Physiological Ovarian Aging Is Associated with Altered Expression of Post-Translational Modifications in Mice
Abstract
:1. Introduction
2. Results
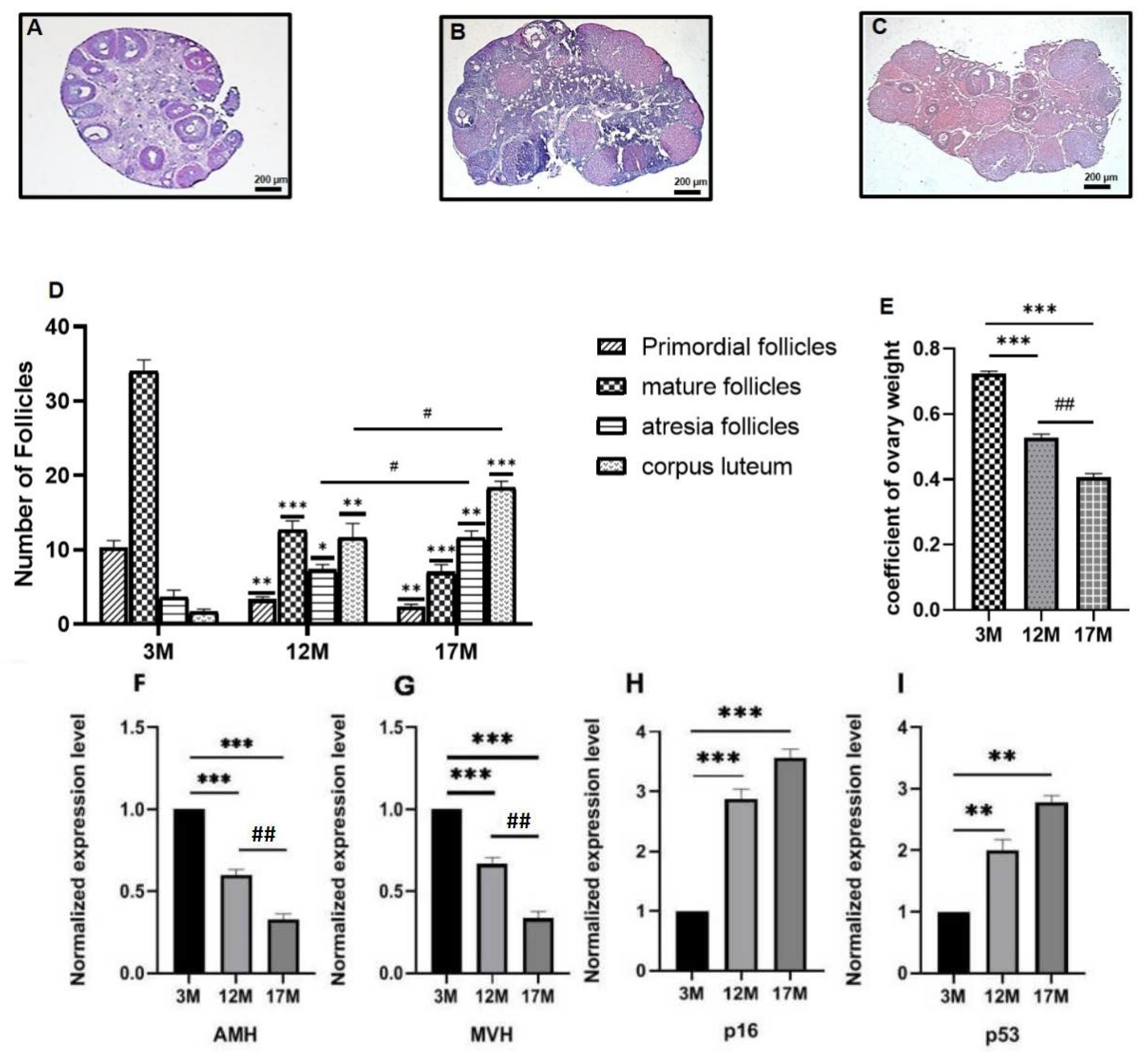
2.1. Ovarian Function of 3M, 12M, and 17M Mice Showed Gradual Decline during the Physiological Aging Process
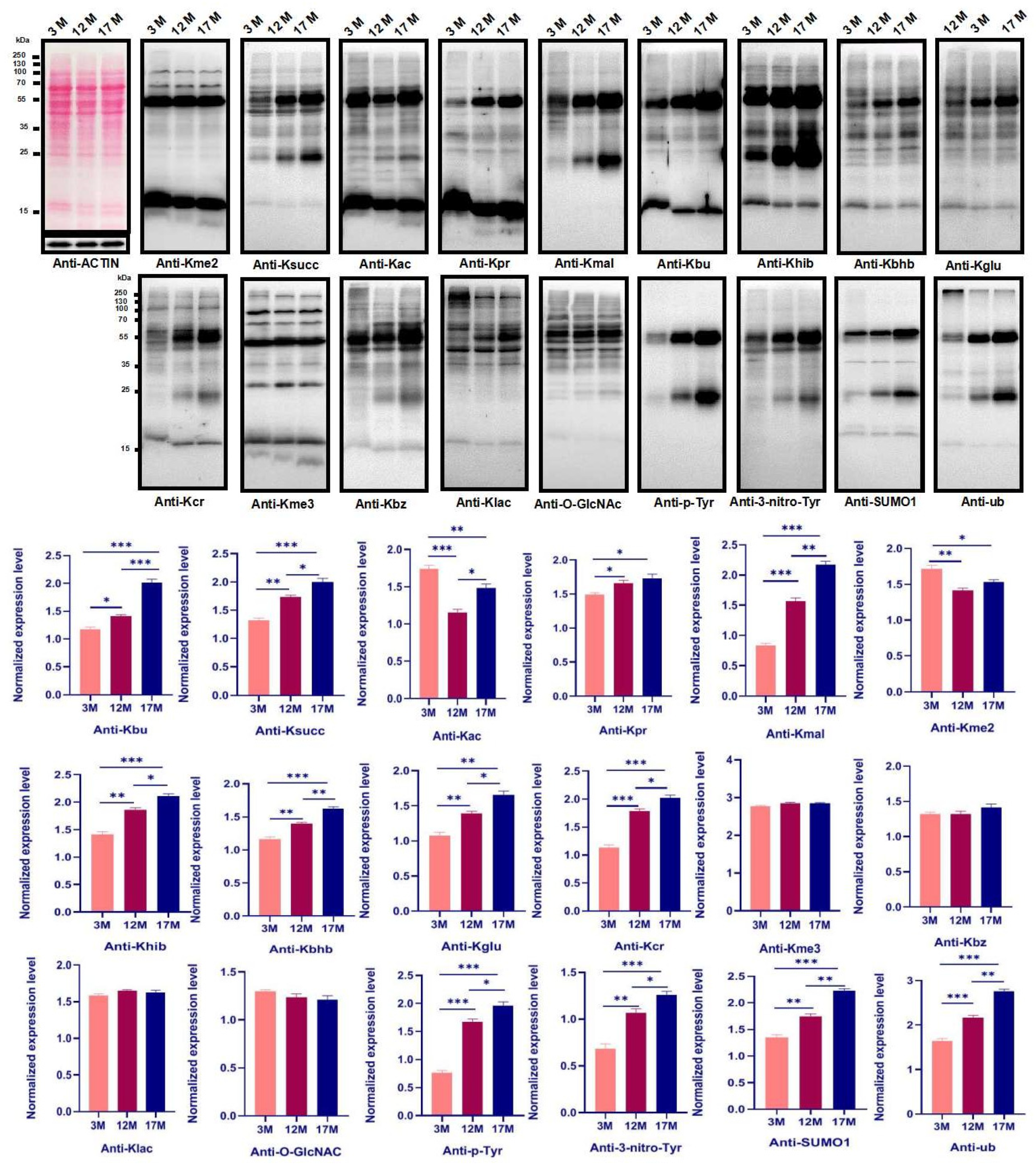
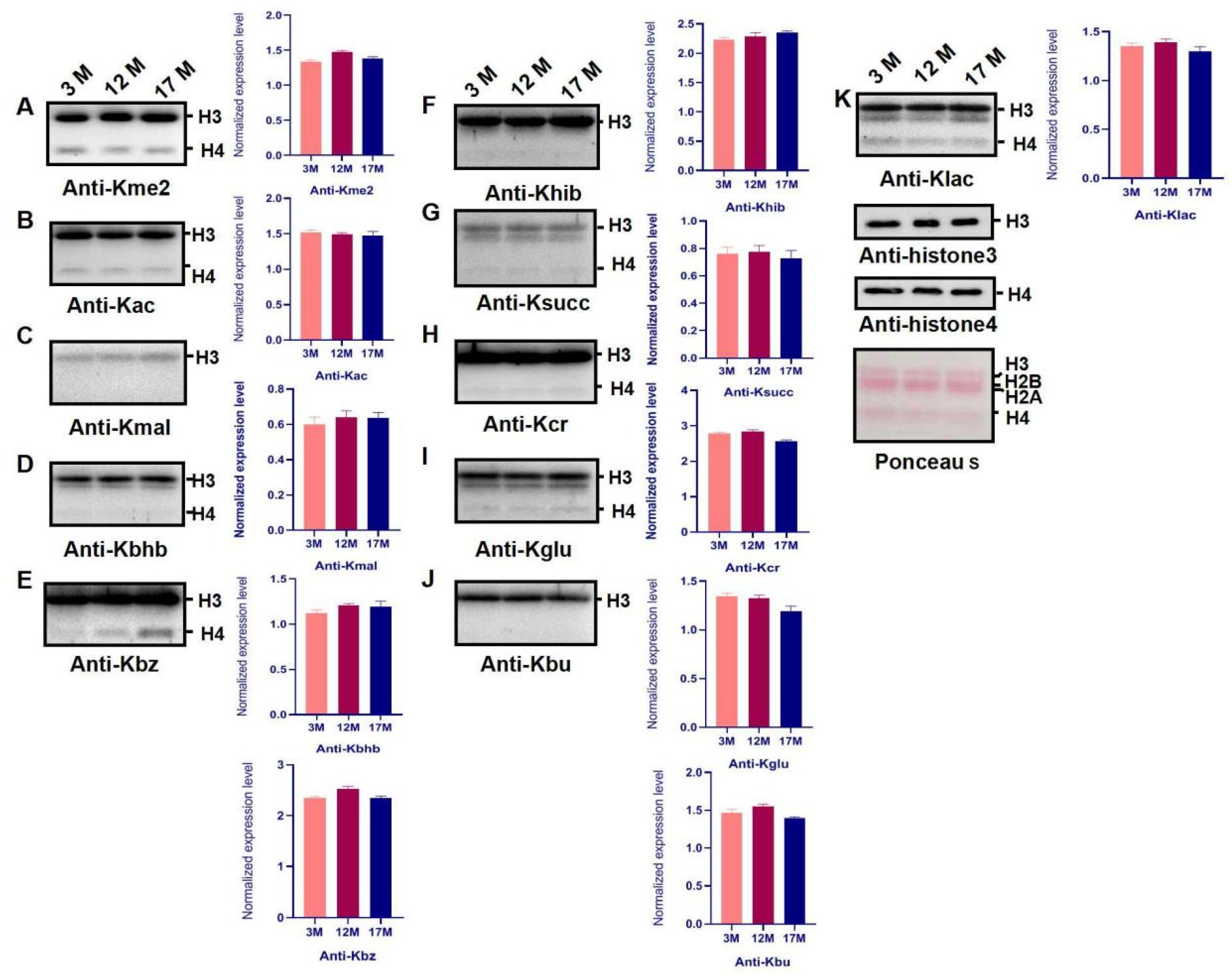
2.2. Ovarian Aging Is Associated with Altered Ovarian Levels of 14 PTMs and Newly Identified Metabolism-Related Lysine Modifications
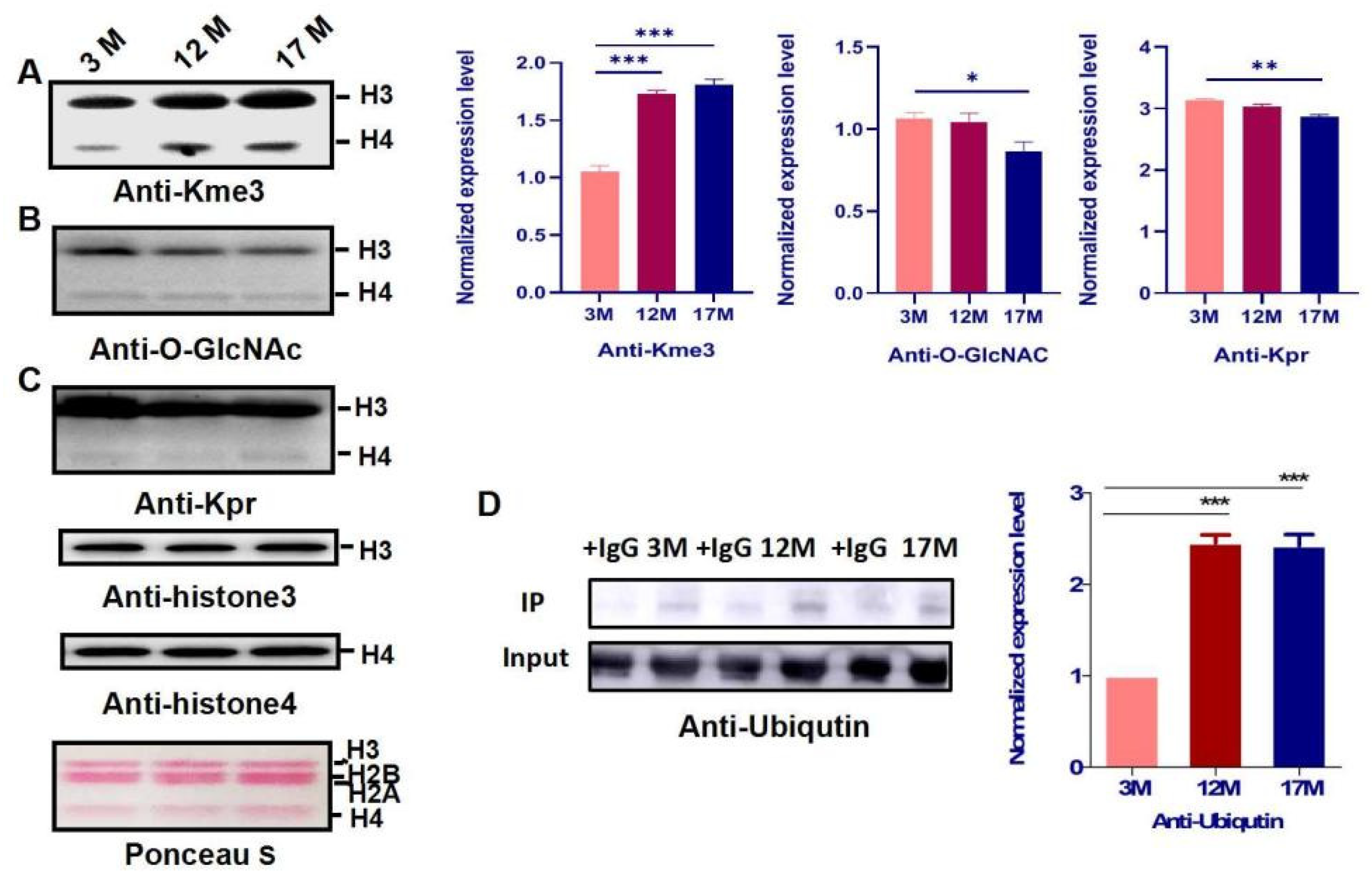
2.3. Histone Modification Levels of Kme3 and Ubiqutin, Kpr, and O-GlcNA Were Altered during Ovarian Aging
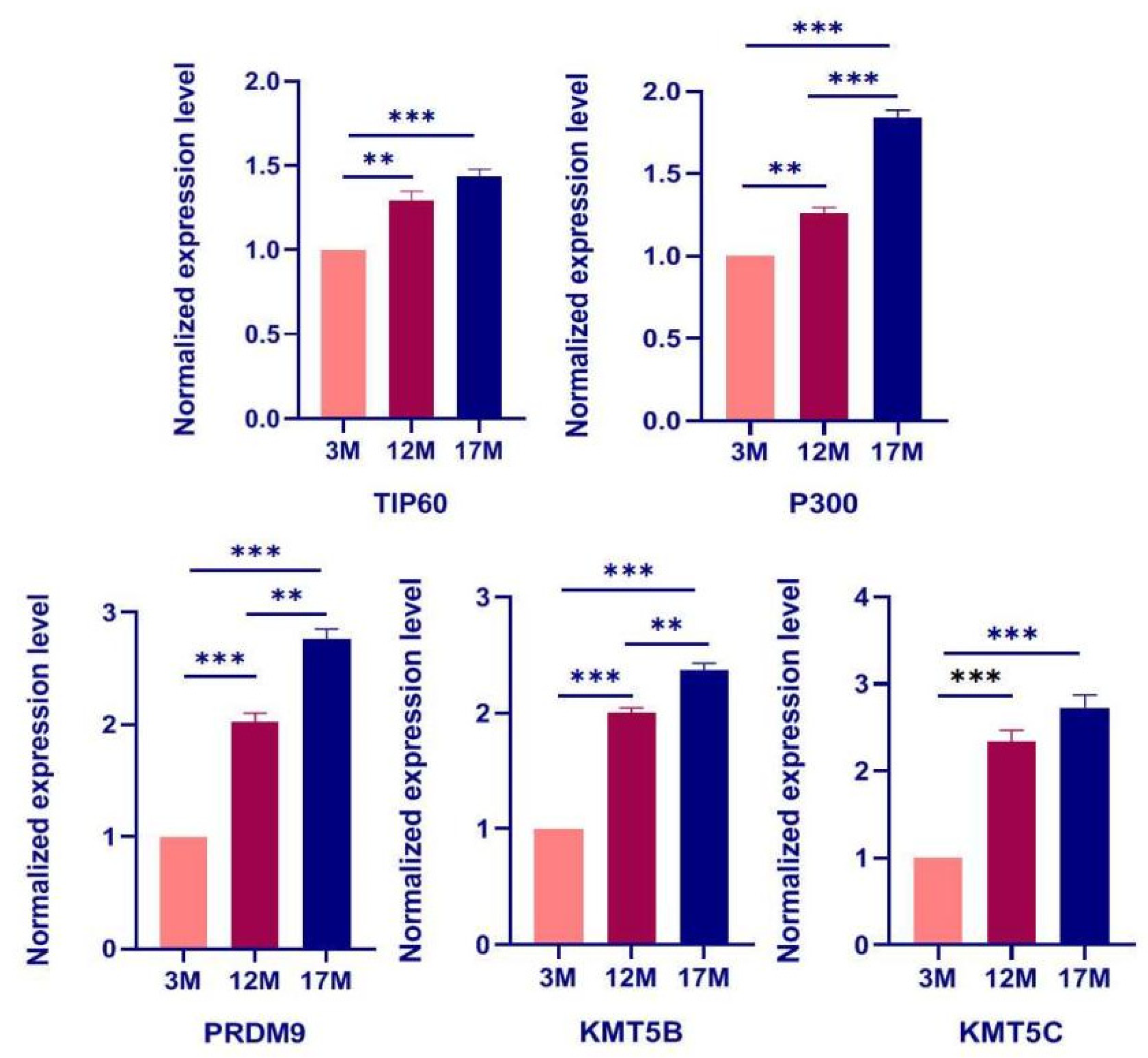
2.4. The Expression of Regulatory Enzyme Genes including TIP60, P300, PRDM9, KMT5B, and KMT5C Increases during Ovarian Aging
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Animals
5.2. Histological Analysis of Ovarian Tissue and Ovarian Follicle Count
5.3. Quantitative Real-Time PCR
5.4. Western Blot Analysis
5.5. Immunoprecipitation
5.6. Statistical Methods
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ruth, K.S.; Day, F.R.; Hussain, J.; Martínez-Marchal, A.; Aiken, C.E.; Azad, A.; Thompson, D.J.; Knoblochova, L.; Abe, H.; Tarry-Adkins, J.L.; et al. Genetic insights into biological mechanisms governing human ovarian ageing. Nature 2021, 596, 393–397. [Google Scholar] [CrossRef]
