Comprehensive Analysis for GRF Transcription Factors in Sacred Lotus (Nelumbo nucifera)
Abstract
:1. Introduction
2. Results
2.1. Identification of GRF Genes
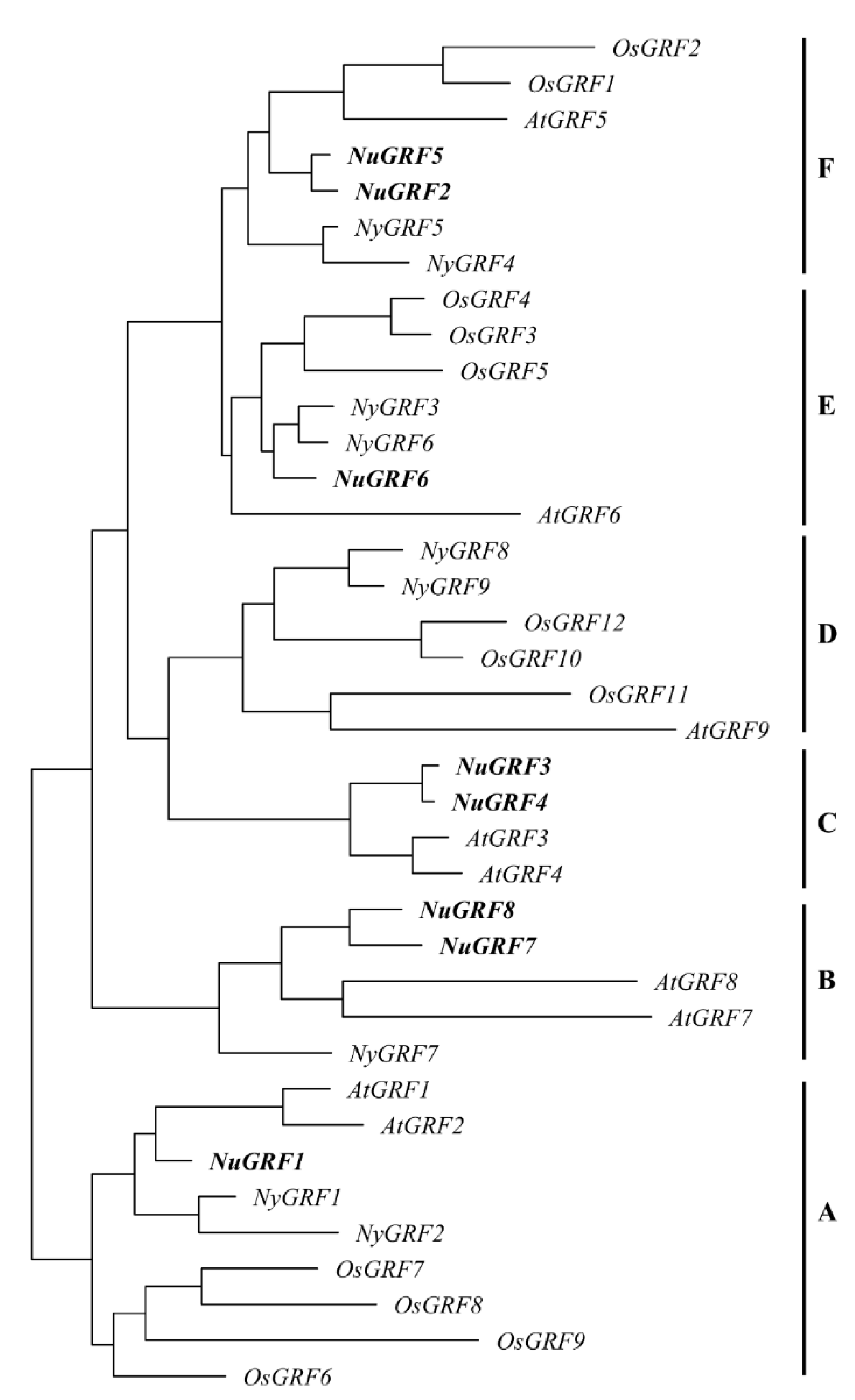
2.2. Phylogenetic Analysis
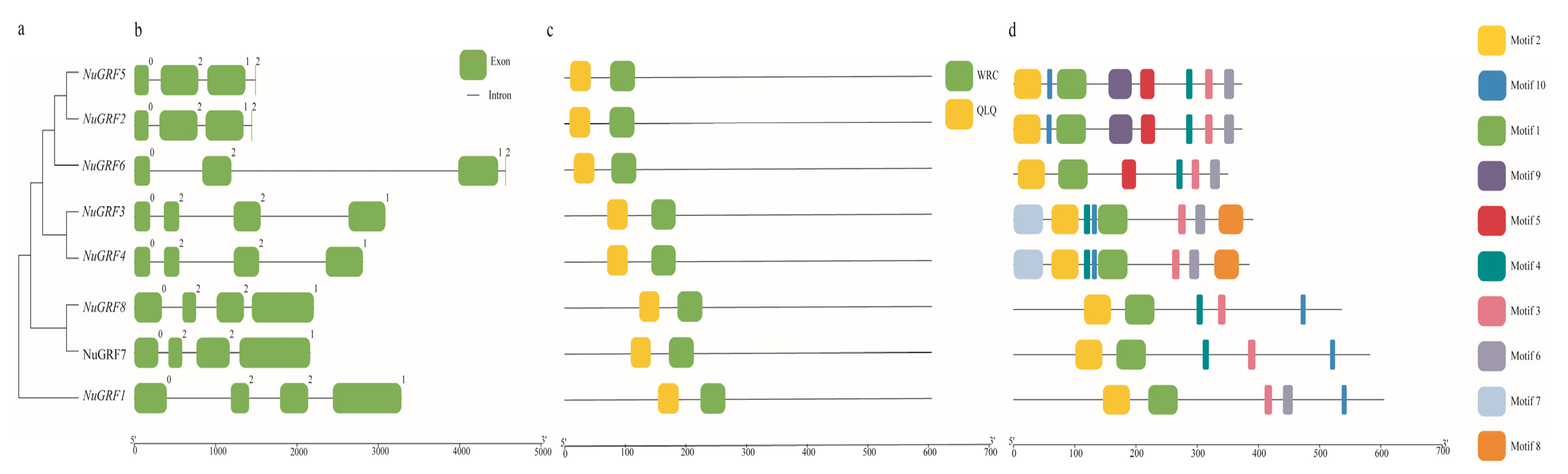
2.3. Gene Structure and Motif Analysis
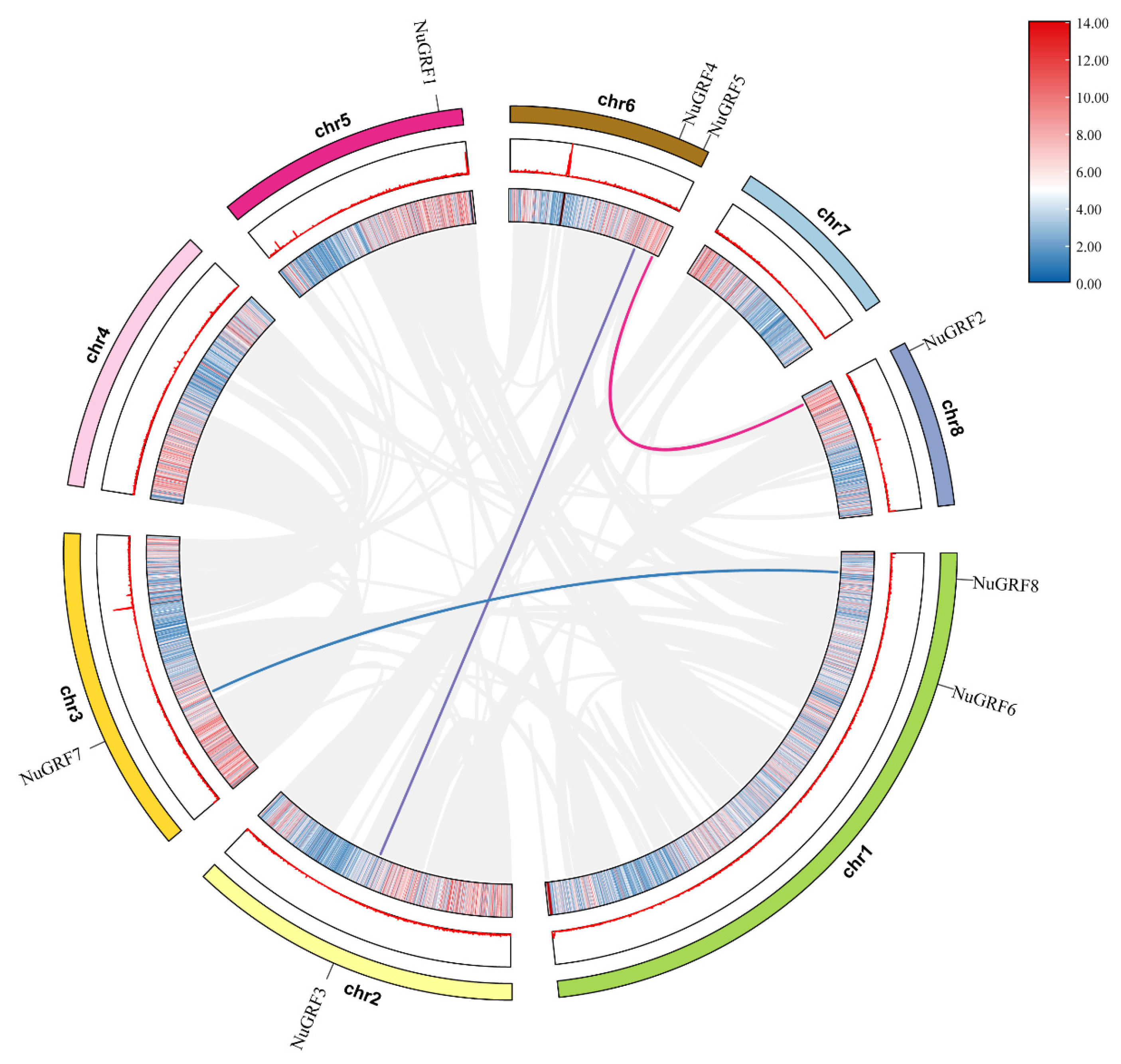
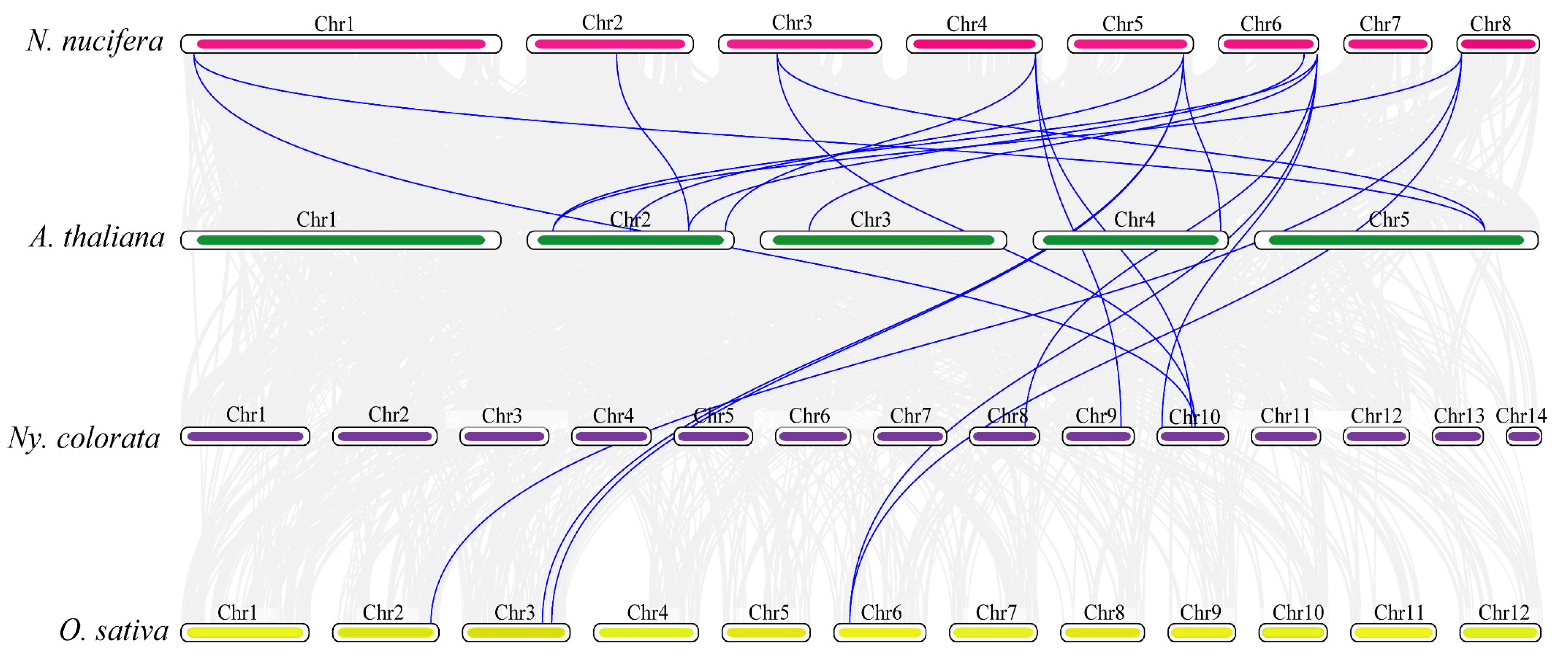
2.4. Chromosome Location and Collinearity Analysis
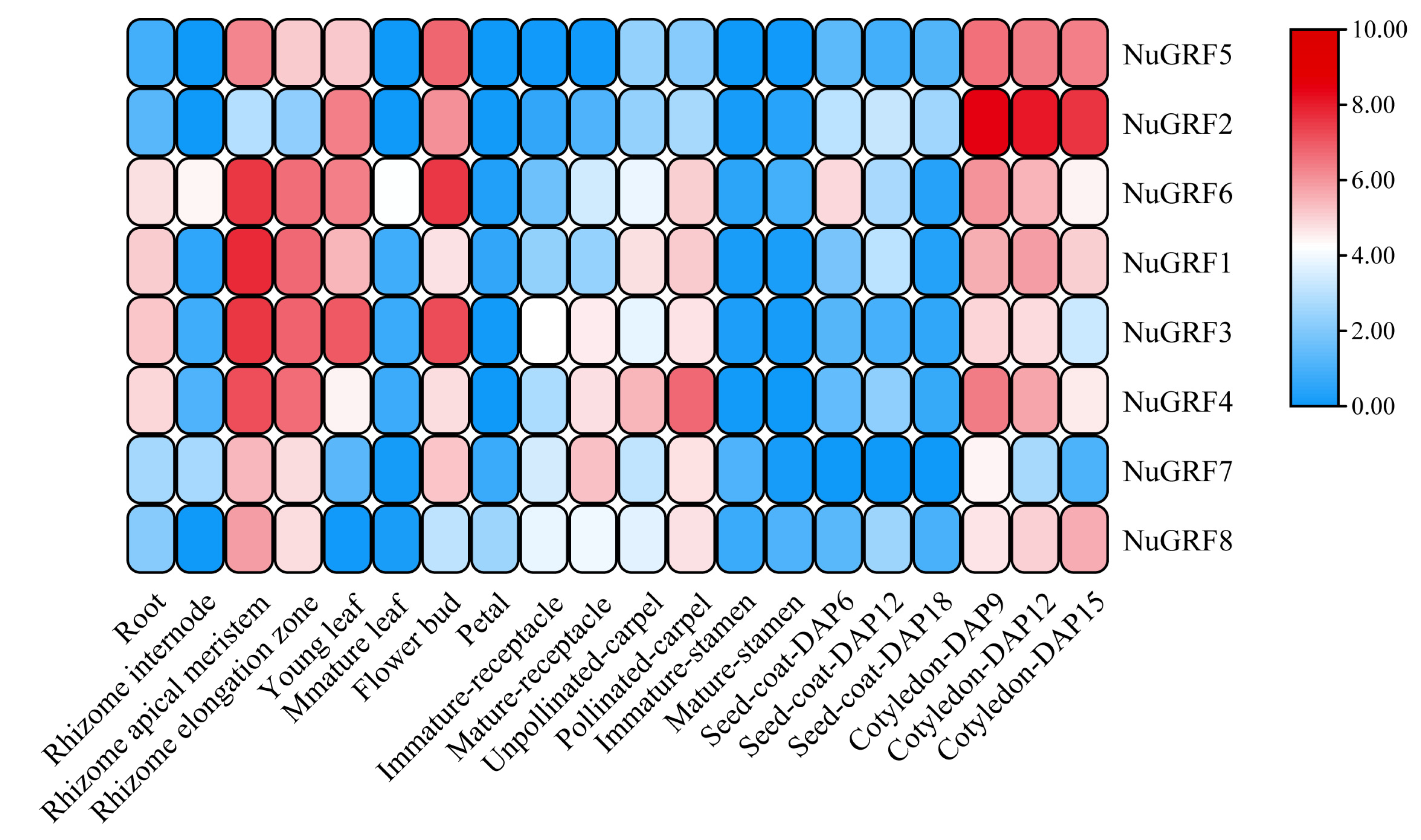
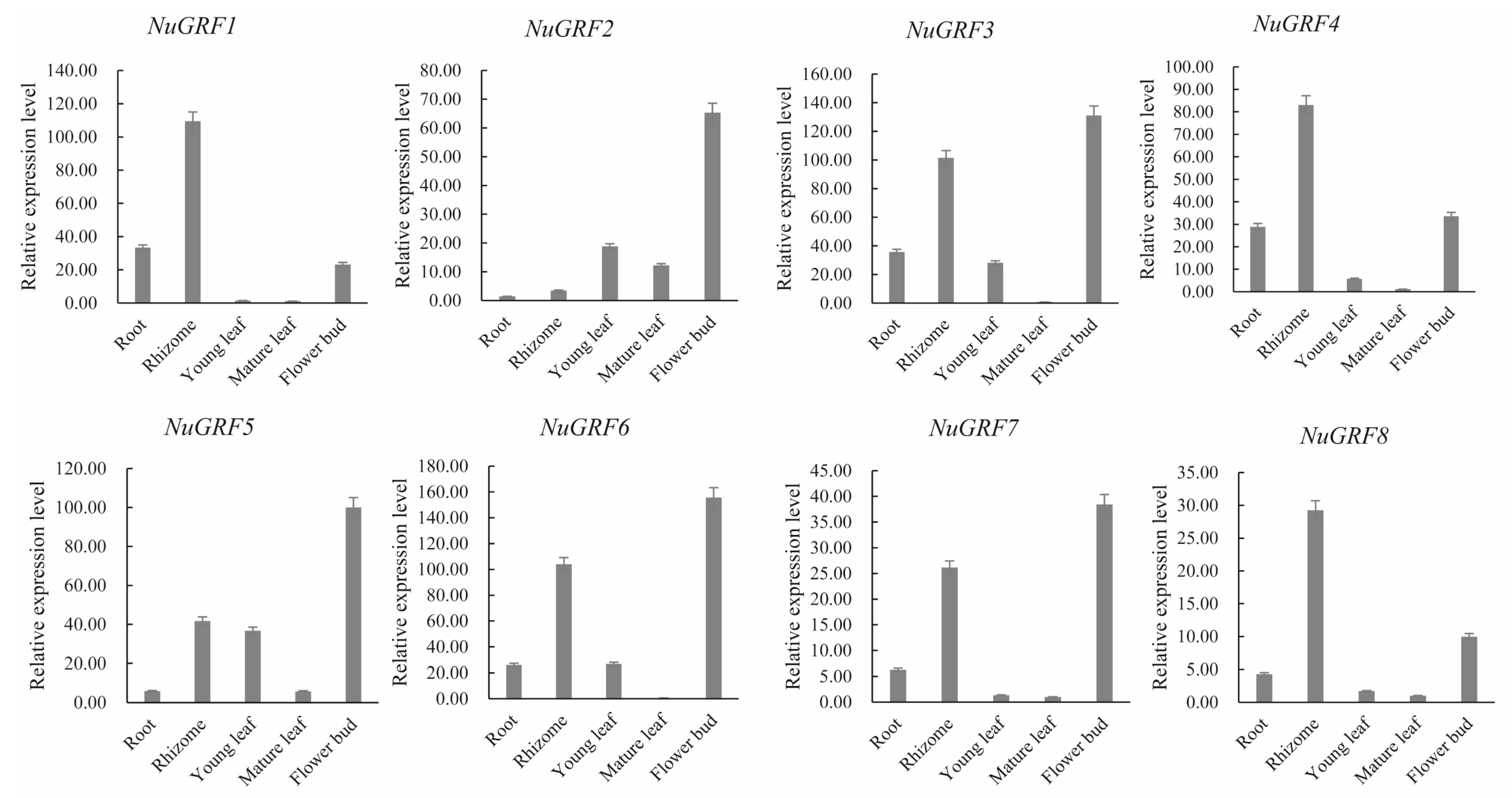
2.5. Expression Pattern of GRF Genes in N. nucifera
2.6. Homology Modeling
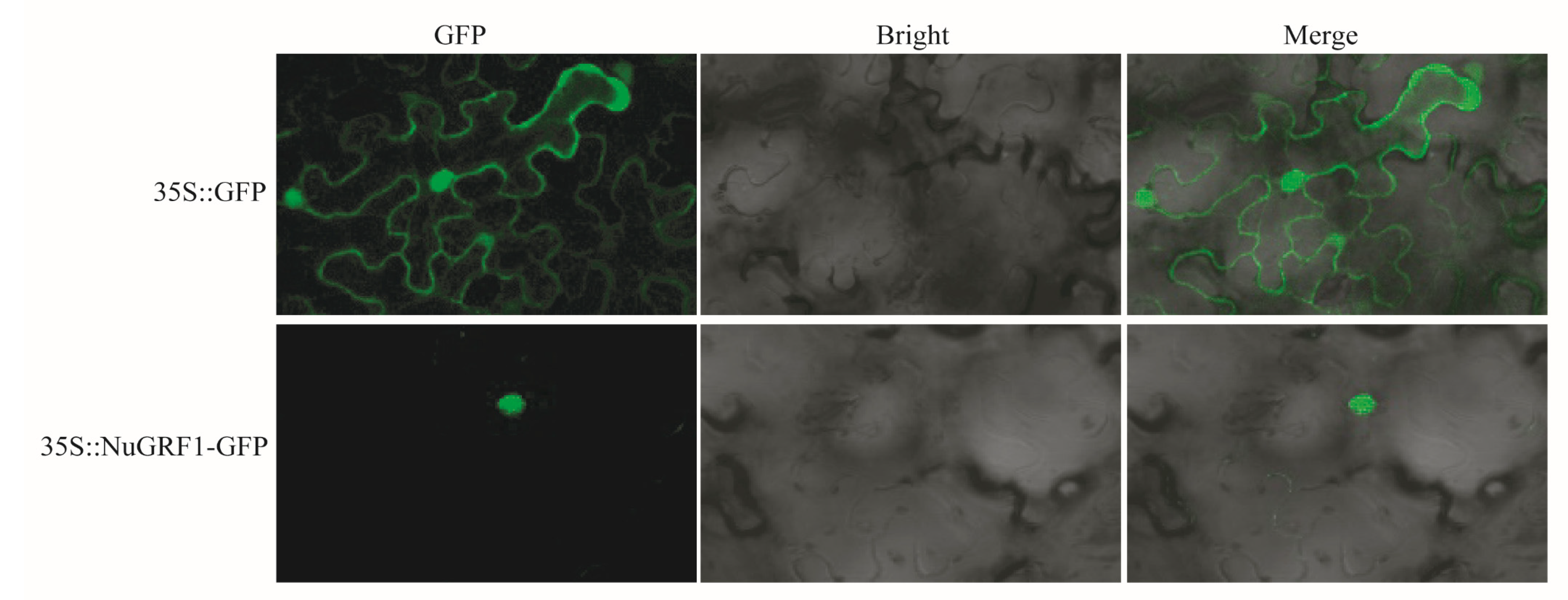
2.7. Subcellular Localization Analysis of NuGRF1
3. Discussion
4. Materials and Methods
4.1. Identification of GRF Gene Family
4.2. Phylogenetic Relationship Analysis
4.3. Gene Structure, Conserved Motifs, and Cis-Acting Element Analysis
4.4. Chromosome Location and Collinearity Analysis
4.5. Expression Pattern Analysis
4.6. Protein Structure Prediction
4.7. Subcellular Localization Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Omidbakhshfard, M.A.; Proost, S.; Fujikura, U.; Mueller-Roeber, B. Growth-regulating factors (GRFs): A small transcription factor family with important functions in plant biology. Mol. Plant 2015, 8, 998–1010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, G.; He, H.; Li, Y.; Wang, F.; Yu, D. Molecular mechanism of microRNA396 mediating pistil development in Arabidopsis. Plant Physiol. 2014, 164, 249–258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.H.; Choi, D.; Kende, H. The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 2003, 36, 94–104. [Google Scholar] [CrossRef] [PubMed]
