Genome-Wide Identification, Expression Patterns and Sugar Transport of the Physic Nut SWEET Gene Family and a Functional Analysis of JcSWEET16 in Arabidopsis
Abstract
:1. Introduction
2. Results
2.1. Identification and Phylogenetic Analysis of SWEET Family Genes in Physic Nut
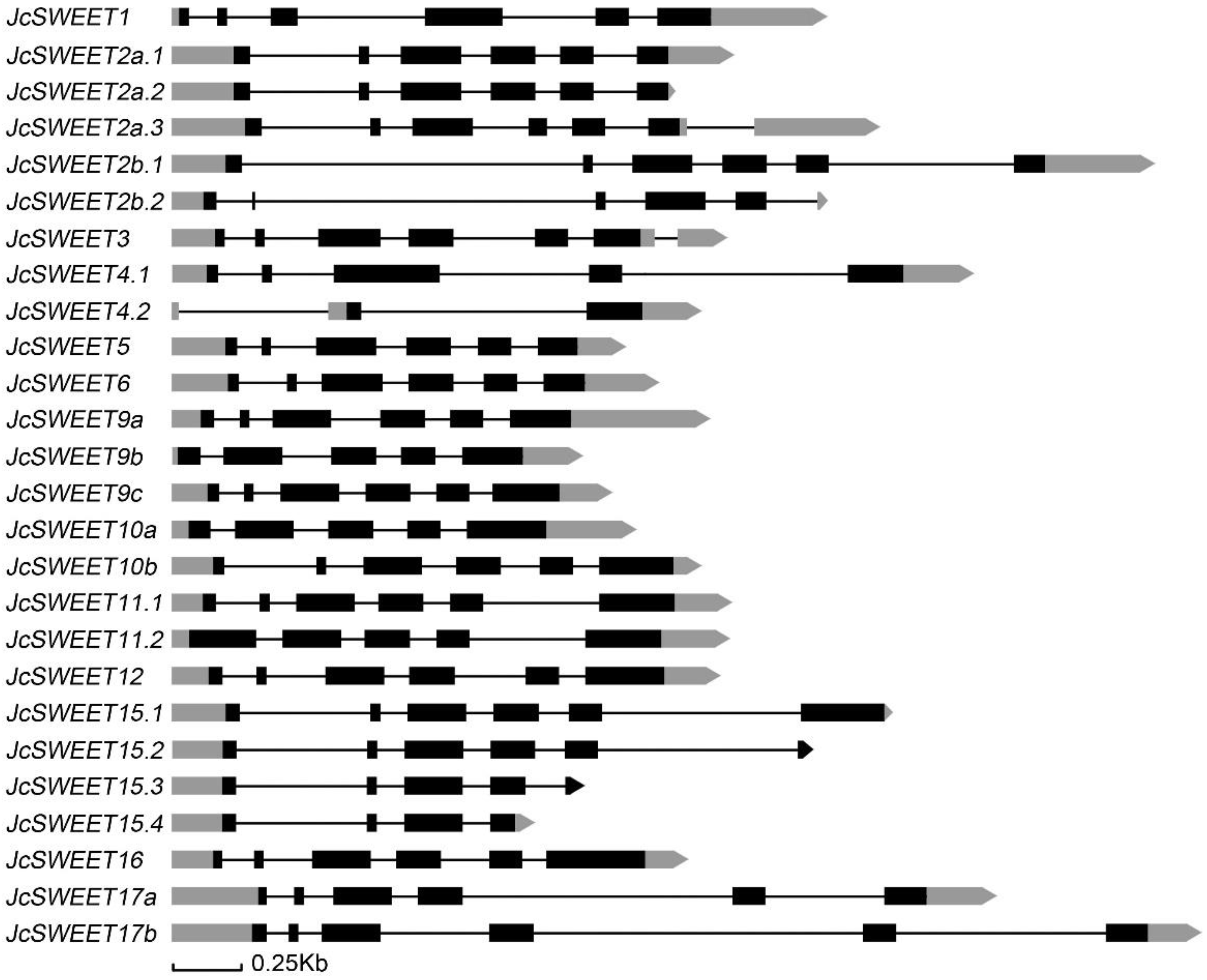
2.2. Gene Structure and Chromosomal Distribution Analysis of SWEET Family Genes in Physic Nut
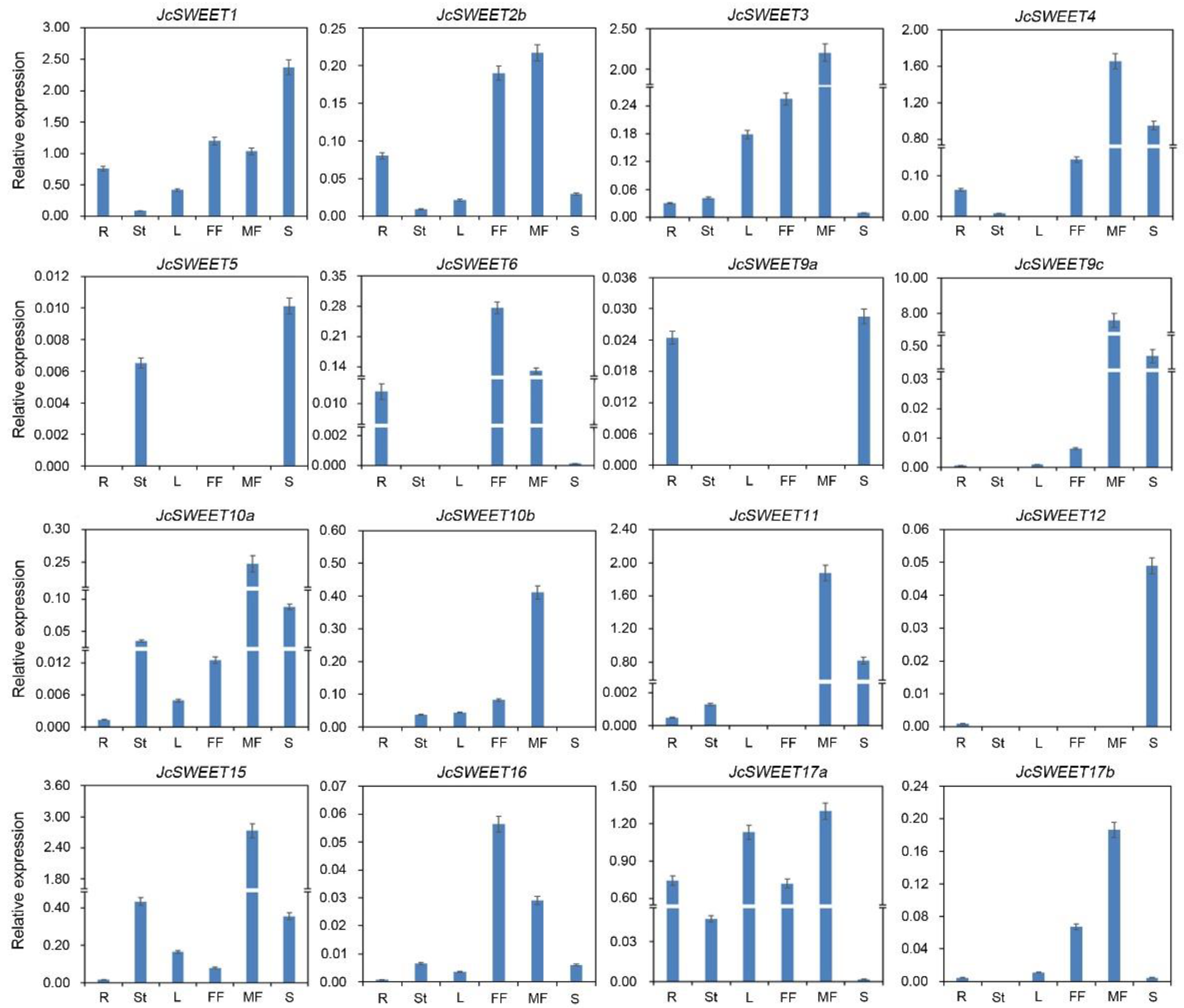
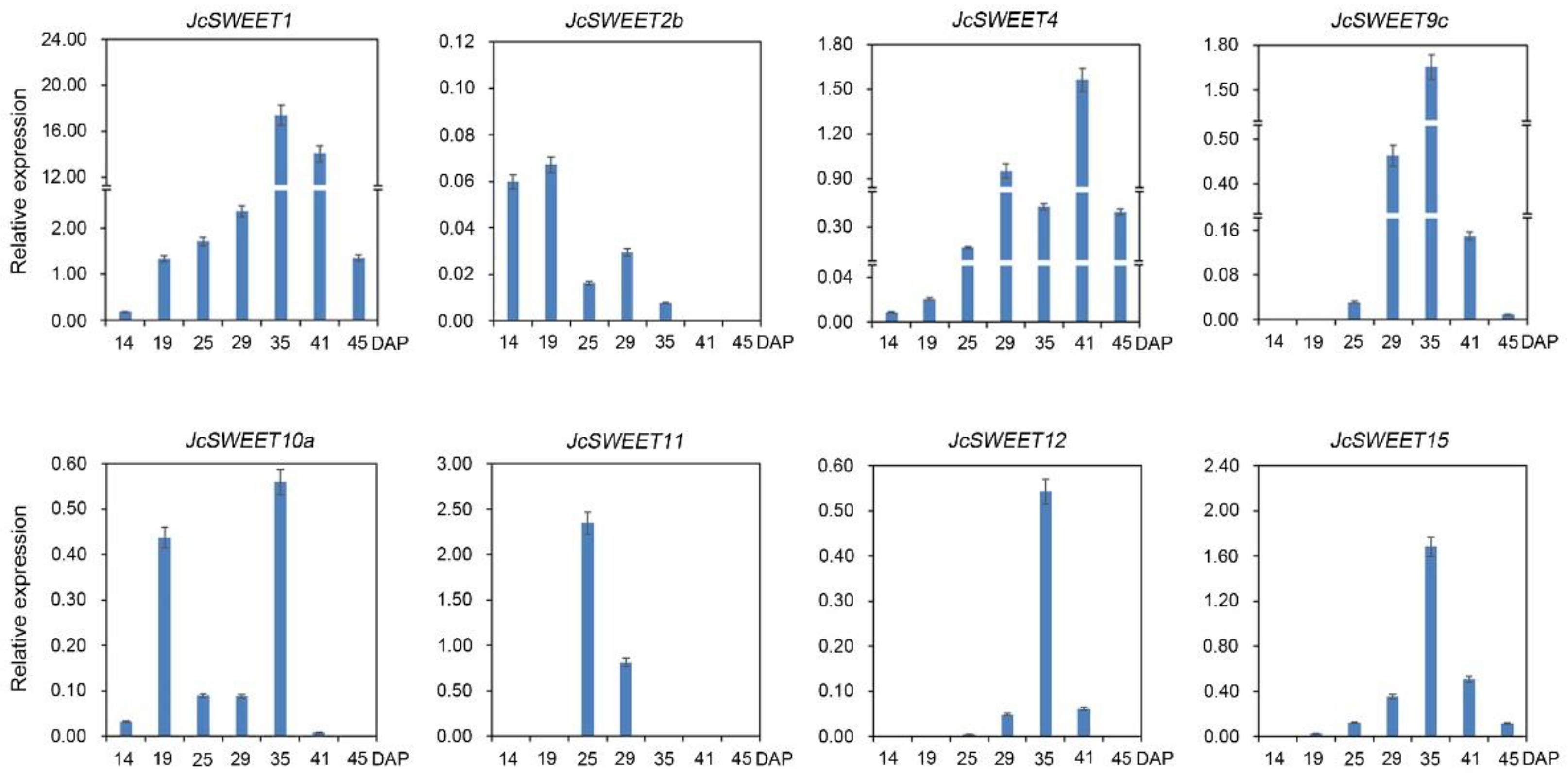
2.3. Expression Profiles of SWEET Genes in Different Physic Nut Tissues
2.4. Expression Profiles of JcSWEET Genes under Drought and Salinity Stress
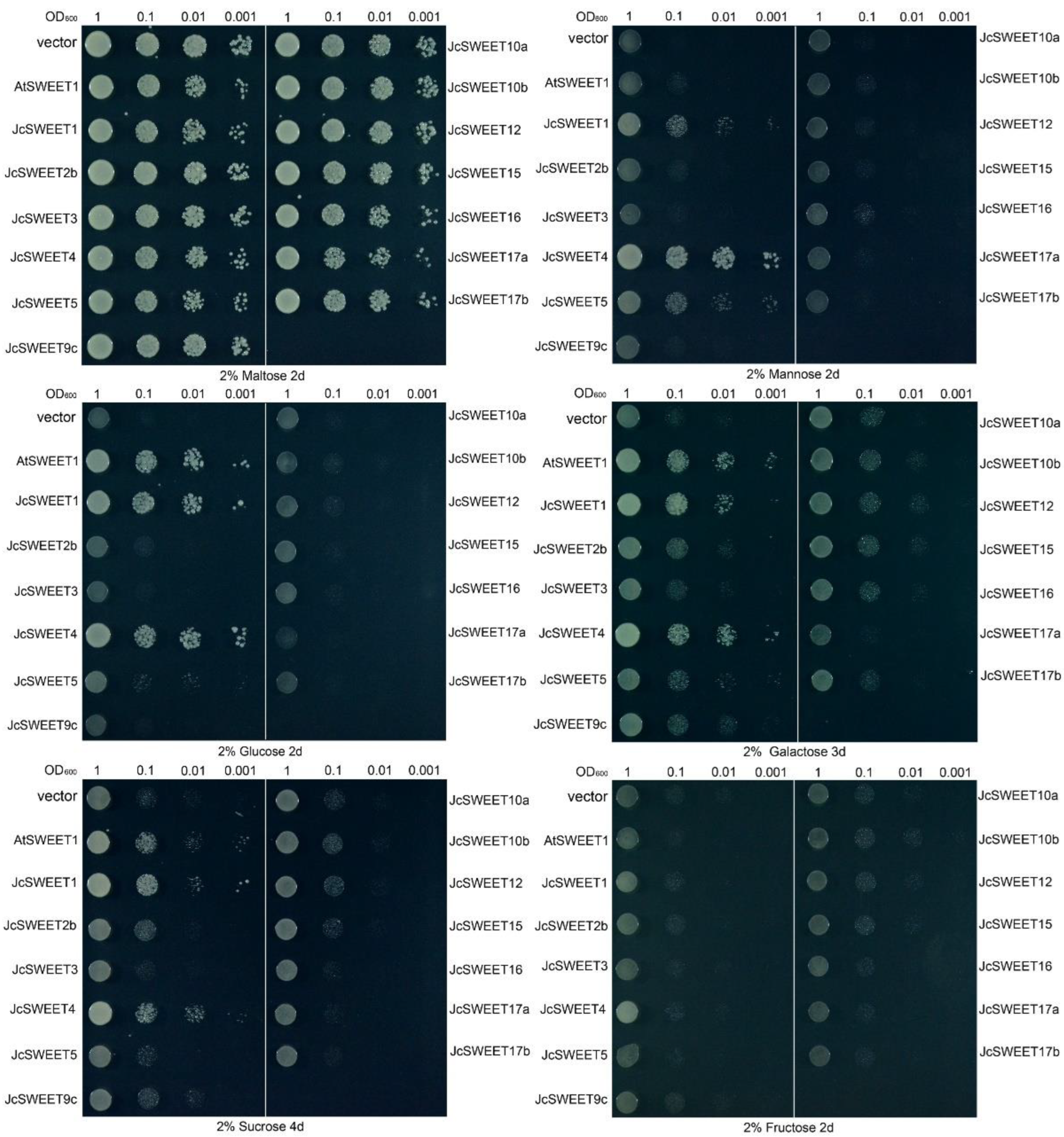
2.5. Transport Activity of JcSWEETs in Yeast
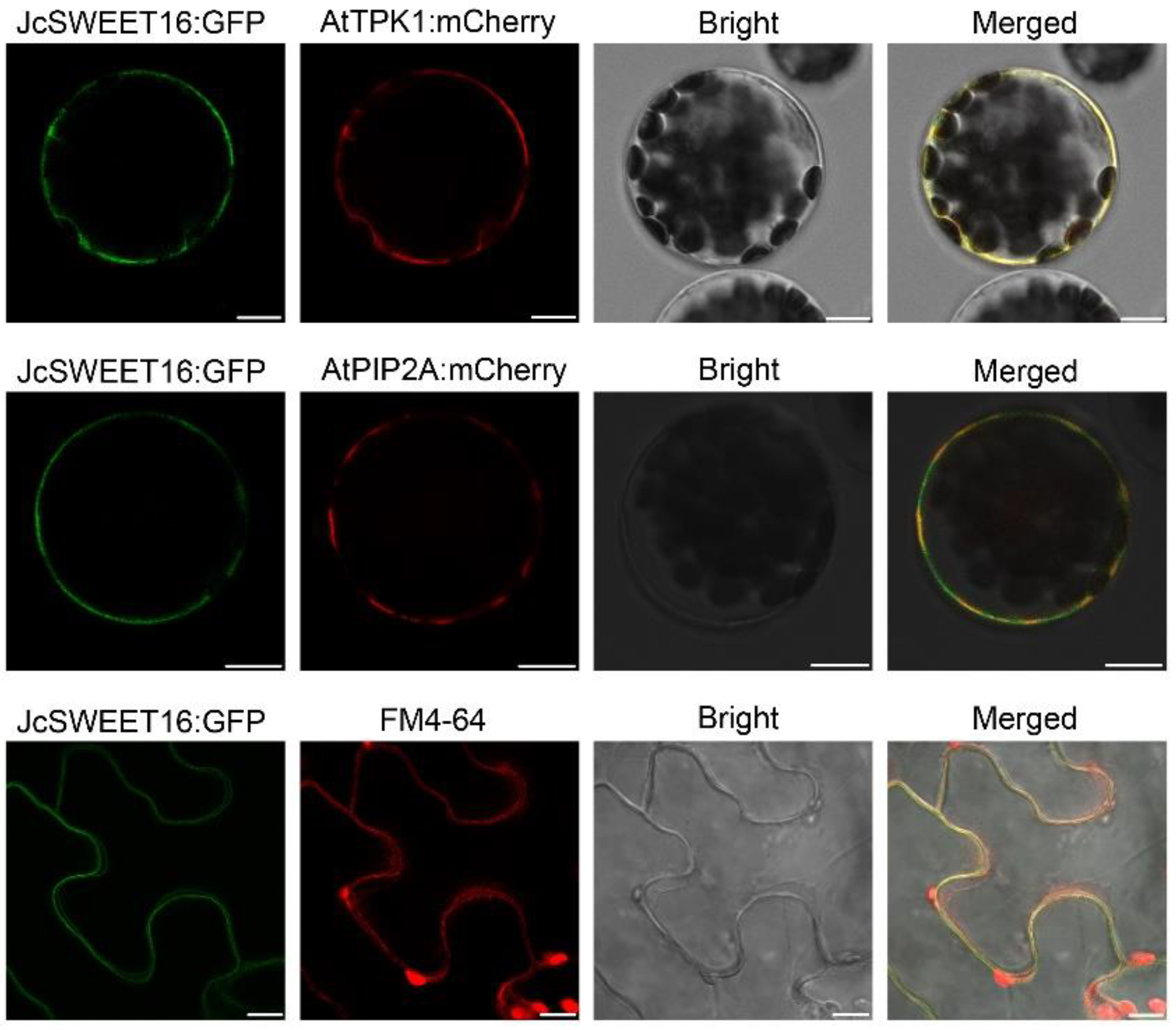
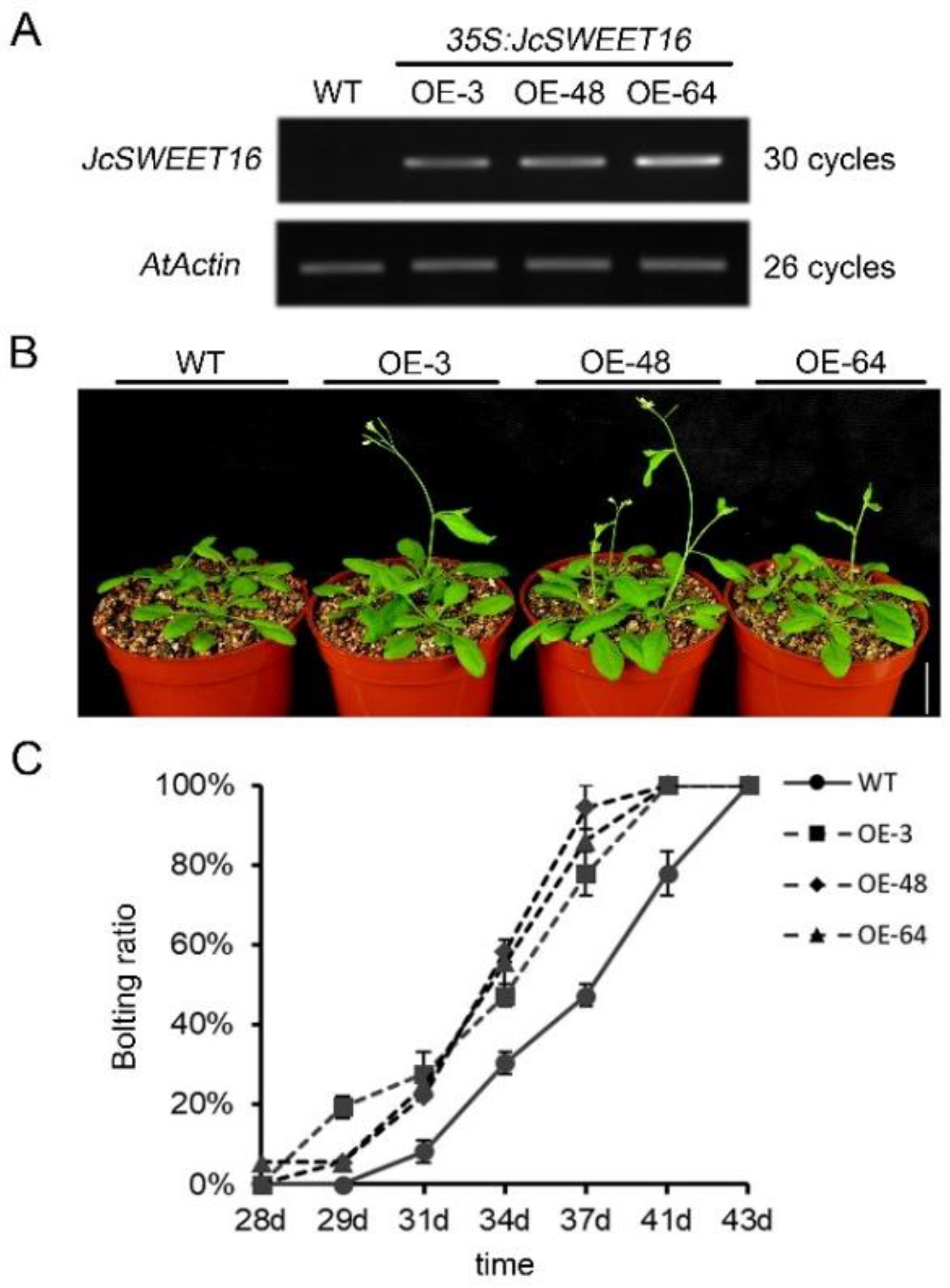
2.6. Overexpression of JcSWEET16 Causes Early Flowering and Increases Salt Stress Tolerance in A. thaliana
3. Discussion
4. Materials and Methods
4.1. Preparation of Plant Materials
4.2. Sequence Database Searches and Gene Cloning
4.3. Sequence Analysis and Phylogenetic Tree Construction
4.4. RNA Isolation and qRT-PCR
4.5. Plasmid Construction and Complementation Assays in Yeast
4.6. Subcellular Localization of JcSWEET16
4.7. Plant Transformation and Salinity Treatment of Transgenic A. thaliana
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ruan, Y.-L. Sucrose Metabolism: Gateway to Diverse Carbon Use and Sugar Signaling. Annu. Rev. Plant Biol. 2014, 65, 33–67. [Google Scholar] [CrossRef] [PubMed]
- Rolland, F.; Moore, B.; Sheen, J. Sugar Sensing and Signaling in Plants. Plant Cell 2002, 14, 185–205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walmsley, A.R.; Barrett, M.P.; Bringaud, F.; Gould, G.W. Sugar transporters from bacteria, parasites and mammals: Structure–activity relationships. Trends Biochem. Sci. 1998, 23, 476–481. [Google Scholar] [CrossRef]
- Chen, L.-Q.; Hou, B.-H.; Lalonde, S.; Takanaga, H.; Hartung, M.L.; Qu, X.-Q.; Guo, W.-J.; Kim, J.-G.; Underwood, W.; Chaudhuri, B.; et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 2010, 468, 527–532. [Google Scholar] [CrossRef] [Green Version]
- Patrick, J.W.; Offler, C.E. Compartmentation of transport and transfer events in developing seeds. J. Exp. Bot. 2001, 52, 551–564. [Google Scholar] [CrossRef]