- Broekmans, F.J.; Soules, M.R.; Fauser, B.C. Ovarian aging: Mechanisms and clinical consequences. Endocr. Rev. 2009, 30, 465–493. [Google Scholar] [CrossRef]
- Gershon, E.; Dekel, N. Newly Identified Regulators of Ovarian Folliculogenesis and Ovulation. Int. J. Mol. Sci. 2020, 21, 4565. [Google Scholar] [CrossRef]
- Hegazy, A.A. Is There any Mean to Postpone the Menopausal Ovarian Senescence? Int. J. Fertil. Steril. 2020, 13, 346–347. [Google Scholar] [PubMed]
- Wang, S.; Zheng, Y.; Li, J.; Yu, Y.; Zhang, W.; Song, M.; Liu, Z.; Min, Z.; Hu, H.; Jing, Y.; et al. Single-Cell Transcriptomic Atlas of Primate Ovarian Aging. Cell 2020, 180, 585–600. [Google Scholar] [CrossRef]
- Takashi, Y.; Takahiro, S.; Go, N.; Haruka, Y.; Mika, I.; Mami, K.; Haruka, K.; Takuya, I.; Kinichi, N.; Ryuichi, N.; et al. Generation of ovarian follicles from mouse pluripotent stem cells. Science 2021, 373, eabe0237. [Google Scholar]
- Shah, J.S.; Sabouni, R.; Vaught, K.C.C.; Owen, C.M.; Albertini, D.F.; Segars, J.H. Biomechanics and mechanical signaling in the ovary: A systematic review. J. Assist. Reprod. Genet. 2018, 35, 1135–1148. [Google Scholar] [CrossRef]
- Tesarik, J.; Galán-Lázaro, M.; Mendoza-Tesarik, R. Ovarian Aging: Molecular Mechanisms and Medical Management. Int. J. Mol. Sci. 2021, 22, 1371. [Google Scholar] [CrossRef] [PubMed]