- Filiz, E.; Koç, İ.; Tombuloğlu, H. Genome-wide identification and analysis of growth regulating factor genes in Brachypodium distachyon: In silico approaches. Turk. J. Biol. 2014, 38, 296–306. [Google Scholar] [CrossRef]
- Kim, J.H.; Kende, H. A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 13374–13379. [Google Scholar] [CrossRef] [Green Version]
- Büyük, I.; Aras, S. Genome-wide in silico identification, characterization and transcriptional analysis of the family of growth-regulating factors in common bean (Phaseolus vulgaris L.) subjected to polyethylene glycol-induced drought Stress. Arch. Biol. Sci. 2017, 69, 5–14. [Google Scholar] [CrossRef]
- Van der Knaap, E.; Kim, J.H.; Kende, H. A novel gibberellin-in-duced gene from rice and its potential regulatory role in stem growth. Plant Physiol. 2000, 122, 695–704. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Qiu, N.W.; Ding, Q.; Li, J.J.; Zhang, Y.H.; Li, H.Y.; Gao, J.W. Genome-wide identification and analysis of the growth-regulating factor family in Chinese cabbage (Brassica rapa L. ssp. pekinensis). BMC Genom. 2014, 15, 807–818. [Google Scholar] [CrossRef] [Green Version]
- Choi, D.; Kim, J.H.; Kende, H. Whole genome analysis of the OsGRF gene family encoding plant-specific putative transcription activators in rice (Oryza sativa L.). Plant Cell Physiol. 2004, 45, 897–904. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.F.; Li, B.; Jia, G.Q.; Zhang, T.F.; Dai, J.R.; Li, J.S.; Wang, S.C. Isolation and characterization of genes encoding GRF transcription factors and GIF transcriptional coactivators in maize (Zea mays L.). Plant Sci. 2008, 175, 809–817. [Google Scholar] [CrossRef]