- Patrick, J.W. Phloem Unloading: Sieve Element Unloading and Post-Sieve Element Transport. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997, 48, 191–222. [Google Scholar] [CrossRef]
- Eom, J.-S.; Chen, L.-Q.; Sosso, D.; Julius, B.T.; Lin, I.; Qu, X.-Q.; Braun, D.M.; Frommer, W.B. SWEETs, transporters for intracellular and intercellular sugar translocation. Curr. Opin. Plant Biol. 2015, 25, 53–62. [Google Scholar] [CrossRef] [Green Version]
- Jeena, G.S.; Kumar, S.; Shukla, R.K. Structure, evolution and diverse physiological roles of SWEET sugar transporters in plants. Plant Mol. Biol. 2019, 100, 351–365. [Google Scholar] [CrossRef]
- Chen, L.-Q.; Cheung, L.S.; Feng, L.; Tanner, W.; Frommer, W.B. Transport of Sugars. Annu. Rev. Biochem. 2015, 84, 865–894. [Google Scholar] [CrossRef]
- Chandran, D. Co-option of developmentally regulated plant SWEET transporters for pathogen nutrition and abiotic stress tolerance. IUBMB Life 2015, 67, 461–471. [Google Scholar] [CrossRef] [Green Version]
- Yuan, M.; Wang, S. Rice MtN3/Saliva/SWEET Family Genes and Their Homologs in Cellular Organisms. Mol. Plant 2013, 6, 665–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wellmer, F.; Alves-Ferreira, M.; Dubois, A.; Riechmann, J.L.; Meyerowitz, E.M. Genome-Wide Analysis of Gene Expression during Early Arabidopsis Flower Development. PLoS Genet. 2006, 2, e117. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.-Q.; Qu, X.-Q.; Hou, B.-H.; Sosso, D.; Osorio, S.; Fernie, A.R.; Frommer, W.B. Sucrose Efflux Mediated by SWEET Proteins as a Key Step for Phloem Transport. Science 2012, 335, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Braun, D.M. Plant science. SWEET! The pathway is complete. Science 2012, 335, 173–174. [Google Scholar] [CrossRef]
- Baker, R.F.; Leach, K.A.; Braun, D.M. SWEET as Sugar: New Sucrose Effluxers in Plants. Mol. Plant 2012, 5, 766–768. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.-Q.; Lin, I.W.; Qu, X.-Q.; Sosso, D.; McFarlane, H.E.; Londoño, A.; Samuels, A.L.; Frommer, W.B. A Cascade of Sequentially Expressed Sucrose Transporters in the Seed Coat and Endosperm Provides Nutrition for the Arabidopsis Embryo. Plant Cell 2015, 27, 607–619. [Google Scholar] [CrossRef] [Green Version]