- Yamada-Fukunaga, T.; Yamada, M.; Hamatani, T.; Chikazawa, N.; Ogawa, S.; Akutsu, H.; Miura, T.; Miyado, K.; Tarín, J.J.; Kuji, N.; et al. Age-associated telomere shortening in mouse oocytes. Reprod. Biol. Endocrinol. 2013, 11, 108. [Google Scholar] [CrossRef]
- Li, Q.; Geng, X.; Zheng, W.; Tang, J.; Xu, B.; Shi, Q. Current understanding of ovarian aging. Sci China Life. Sci. 2012, 55, 659–669. [Google Scholar] [CrossRef]
- Heo, K.S. Regulation of post-translational modification in breast cancer treatment. BMB Rep. 2019, 52, 113–118. [Google Scholar] [CrossRef]
- Zheng, L.P.; Luo, R.C.; Su, T.; Hu, L.L.; Gao, F.X.; Zhang, X.N. Differentially expressed lncRNAs after the activation of primordial follicles in mouse. Reprod. Sci. 2019, 26, 1094–1104. [Google Scholar] [CrossRef]
- García-Giménez, J.L.; Garcés, C.; Romá-Mateo, C.; Pallardó, F.V. Oxidative stress-mediated alterations in histone post-translational modifications. Free. Radic. Biol. Med. 2021, 170, 6–18. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Shang, R.; Gong, D.; Xu, X.; Liu, S. Characterization of H3 methylation in regulating oocyte development in cyprinid fish. Sci. China Life Sci. 2018, 62, 829–837. [Google Scholar] [CrossRef]