- Liu, X.; Guo, L.X.; Jin, L.F.; Liu, Y.Z.; Liu, T.; Fan, Y.H.; Peng, S.A. Identification and transcript profiles of citrus growth-regulating factor genes involved in the regulation of leaf and fruit development. Mol. Biol. Rep. 2016, 43, 1059–1067. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.Q.; Jian, H.J.; Yang, B.; Lu, K.; Zhang, A.X.; Liu, P.; Li, J.N. Genome-wide analysis and expression profiling of the GRF gene family in oil seed rape (Brassica napus L.). Gene 2017, 620, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Ge, L.; Li, G.; Jiang, L.; Yang, Y. Characterization and expression analysis of growth regulating factor (GRF) family genes in cucumber. Arch. Biol. Sci. 2018, 70, 629–637. [Google Scholar] [CrossRef] [Green Version]
- Cao, D.; Wang, J.; Ju, Z.; Liu, Q.; Li, S.; Tian, H.; Fu, D.; Zhu, H.; Luo, Y.; Zhu, B. Regulations on growth and development in tomato cotyledon, flower and fruit via destruction of miR396 with short tandem target mimic. Plant Sci. 2016, 247, 1–12. [Google Scholar] [CrossRef]
- Zhang, J.; Li, Z.; Jin, J.; Xie, X.; Zhang, H.; Chen, Q.; Luo, Z.; Yang, J. Genome-wide identification and analysis of the growth-regulating factor family in tobacco (Nicotiana tabacum). Gene 2018, 639, 117–127. [Google Scholar] [CrossRef]
- Horiguchi, G.; Kim, G.T.; Tsukaya, H. The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J. 2005, 43, 68–78. [Google Scholar] [CrossRef]
- Lee, S.J.; Lee, B.H.; Jung, J.H.; Park, S.K.; Song, J.T.; Kim, J.H. GROWTH-REGULATING F ACTOR And GRF-IN-TERACTING FACTOR specify meristematic cells of gynoecia and anthers. Plant Physiol. 2018, 176, 717–729. [Google Scholar] [CrossRef] [Green Version]