- Guan, Y.-F.; Huang, X.-Y.; Zhu, J.; Gao, J.-F.; Zhang, H.-X.; Yang, Z.-N. RUPTURED POLLEN GRAIN1, a Member of the MtN3/saliva Gene Family, Is Crucial for Exine Pattern Formation and Cell Integrity of Microspores in Arabidopsis. Plant Physiol. 2008, 147, 852–863. [Google Scholar] [CrossRef] [Green Version]
- Lin, I.W.; Sosso, D.; Chen, L.-Q.; Gase, K.; Kim, S.-G.; Kessler, D.; Klinkenberg, P.M.; Gorder, M.K.; Hou, B.-H.; Qu, X.-Q.; et al. Nectar secretion requires sucrose phosphate synthases and the sugar transporter SWEET9. Nature 2014, 508, 546–549. [Google Scholar] [CrossRef]
- Kanno, Y.; Oikawa, T.; Chiba, Y.; Ishimaru, Y.; Shimizu, T.; Sano, N.; Koshiba, T.; Kamiya, Y.; Ueda, M.; Seo, M. AtSWEET13 and AtSWEET14 regulate gibberellin-mediated physiological processes. Nat. Commun. 2016, 7, 13245. [Google Scholar] [CrossRef]
- Yang, J.; Luo, D.; Yang, B.; Frommer, W.B.; Eom, J.S. SWEET 11 and 15 as key players in seed filling in rice. New Phytol. 2018, 218, 604–615. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Zhang, C.; Han, X.; Wang, Z.Y.; Ma, L.; Yuan, P.; Wu, J.N.; Zhu, X.F.; Liu, J.M.; Li, D.P.; et al. Inhibition of OsSWEET11 function in mesophyll cells improves resistance of rice to sheath blight disease. Mol. Plant Pathol. 2018, 19, 2149–2161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, L.; Zhang, D.; Miao, Q.; Yang, J.; Xuan, Y.; Hu, Y. Essential Role of Sugar Transporter OsSWEET11 During the Early Stage of Rice Grain Filling. Plant Cell Physiol. 2017, 58, 863–873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Yokosho, K.; Guo, R.; Whelan, J.; Ruan, Y.-L.; Ma, J.F.; Shou, H. The Soybean Sugar Transporter GmSWEET15 Mediates Sucrose Export from Endosperm to Early Embryo. Plant Physiol. 2019, 180, 2133–2141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patil, G.; Valliyodan, B.; Deshmukh, R.; Prince, S.; Nicander, B.; Zhao, M.; Sonah, H.; Song, L.; Lin, L.; Chaudhary, J.; et al. Soybean (Glycine max) SWEET gene family: Insights through comparative genomics, transcriptome profiling and whole genome re-sequence analysis. BMC Genom. 2015, 16, 520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bezrutczyk, M.; Hartwig, T.; Horschman, M.; Char, S.N.; Yang, J.; Yang, B.; Frommer, W.B.; Sosso, D. Impaired phloem loading in zmsweet13a,b,c sucrose transporter triple knock-out mutants in Zea mays. New Phytol. 2018, 218, 594–603. [Google Scholar] [CrossRef] [Green Version]
