The Xanthophyll Carotenoid Lutein Reduces the Invasive Potential of Pseudomonas aeruginosa and Increases Its Susceptibility to Tobramycin
Abstract
:1. Introduction
2. Results
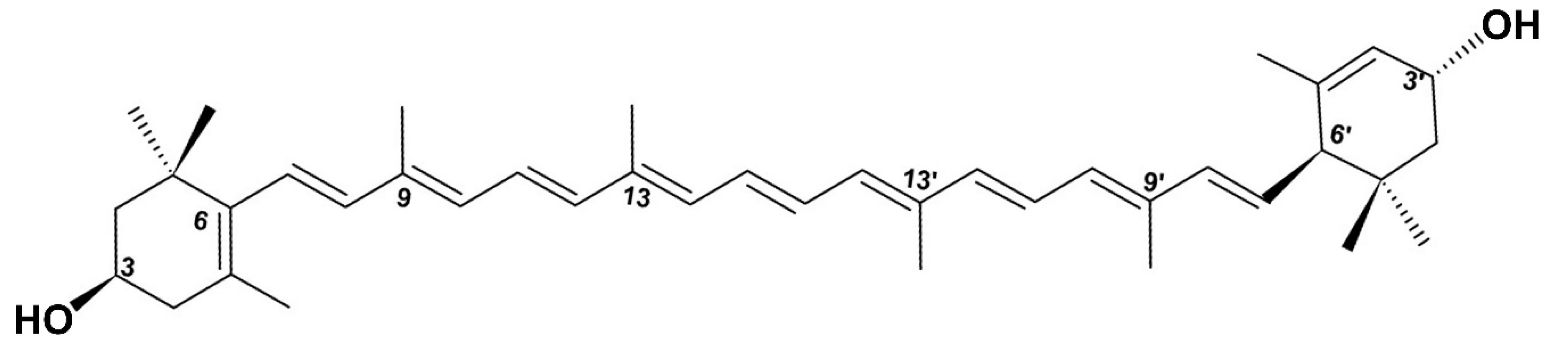
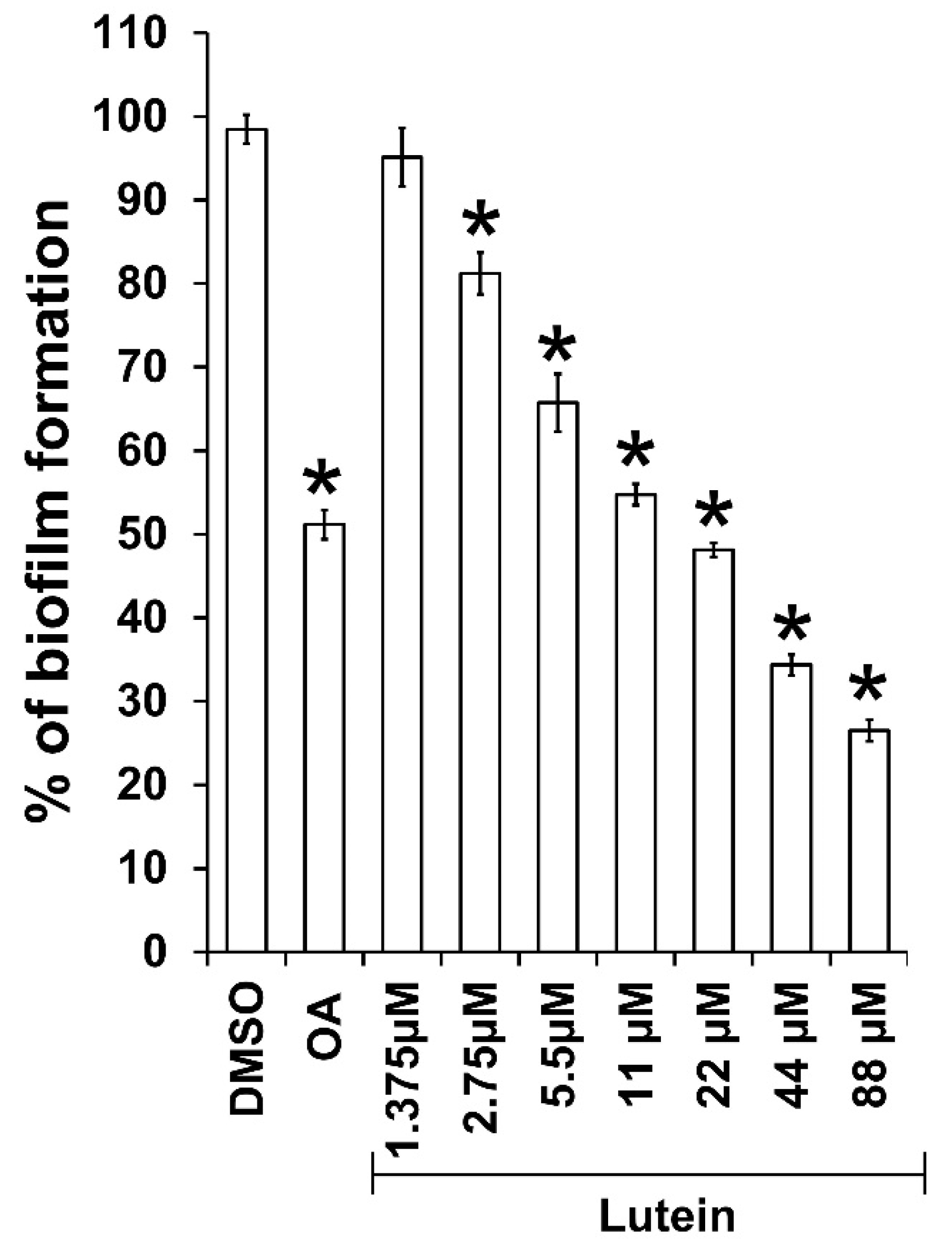
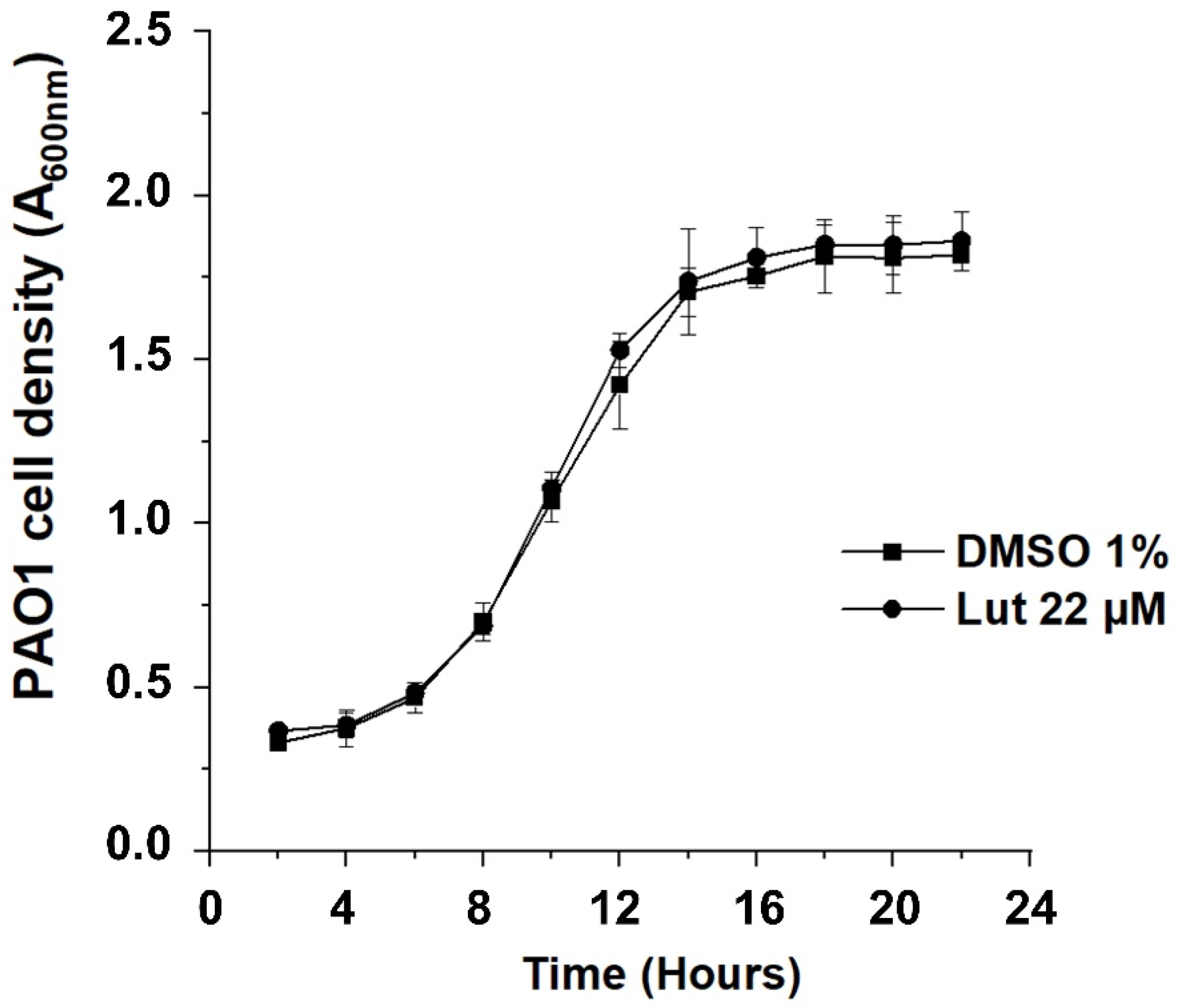