- Li, S.C.; Gao, F.Y.; Xie, K.L.; Zeng, X.H.; Cao, Y.; Zeng, J.; He, Z.S.; Ren, Y.; Li, W.B.; Deng, Q.M.; et al. The OsmiR396c-OsGRF4-OsGIF1 regulatory module determines grain size and yield in rice. Plant Biotechnol. J. 2016, 14, 2134–2146. [Google Scholar] [CrossRef]
- Che, R.H.; Tong, H.N.; Shi, B.H.; Liu, Y.Q.; Fang, S.R.; Liu, D.P.; Xiao, Y.H.; Hu, B.; Liu, L.C.; Wang, H.R.; et al. Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat. Plants 2015, 2, 15195. [Google Scholar] [CrossRef]
- Liu, J.; Hua, W.; Yang, H.L.; Zhan, G.M.; Li, R.J.; Deng, L.B.; Wang, X.F.; Liu, G.H.; Wang, H.Z. The BnGRF2 gene (GRF2-like gene from Brassica napus) enhances seed oil production through regulating cell number and plant photosynthesis. J. Exp. Bot. 2012, 63, 3727–3740. [Google Scholar] [CrossRef] [Green Version]
- Omidbakhshfard, M.A.; Fujikura, U.; Olas, J.J.; Xue, G.P.; Balazadeh, S.; Mueller-Roeber, B. Growth-Regulating Factor 9 negatively regulates Arabidopsis leaf growth by controlling ORG3 and restricting cell proliferation in leaf primordia. PLoS Genet. 2018, 14, e1007484. [Google Scholar] [CrossRef] [PubMed]
- Zheng, P.; Sun, H.; Liu, J.; Lin, J.S.; Zhang, X.T.; Qin, Y.; Zhang, W.P.; Xu, X.M.; Deng, X.B.; Yang, D.; et al. Comparative analyses of American and Asian lotus genomes reveal insights into petal color, carpel thermogenesis and domestication. Plant J. 2022, 110, 1498–1515. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, P.K.; Mukherjee, D.; Maji, A.K.; Rai, S.; Heinrich, M. The sacred lotus (Nelumbo nucifera)—phytochemical and therapeutic profile. J. Pharm. Pharmacol. 2009, 61, 407–422. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Cao, D.; Damaris, R.N.; Yang, P. Genome-wide identification of MADS-box gene family in sacred lotus (Nelumbo nucifera) identifies a SEPALLATA homolog gene involved in floral development. BMC Plant Biol. 2020, 20, 497. [Google Scholar] [CrossRef]
- Shi, T.; Chen, J. A reappraisal of the phylogenetic placement of the Aquilegia whole-genome duplication. Genome Biol. 2020, 21, 295. [Google Scholar] [CrossRef]
- Shi, T.; Rahmani, R.S.; Gugger, P.F.; Wang, M.; Li, H.; Zhang, Y.; Li, Z.Z.; Wang, Q.F.; Peer, Y.V.D.; Marchal, K.; et al. Distinct expression and methylation patterns for genes with different fates following a single whole-genome duplication in flowering plants. Mol. Biol. Evol. 2020, 37, 2394–2413. [Google Scholar] [CrossRef]
- Li, J.; Xiong, Y.C.; Li, Y.; Ye, S.Q.; Yin, Q.; Gao, S.Q.; Yang, D.; Yang, M.; Palva, E.T.; Deng, X.B. Comprehensive analysis and functional studies of WRKY transcription factors in Nelumbo nucifera. Int. J. Mol. Sci. 2019, 20, 5006. [Google Scholar] [CrossRef] [Green Version]
- Deng, J.; Li, M.; Huang, L.Y.; Yang, M.; Yang, P.F. Genome-wide analysis of the R2R3 MYB subfamily genes in lotus (Nelumbo nucifera). Plant Mol. Biol. Rep. 2016, 34, 1016–1026. [Google Scholar] [CrossRef]
- Wang, Y.; Shi, S.L.; Zhou, Y.; Zhou, Y.; Yang, J.; Tang, X.Q. Genome-wide identification and characterization of gras transcription factors in sacred lotus (Nelumbo nucifera). PeerJ 2016, 4, e2388. [Google Scholar] [CrossRef] [Green Version]
- Pertl, H.; Himly, M.; Gehwolf, R.; Kriechbaumer, R.; Strasser, D.; Michalke, W.; Richter, K.; Ferreira, F.; Obermeyer, G. Molecular and physiological characterisation of a 14-3-3 protein from lily pollen grains regulating the activity of the plasma membrane H+ ATPase during pollen grain germination and tube growth. Planta 2001, 213, 132–141. [Google Scholar] [CrossRef]