- Tan, M.; Luo, H.; Lee, S.; Jin, F.; Yang, J.S.; Montellier, E.; Buchou, T.; Cheng, Z.; Rousseaux, S.; Rajagopal, N.; et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 2011, 146, 1016–1028. [Google Scholar] [CrossRef]
- Meng, T.G.; Zhou, Q.; Ma, X.S.; Liu, X.Y.; Meng, Q.R.; Huang, X.J.; Liu, H.L.; Lei, W.L.; Zhao, Z.H.; Sun, Q.Y.; et al. PRC2 and EHMT1 regulate H3K27me2 and H3K27me3 establishment across the zygote genome. Nat. Commun. 2020, 11, 6354. [Google Scholar] [CrossRef] [PubMed]
- Hirschey, M.D.; Zhao, Y.M. Metabolic regulation by lysinemalonylation, succinylation, and glutarylation. Mol. Cell. Proteom. 2015, 14, 2308–2315. [Google Scholar] [CrossRef]
- Greer, E.L.; Shi, Y. Histone methylation: A dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012, 13, 343–357. [Google Scholar] [CrossRef] [PubMed]
- Bui, H.T.; Van, T.N.; Kishigami, S.; Wakayama, S.; Hikichi, T.; Ohta, H.; Mizutani, E.; Yamaoka, E.; Wakayama, T.; Miyano, T. Regulation of chromatin and chromosome morphology by histone H3 modifications in pig oocytes. Reproduction 2007, 133, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Tang, S.; Ji, M.; Tang, Z.; Shimada, M.; Liu, X.; Qi, S.; Locasale, J.W.; Roeder, R.G.; Zhao, Y.; et al. Share p300-Mediated Lysine 2-Hydroxyisobutyrylation Regulates Glycolysis. Mol. Cell 2018, 70, 663–678. [Google Scholar] [CrossRef]