- Valifard, M.; Le Hir, R.; Müller, J.; Scheuring, D.; Neuhaus, H.E.; Pommerrenig, B. Vacuolar fructose transporter SWEET17 is critical for root development and drought tolerance. Plant Physiol. 2021, 187, 2716–2730. [Google Scholar] [CrossRef]
- Klemens, P.A.; Patzke, K.; Deitmer, J.; Spinner, L.; Le Hir, R.; Bellini, C.; Bedu, M.; Chardon, F.; Krapp, A.; Neuhaus, H.E. Overexpression of the Vacuolar Sugar Carrier AtSWEET16 Modifies Germination, Growth, and Stress Tolerance in Arabidopsis. Plant Physiol. 2013, 163, 1338–1352. [Google Scholar] [CrossRef] [Green Version]
- Seo, P.J.; Park, J.-M.; Kang, S.K.; Kim, S.-G.; Park, C.-M. An Arabidopsis senescence-associated protein SAG29 regulates cell viability under high salinity. Planta 2011, 233, 189–200. [Google Scholar] [CrossRef]
- Le Hir, R.; Spinner, L.; Klemens, P.A.W.; Chakraborti, D.; de Marco, F.; Vilaine, F.; Wolff, N.; Lemoine, R.; Porcheron, B.; Géry, C.; et al. Disruption of the Sugar Transporters AtSWEET11 and AtSWEET12 Affects Vascular Development and Freezing Tolerance in Arabidopsis. Mol. Plant 2015, 8, 1687–1690. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Zhang, Y.; Yang, C.; Tian, Z.; Li, J. AtSWEET4, a hexose facilitator, mediates sugar transport to axial sinks and affects plant development. Sci. Rep. 2016, 6, 24563. [Google Scholar] [CrossRef] [Green Version]
- Mathan, J.; Singh, A.; Ranjan, A. Sucrose transport in response to drought and salt stress involves ABA-mediated induction of OsSWEET13 and OsSWEET15 in rice. Physiol. Plant. 2020, 171, 620–637. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Zhang, F.; Song, S.; Yu, X.; Ren, Y.; Zhao, X.; Liu, H.; Liu, G.; Wang, Y.; He, H. CsSWEET2, a Hexose Transporter from Cucumber (Cucumis sativus L.), Affects Sugar Metabolism and Improves Cold Tolerance in Arabidopsis. Int. J. Mol. Sci. 2022, 23, 3886. [Google Scholar] [CrossRef] [PubMed]