- Chang, I.F.; Curran, A.; Woolsey, R.; Quilici, D.; Cushman, J.C.; Mittler, R.; Harmon, A.; Harper, J.F. Proteomic profiling of tandem affinity purified 14-3-3 protein complexes in Arabidopsis thaliana. Proteomics 2009, 9, 2967–2985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, S.; Yam-aguchi-Shinozaki, K.; Shinozaki, K. Two transcription factors, DREB1and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 1998, 10, 1391–1406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakuma, Y.; Liu, Q.; Dubouzet, J.G.; Abe, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 2002, 290, 998–1009. [Google Scholar] [CrossRef] [PubMed]
- Baucher, M.; Moussawi, J.; Vandeputte, O.M.; Monteyne, D.; Mol, A.; Pérez-Morga, D.; Jaziri, M.E. A role for the miR396/GRF network in specification of organ type during flower development, as supported by ectopic expression of Populus trichocarpa miR396c in transgenic tobacco. Plant Biol. 2013, 15, 892–898. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yang, X.Y.; Zhang, Y.; Gao, Z.Y.; Liang, Y.T.; Chen, J.M.; Shi, T. Nelumbo genome database, an integrative resource for gene expression and variants of Nelumbo nucifera. Sci. Data 2021, 8, 38. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. Mega7: Molecular Evolutionary Genetics AnalysisVersion 7.0 ForBigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Stamatakis, A.; Hoover, P.; Rougemont, J. A rapid bootstrap algorithm for the RAxML web–servers. Syst. Biol. 2008, 57, 758–771. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Deléage, G. ALIGNSEC: Viewing protein secondary structure predictions within large multiple sequence alignments. Bioinformatics 2017, 33, 3991–3992. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed] [Green Version]

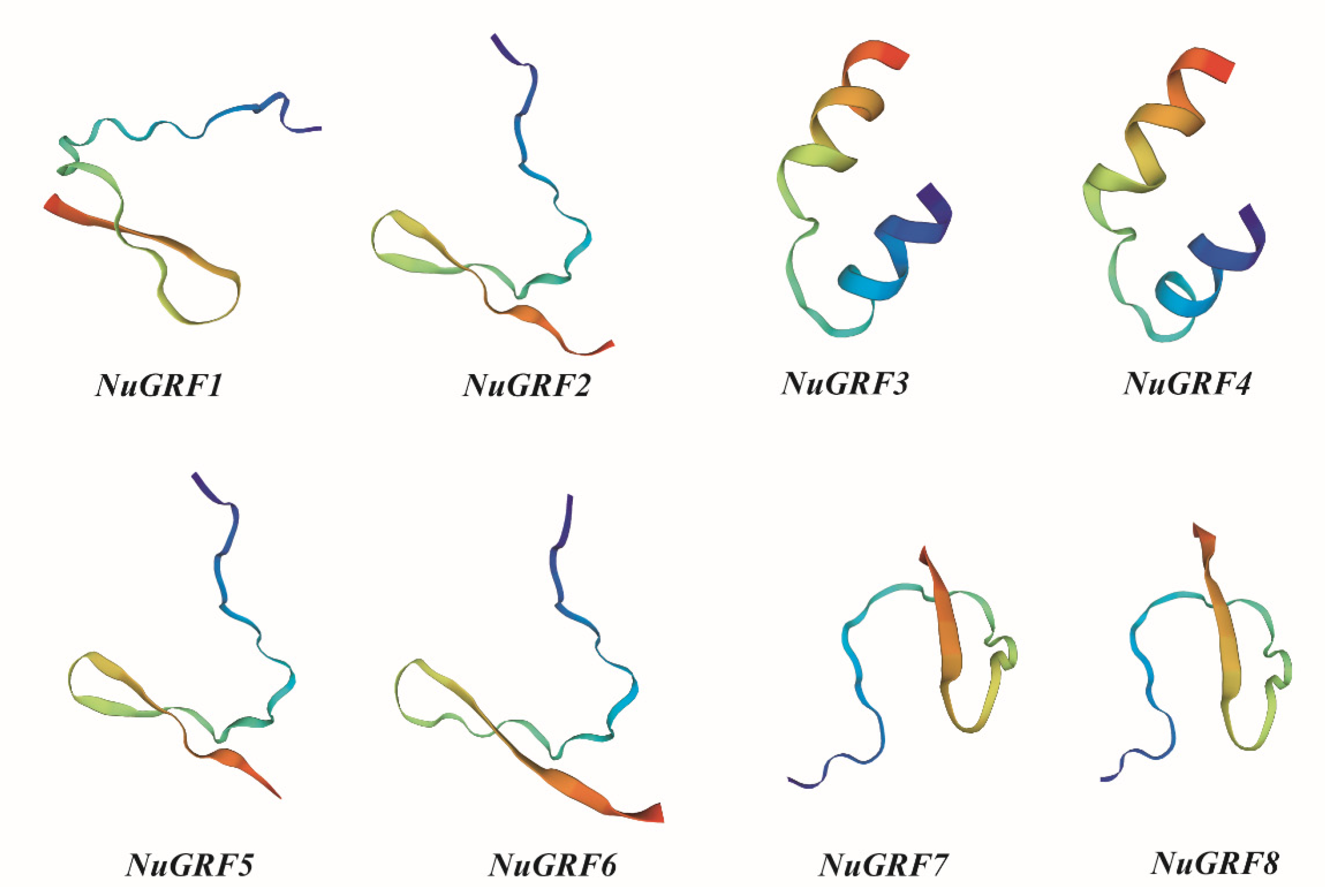
| Gene | Number of Amino Acids | Molecular Weight (Average) | PI | Instability Index | Aliphatic Index | Grand Average of Hydropathicity (GRAVY) | Subcellular Localization |
|---|---|---|---|---|---|---|---|
| NuGRF1 | 604 | 65,565 | 8.14 | 58.7 | 65.71 | −0.579 | Nuclear |
| NuGRF2 | 372 | 42,187.6 | 8.61 | 64.37 | 50.4 | −0.881 | Nuclear |
| NuGRF3 | 390 | 42,189.9 | 8.7 | 55.67 | 62.59 | −0.602 | Nuclear |
| NuGRF4 | 384 | 41,746.4 | 8.92 | 55.27 | 63.52 | −0.605 | Nuclear |
| NuGRF5 | 372 | 42,107.57 | 8.95 | 57.59 | 52.98 | −0.902 | Nuclear |
| NuGRF6 | 349 | 38,940.19 | 8.47 | 59.49 | 47.28 | −0.762 | Nuclear |
| NuGRF7 | 581 | 61,707.56 | 6.36 | 57 | 53.96 | −0.492 | Nuclear |
| NuGRF8 | 535 | 535 | 8.39 | 65.39 | 65.39 | −0.455 | Nuclear |
| Gene | Alpha Helix (Hh) | 310 Helix (Gg) | Pi Helix (Ii) | Beta bridge (Bb) | Extended Strand (Ee) | Beta Turn (Tt) | Bend Region (Ss) | Random Coil (Cc) | AMBIGUOUS States (?) | Other States |
|---|---|---|---|---|---|---|---|---|---|---|
| NuGRF1 | 14.74% | 0.00% | 0.00% | 0.00% | 9.11% | 0.99% | 0.00% | 75.17% | 0.00% | 0.00% |
| NuGRF2 | 15.59% | 0.00% | 0.00% | 0.00% | 10.75% | 2.96% | 0.00% | 70.70% | 0.00% | 0.00% |
| NuGRF3 | 18.46% | 0.00% | 0.00% | 0.00% | 11.03% | 2.05% | 0.00% | 68.46% | 0.00% | 0.00% |
| NuGRF4 | 17.19% | 0.00% | 0.00% | 0.00% | 10.42% | 2.08% | 0.00% | 70.31% | 0.00% | 0.00% |
