications in clinical cancer. Biochimie 2013, 95, 1466–1475. [Google Scholar] [CrossRef]
- Orosa, B.; Yates, G.; Verma, V.; Srivastava, A.K.; Srivastava, M.; Campanaro, A.; de Vega, D.; Fernandes, A.; Zhang, C.; Lee, J.; et al. SUMO conjugation to the pattern recognition receptor FLS2 triggers intracellular signalling in plant innate immunity. Nat. Commun. 2018, 9, 5185. [Google Scholar] [CrossRef]
- Wang, L.; Xu, Y.; Zhang, C.; Ma, Q.; Joo, S.H.; Kim, S.K.; Xu, Z.; Chong, K. OsLIC, a novel CCCH-type zinc finger protein with transcription activation, mediates rice architecture via brassinosteroids signaling. PLoS ONE 2008, 3, e3521. [Google Scholar] [CrossRef]
- Beemster, G.T.; Baskin, T.I. Analysis of cell division and elongation underlying the developmental acceleration of root growth in Arabidopsis thaliana. Plant Physiol. 1998, 116, 1515–1526. [Google Scholar] [CrossRef]
- Inze, A.; Vanderauwera, S.; Hoeberichts, F.A.; Vandorpe, M.; van Gaever, T.; van Breusegem, F. A subcellular localization compendium of hydrogen peroxide-induced proteins. Plant Cell Environ. 2012, 35, 308–320. [Google Scholar] [CrossRef]
- Ju, Y.; Guo, L.; Cai, Q.; Ma, F.; Zhu, Q.Y.; Zhang, Q.; Sodmergen. Arabidopsis JINGUBANG is a negative regulator of pollen germination that prevents pollination in moist environments. Plant Cell 2016, 28, 2131–2146. [Google Scholar] [CrossRef]
- Guo, R.; Sun, W. Sumoylation stabilizes RACK1B and enhance its interaction with RAP2.6 in the abscisic acid response. Sci. Rep. 2017, 7, 44090. [Google Scholar] [CrossRef]
- Zhao, Q.; Xie, Y.; Zheng, Y.; Jiang, S.; Liu, W.; Mu, W.; Liu, Z.; Zhao, Y.; Xue, Y.; Ren, J. GPS-SUMO: A tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res. 2014, 42, W325–W330. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Park, H.Y.; Lee, S.Y.; Seok, H.Y.; Kim, S.H.; Sung, Z.R.; Moon, Y.H. EMF1 interacts with EIP1, EIP6 or EIP9 involved in the regulation of flowering time in Arabidopsis. Plant Cell Physiol. 2011, 52, 1376–1388. [Google Scholar] [CrossRef]
- Hofgen, R.; Willmitzer, L. Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res. 1988, 16, 9877. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Seok, H.Y.; Ha, J.; Lee, S.Y.; Bae, H.; Moon, Y.H. Two alternative splicing variants of AtERF73/HRE1, HRE1α and HRE1β, have differential transactivation activities in Arabidopsis. Int. J. Mol. Sci. 2020, 21, 6984. [Google Scholar] [CrossRef]
- Yoo, S.D.; Cho, Y.H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef]
- Miller, K.Y.; Wu, J.; Miller, B.L. StuA is required for cell pattern formation in Aspergillus. Genes Dev. 1992, 6, 1770–1782. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seok, H.-Y.; Bae, H.; Kim, T.; Mehdi, S.M.M.; Nguyen, L.V.; Lee, S.-Y.; Moon, Y.-H. Non-TZF Protein AtC3H59/ZFWD3 Is Involved in Seed Germination, Seedling Development, and Seed Development, Interacting with PPPDE Family Protein Desi1 in Arabidopsis. Int. J. Mol. Sci. 2021, 22, 4738. https://doi.org/10.3390/ijms22094738
Seok H-Y, Bae H, Kim T, Mehdi SMM, Nguyen LV, Lee S-Y, Moon Y-H. Non-TZF Protein AtC3H59/ZFWD3 Is Involved in Seed Germination, Seedling Development, and Seed Development, Interacting with PPPDE Family Protein Desi1 in Arabidopsis. International Journal of Molecular Sciences. 2021; 22(9):4738. https://doi.org/10.3390/ijms22094738
Chicago/Turabian StyleSeok, Hye-Yeon, Hyungjoon Bae, Taehyoung Kim, Syed Muhammad Muntazir Mehdi, Linh Vu Nguyen, Sun-Young Lee, and Yong-Hwan Moon. 2021. "Non-TZF Protein AtC3H59/ZFWD3 Is Involved in Seed Germination, Seedling Development, and Seed Development, Interacting with PPPDE Family Protein Desi1 in Arabidopsis" International Journal of Molecular Sciences 22, no. 9: 4738. https://doi.org/10.3390/ijms22094738
APA StyleSeok, H.-Y., Bae, H., Kim, T., Mehdi, S. M. M., Nguyen, L. V., Lee, S.-Y., & Moon, Y.-H. (2021). Non-TZF Protein AtC3H59/ZFWD3 Is Involved in Seed Germination, Seedling Development, and Seed Development, Interacting with PPPDE Family Protein Desi1 in Arabidopsis. International Journal of Molecular Sciences, 22(9), 4738. https://doi.org/10.3390/ijms22094738