- Sabari, B.R.; Zhangm, D.; Allis, C.D.; Zhao, Y. Metabolic regulation of gene expression through histone acylations. Nat. Rev. Mol. Cell Biol. 2017, 18, 90–101. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Zhang, D.; Chung, D.; Tang, Z.; Huang, H.; Dai, L.; Dai, L.; Qi, S.; Li, J.; Zhao, Y.; et al. Metabolic Regulation of Gene Expression by Histone Lysine β-Hydroxybutyrylation. Mol. Cell 2016, 62, 194–206. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Xu, X.; Ding, J.; Yang, L.; Doan, M.T.; Karmaus, P.W.F.; Snyder, N.W.; Zhao, Y.; Li, J.L.; Li, X. Histone crotonylation promotes mesoendodermal commitment of human embryonic stem cells. Cell Stem Cell 2021, 28, 748–763. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Liu, X.; Chen, J.; Gao, S.; Lu, L.; Zhang, H.; Ding, G.; Wang, Z.; Chen, Z.; Shi, T.; et al. Class I histone deacetylases are major histone decrotonylases: Evidence for critical and broad function of histone crotonylation in transcription. Cell Res. 2017, 27, 898–915. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Sun, Q.; Sun, N.; Liang, M.; Tian, Z. Mitochondrial Dysfunction and Altered Renal Metabolism in Dahl Salt-Sensitive Rats. Kidney Blood Press Res. 2017, 42, 587–597. [Google Scholar] [CrossRef]