- Xuan, C.; Lan, G.; Si, F.; Zeng, Z.; Wang, C.; Yadav, V.; Wei, C.; Zhang, X. Systematic Genome-Wide Study and Expression Analysis of SWEET Gene Family: Sugar Transporter Family Contributes to Biotic and Abiotic Stimuli in Watermelon. Int. J. Mol. Sci. 2021, 22, 8407. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yao, L.; Hao, X.; Li, N.; Qian, W.; Yue, C.; Ding, C.; Zeng, J.; Yang, Y.; Wang, X. Tea plant SWEET transporters: Expression profiling, sugar transport, and the involvement of CsSWEET16 in modifying cold tolerance in Arabidopsis. Plant Mol. Biol. 2018, 96, 577–592. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.; Singh, K.; Rao, G.R.; Chikara, J.; Kumar, D.; Mishra, D.K.; Saikia, S.P.; Pathre, U.V.; Raghuvanshi, N.; Rahi, T.S.; et al. Agro-technology of Jatropha curcas for diverse environmental conditions in India. Biomass Bioenergy 2013, 48, 191–202. [Google Scholar] [CrossRef]
- Wu, P.; Zhou, C.; Cheng, S.; Wu, Z.; Lu, W.; Han, J.; Chen, Y.; Chen, Y.; Ni, P.; Wang, Y.; et al. Integrated genome sequence and linkage map of physic nut (Jatropha curcas L.), a biodiesel plant. Plant J. 2015, 81, 810–821. [Google Scholar] [CrossRef]
- Sato, S.; Hirakawa, H.; Isobe, S.; Fukai, E.; Watanabe, A.; Kato, M.; Kawashima, K.; Minami, C.; Muraki, A.; Nakazaki, N.; et al. Sequence Analysis of the Genome of an Oil-Bearing Tree, Jatropha curcas L. DNA Res. 2011, 18, 65–76. [Google Scholar] [CrossRef]
- Natarajan, P.; Parani, M. De novo assembly and transcriptome analysis of five major tissues of Jatropha curcas L. using GS FLX titanium platform of 454 pyrosequencing. BMC Genom. 2011, 12, 191. [Google Scholar] [CrossRef] [Green Version]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Zhang, L.; Zhang, S.; Zhu, S.; Wu, P.; Chen, Y.; Li, M.; Jiang, H.; Wu, G. Global analysis of gene expression profiles in physic nut (Jatropha curcas L.) seedlings exposed to drought stress. BMC Plant Biol. 2015, 15, 17. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Zhang, C.; Wu, P.; Chen, Y.; Li, M.; Jiang, H.; Wu, G. Global Analysis of Gene Expression Profiles in Physic Nut (Jatropha curcas L.) Seedlings Exposed to Salt Stress. PLoS ONE 2014, 9, e97878. [Google Scholar] [CrossRef] [PubMed]