| NuGRF5 | 15.59% | 0.00% | 0.00% | 0.00% | 10.75% | 2.96% | 0.00% | 70.70% | 0.00% | 0.00% |
| NuGRF6 | 17.19% | 0.00% | 0.00% | 0.00% | 5.44% | 3.15% | 0.00% | 74.21% | 0.00% | 0.00% |
| NuGRF7 | 17.38% | 0.00% | 0.00% | 0.00% | 11.19% | 2.93% | 0.00% | 68.50% | 0.00% | 0.00% |
| NuGRF8 | 17.57% | 0.00% | 0.00% | 0.00% | 11.96% | 3.18% | 0.00% | 67.29% | 0.00% | 0.00% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, G.-Z.; Huang, J.; Zhou, X.-Q.; Hao, Y.; Chen, J.-L.; Zhou, Y.-Z.; Ahmad, S.; Lan, S.; Liu, Z.-J.; Peng, D.-H. Comprehensive Analysis for GRF Transcription Factors in Sacred Lotus (Nelumbo nucifera). Int. J. Mol. Sci. 2022, 23, 6673. https://doi.org/10.3390/ijms23126673
Chen G-Z, Huang J, Zhou X-Q, Hao Y, Chen J-L, Zhou Y-Z, Ahmad S, Lan S, Liu Z-J, Peng D-H. Comprehensive Analysis for GRF Transcription Factors in Sacred Lotus (Nelumbo nucifera). International Journal of Molecular Sciences. 2022; 23(12):6673. https://doi.org/10.3390/ijms23126673
Chicago/Turabian StyleChen, Gui-Zhen, Jie Huang, Xiao-Qin Zhou, Yang Hao, Jin-Liao Chen, Yu-Zhen Zhou, Sagheer Ahmad, Siren Lan, Zhong-Jian Liu, and Dong-Hui Peng. 2022. "Comprehensive Analysis for GRF Transcription Factors in Sacred Lotus (Nelumbo nucifera)" International Journal of Molecular Sciences 23, no. 12: 6673. https://doi.org/10.3390/ijms23126673
APA StyleChen, G.-Z., Huang, J., Zhou, X.-Q., Hao, Y., Chen, J.-L., Zhou, Y.-Z., Ahmad, S., Lan, S., Liu, Z.-J., & Peng, D.-H. (2022). Comprehensive Analysis for GRF Transcription Factors in Sacred Lotus (Nelumbo nucifera). International Journal of Molecular Sciences, 23(12), 6673. https://doi.org/10.3390/ijms23126673