2.1. All-Trans Lutein Reduces PAO1 Biofilm Formation in a Dose-Dependent Manner
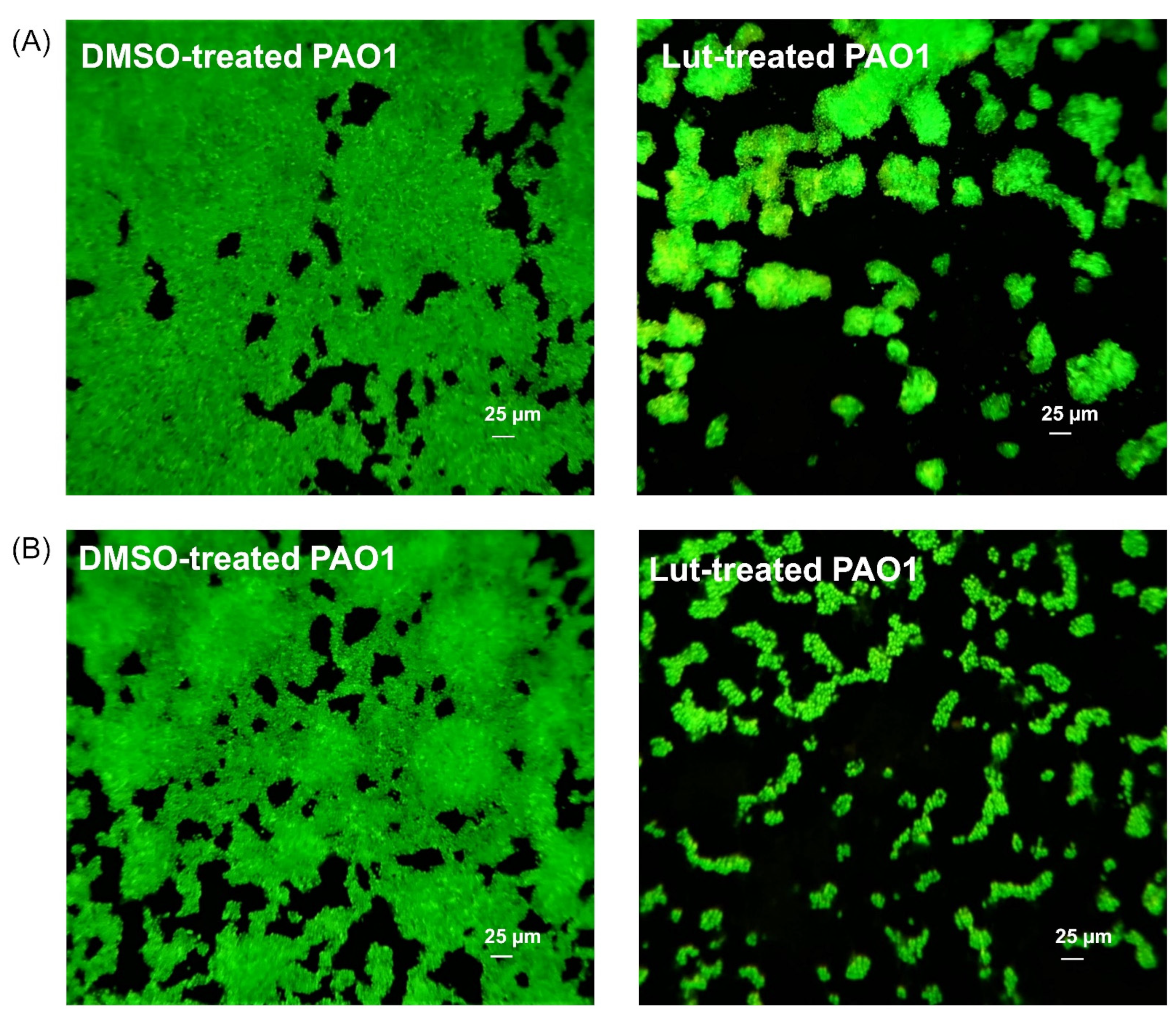
2.2. Lut Affects the Biofilm Phenotype of PAO1