- Wieczorke, R.; Krampe, S.; Weierstall, T.; Freidel, K.; Hollenberg, C.P.; Boles, E. Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. FEBS Lett. 1999, 464, 123–128. [Google Scholar] [CrossRef] [Green Version]
- Ko, H.-Y.; Ho, L.-H.; Neuhaus, H.E.; Guo, W.-J. Transporter SlSWEET15 unloads sucrose from phloem and seed coat for fruit and seed development in tomato. Plant Physiol. 2021, 187, 2230–2245. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.-P.; Zhang, F.; Song, S.-H.; Tang, X.-W.; Xu, H.; Liu, G.-M.; Wang, Y.; He, H.-J. Genome-wide identification, characterization, and expression analysis of the SWEET gene family in cucumber. J. Integr. Agric. 2017, 16, 1486–1501. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Wang, Z.Y.; Kumar, V.; Xu, X.F.; Yuan, D.P.; Zhu, X.F.; Li, T.Y.; Jia, B.; Xuan, Y.H. Genome-wide identification of the SWEET gene family in wheat. Gene 2018, 642, 284–292. [Google Scholar] [CrossRef]
- Soltis, D.E.; Visger, C.J.; Soltis, P.S. The polyploidy revolution then...and now: Stebbins revisited. Am. J. Bot. 2014, 101, 1057–1078. [Google Scholar] [CrossRef] [Green Version]
- Severin, A.J.; Cannon, S.B.; Graham, M.M.; Grant, D.; Shoemaker, R.C. Changes in Twelve Homoeologous Genomic Regions in Soybean following Three Rounds of Polyploidy. Plant Cell 2011, 23, 3129–3136. [Google Scholar] [CrossRef] [Green Version]
- Vision, T.J.; Brown, D.G.; Tanksley, S.D. The Origins of Genomic Duplications in Arabidopsis. Science 2000, 290, 2114–2117. [Google Scholar] [CrossRef] [Green Version]
- Long, M.; Rosenberg, C.; Gilbert, W. Intron phase correlations and the evolution of the intron/exon structure of genes. Proc. Natl. Acad. Sci. USA 1995, 92, 12495–12499. [Google Scholar] [CrossRef] [Green Version]
- Chardon, F.; Bedu, M.; Calenge, F.; Klemens, P.A.; Spinner, L.; Clement, G.; Chietera, G.; Léran, S.; Ferrand, M.; Lacombe, B.; et al. Leaf Fructose Content Is Controlled by the Vacuolar Transporter SWEET17 in Arabidopsis. Curr. Biol. 2013, 23, 697–702. [Google Scholar] [CrossRef]
- Yao, L.; Ding, C.; Hao, X.; Zeng, J.; Yang, Y.; Wang, X.; Wang, L. CsSWEET1a and CsSWEET17 Mediate Growth and Freezing Tolerance by Promoting Sugar Transport across the Plasma Membrane. Plant Cell Physiol. 2020, 61, 1669–1682. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Sun, M.-H.; Ma, Q.-J.; Kang, H.; Liu, Y.-J.; Hao, Y.-J.; You, C.-X. MdSWEET17, a sugar transporter in apple, enhances drought tolerance in tomato. J. Integr. Agric. 2019, 18, 2041–2051. [Google Scholar] [CrossRef]