- Zhou, L.; Wang, F.; Sun, R.; Chen, X.; Zhang, M.; Xu, Q.; Wang, Y.; Wang, S.; Xiong, Y.; Guan, K.L.; et al. SIRT5 promotes IDH2 desuccinylation and G6PD deglutarylation to enhance cellular antioxidant defense. EMBO Rep. 2016, 17, 811–822. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Li, M.; Yang, M. Post-Translational Modifications in Oocyte Maturation and Embryo Development. Front. Cell Dev. Biol. 2021, 9, 645318. [Google Scholar] [CrossRef]
- Clarke, H.J.; Vieux, K.F. Epigenetic inheritance through the female germ-line: The known, the unknown, and the possible. Semin. Cell Dev. Biol. 2015, 43, 106–116. [Google Scholar] [CrossRef]
- Moritz, B.; Woltering, L.; Becker, P.B.; Gopfert, U. High levels of histone H3 acetylation at the CMV promoter are predictive of stable expression in Chinese hamster ovary cells. Biotechnol. Prog. 2016, 32, 776–786. [Google Scholar] [CrossRef]
- Cheung, P.; Vallania, F.; Warsinske, H.C.; Donato, M.; Schaffert, S.; Chang, S.E.; Dvorak, M.; Dekker, C.L.; Davis, M.M.; Utz, P.J.; et al. Single-Cell Chromatin Modification Profiling Reveals Increased Epigenetic Variations with Aging. Cell 2018, 173, 1385–1397. [Google Scholar] [CrossRef]
- Wu, L.; Wei, Y.; Li, H.; Li, W.; Gu, C.; Sun, J.; Xia, H.; Zhang, J.; Chen, F.; Liu, Q. The ubiquitination and acetylation of histones are associated with male reproductive disorders induced by chronic exposure to arsenite. Toxicol. Appl. Pharmacol. 2020, 408, 115253. [Google Scholar] [CrossRef] [PubMed]