- Zhou, A.; Ma, H.; Feng, S.; Gong, S.; Wang, J. DsSWEET17, a Tonoplast-Localized Sugar Transporter from Dianthus spiculifolius, Affects Sugar Metabolism and Confers Multiple Stress Tolerance in Arabidopsis. Int. J. Mol. Sci. 2018, 19, 1564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Durand, M.; Mainson, D.; Porcheron, B.; Maurousset, L.; Lemoine, R.; Pourtau, N. Carbon source–sink relationship in Arabidopsis thaliana: The role of sucrose transporters. Planta 2018, 247, 587–611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lemoine, R.; La Camera, S.; Atanassova, R.; Dédaldéchamp, F.; Allario, T.; Pourtau, N.; Bonnemain, J.L.; Laloi, M.; Coutos-Thévenot, P.; Maurousset, L.; et al. Source-to-sink transport of sugar and regulation by environmental factors. Front. Plant Sci. 2013, 4, 272. [Google Scholar] [CrossRef] [Green Version]
- Corbesier, L.; Lejeune, P.; Bernier, G. The role of carbohydrates in the induction of flowering in Arabidopsis thaliana: Comparison between the wild type and a starchless mutant. Planta 1998, 206, 131–137. [Google Scholar] [CrossRef]
- Micallef, B.J.; Haskins, K.A.; Vanderveer, P.J.; Roh, K.-S.; Shewmaker, C.K.; Sharkey, T.D. Altered photosynthesis, flowering, and fruiting in transgenic tomato plants that have an increased capacity for sucrose synthesis. Planta 1995, 196, 327–334. [Google Scholar] [CrossRef]
- Müller-Röber, B.; Sonnewald, U.; Willmitzer, L. Inhibition of the ADP-glucose pyrophosphorylase in transgenic potatoes leads to sugar-storing tubers and influences tuber formation and expression of tuber storage protein genes. EMBO J. 1992, 11, 1229–1238. [Google Scholar] [CrossRef]
- Jiang, H.; Wu, P.; Zhang, S.; Song, C.; Chen, Y.; Li, M.; Jia, Y.; Fang, X.; Chen, F.; Wu, G. Global Analysis of Gene Expression Profiles in Developing Physic Nut (Jatropha curcas L.) Seeds. PLoS ONE 2012, 7, e36522. [Google Scholar] [CrossRef] [Green Version]
- da Silva, E.N.; Gomes Silveira, J.A.; Rodrigues Fernandes, C.R.; Batista Dutra, A.T.; de Aragao, R.M. Ion uptake and growth of physic nut under different salinity levels. Rev. Cienc. Agron. 2009, 40, 240–246. [Google Scholar] [CrossRef]
- Zhao, Q.; Zhang, L.; Zhu, S.; Zhang, S.; Wu, P.; Chen, Y.; Jiang, H.; Wu, G.; Li, M. Effects of several abiotic stresses on photosynthetic rate and other physiological indexes in Jatropha curcas L. seedlings. J. Trop. Subtrop. Bot. 2012, 20, 432–438. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, A.-Y.; Zhu, Q.H.; Chen, X.; Luo, J.C. GSDS: A gene structure display server. Yi Chuan Hered. 2007, 29, 1023–1026. [Google Scholar] [CrossRef]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiong, W.; Xu, X.; Zhang, L.; Wu, P.; Chen, Y.; Li, M.; Jiang, H.; Wu, G. Genome-wide analysis of the WRKY gene family in physic nut (Jatropha curcas L.). Gene 2013, 524, 124–132. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.-D.; Cho, Y.-H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef] [Green Version]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Zhao, H.; Gao, F.; Yao, P.; Deng, R.; Li, C.; Chen, H.; Wu, Q. A R2R3-MYB transcription factor gene, FtMYB13, from Tartary buckwheat improves salt/drought tolerance in Arabidopsis. Plant Physiol. Biochem. 2018, 132, 238–248. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Y.; Wu, P.; Xu, S.; Chen, Y.; Li, M.; Wu, G.; Jiang, H. Genome-Wide Identification, Expression Patterns and Sugar Transport of the Physic Nut SWEET Gene Family and a Functional Analysis of JcSWEET16 in Arabidopsis. Int. J. Mol. Sci. 2022, 23, 5391. https://doi.org/10.3390/ijms23105391
Wu Y, Wu P, Xu S, Chen Y, Li M, Wu G, Jiang H. Genome-Wide Identification, Expression Patterns and Sugar Transport of the Physic Nut SWEET Gene Family and a Functional Analysis of JcSWEET16 in Arabidopsis. International Journal of Molecular Sciences. 2022; 23(10):5391. https://doi.org/10.3390/ijms23105391
Chicago/Turabian StyleWu, Youting, Pingzhi Wu, Shaoming Xu, Yaping Chen, Meiru Li, Guojiang Wu, and Huawu Jiang. 2022. "Genome-Wide Identification, Expression Patterns and Sugar Transport of the Physic Nut SWEET Gene Family and a Functional Analysis of JcSWEET16 in Arabidopsis" International Journal of Molecular Sciences 23, no. 10: 5391. https://doi.org/10.3390/ijms23105391