- 33Schotta, G.; Lachner, M.; Sarma, K.; Ebert, A.; Sengupta, R.; Reuter, G.; Reinberg, D.; Jenuwein, T. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004, 18, 1251–1262. [Google Scholar]
- Lyu, G.; Guan, Y.; Zhang, C.; Zong, L.; Sun, L.; Huang, X.; Huang, L.; Zhang, L.; Tian, X.L.; Zhou, Z.; et al. Addendum: TGF-β signaling alters H4K20me3 status via miR-29 and contributes to cellular senescence and cardiac aging. Nat. Commun. 2018, 9, 4134. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Lyu, T.; Che, X.; Jia, N.; Li, Q.; Feng, W. Overexpression of SMYD3 in Ovarian Cancer is Associated with Ovarian Cancer Proliferation and Apoptosis via Methylating H3K4 and H4K20. J. Cancer 2019, 10, 4072–4084. [Google Scholar] [CrossRef] [PubMed]
- Yokoyamac, Y.; Matsumoto, A.; Hieda, M.; Shinchi, Y.; Ogihara, E.; Hamada, M.; Nishioka, Y.; Kimura, H.; Yoshidome, K.; Tsujimoto, M.; et al. Loss of histone H4K20 trimethylation predicts poor prognosis in breast cancer and is associated with invasive activity. Breast Cancer Res. 2014, 16, R66. [Google Scholar] [CrossRef]
- Fraga, M.F.; Ballestar, E.; Villar-Garea, A.; Boix-Chornet, M.; Espada, J.; Schotta, G.; Bonaldi, T.; Haydon, C.; Ropero, S.; Petrie, K.; et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat. Genet 2005, 37, 391–400. [Google Scholar] [CrossRef]
- Fritz, K.S.; Green, M.F.; Petersen, D.R.; Hirschey, M.D. Ethanol metabolism modifies hepatic protein acylation in mice. PLoS ONE 2013, 8, e75868. [Google Scholar] [CrossRef] [PubMed]
- Greer, E.L.; Maures, T.J.; Hauswirth, A.G.; Green, E.M.; Leeman, D.S.; Maro, G.S.; Han, S.; Banko, M.R.; Gozani, O.; Brunet, A. Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature 2010, 466, 383–387. [Google Scholar] [CrossRef]
- Tilly, J.L. Ovarian follicle counts—Not as simple as 1, 2, 3. Reprod. Biol. Endocrinol. 2003, 1, 11. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, M.; Li, J.; Yan, H.; Luo, T.; Huang, J.; Yuan, Y.; Hu, L.; Zheng, L. Physiological Ovarian Aging Is Associated with Altered Expression of Post-Translational Modifications in Mice. Int. J. Mol. Sci. 2022, 23, 2. https://doi.org/10.3390/ijms23010002
Wei M, Li J, Yan H, Luo T, Huang J, Yuan Y, Hu L, Zheng L. Physiological Ovarian Aging Is Associated with Altered Expression of Post-Translational Modifications in Mice. International Journal of Molecular Sciences. 2022; 23(1):2. https://doi.org/10.3390/ijms23010002
Chicago/Turabian StyleWei, Minli, Jia Li, Huilin Yan, Tao Luo, Jiang Huang, Yangyang Yuan, Liaoliao Hu, and Liping Zheng. 2022. "Physiological Ovarian Aging Is Associated with Altered Expression of Post-Translational Modifications in Mice" International Journal of Molecular Sciences 23, no. 1: 2. https://doi.org/10.3390/ijms23010002