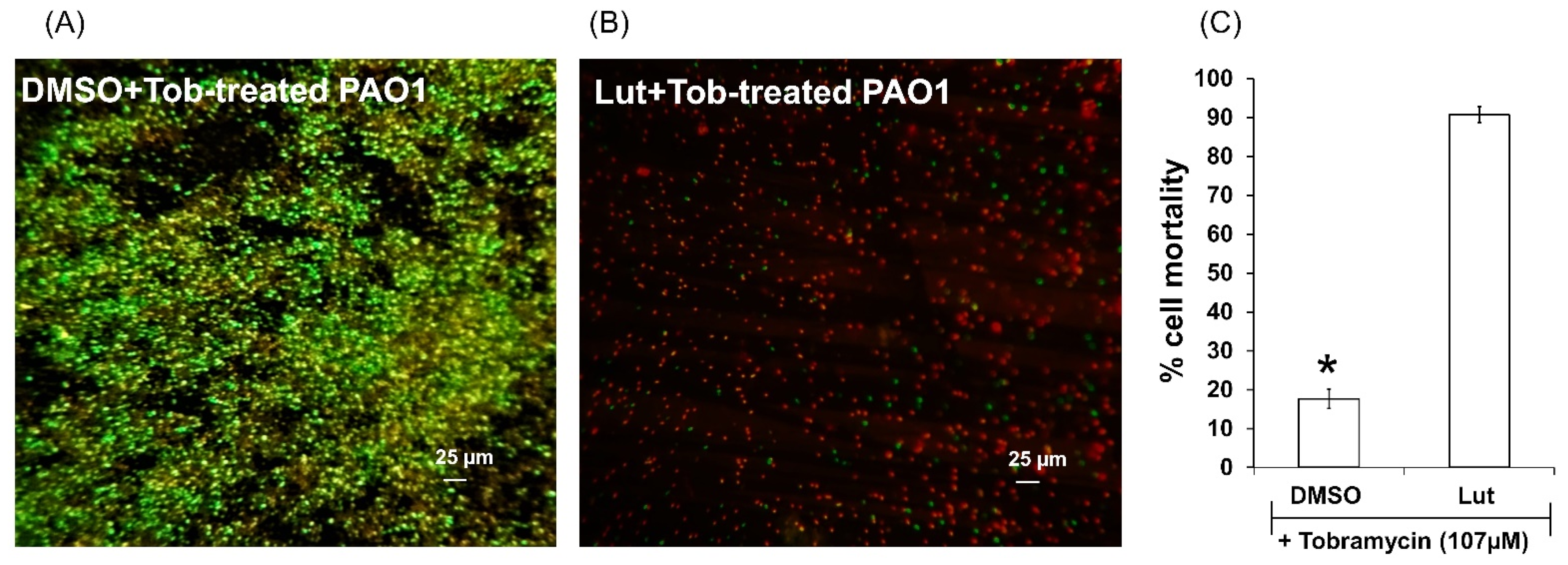
2.3. Lut Exhibits Antibiotic-Synergizing Activities in One-Day-Old PAO1 Biofilm
2.4. Lut Affects PAO1 Swarming and Twitching Motilities
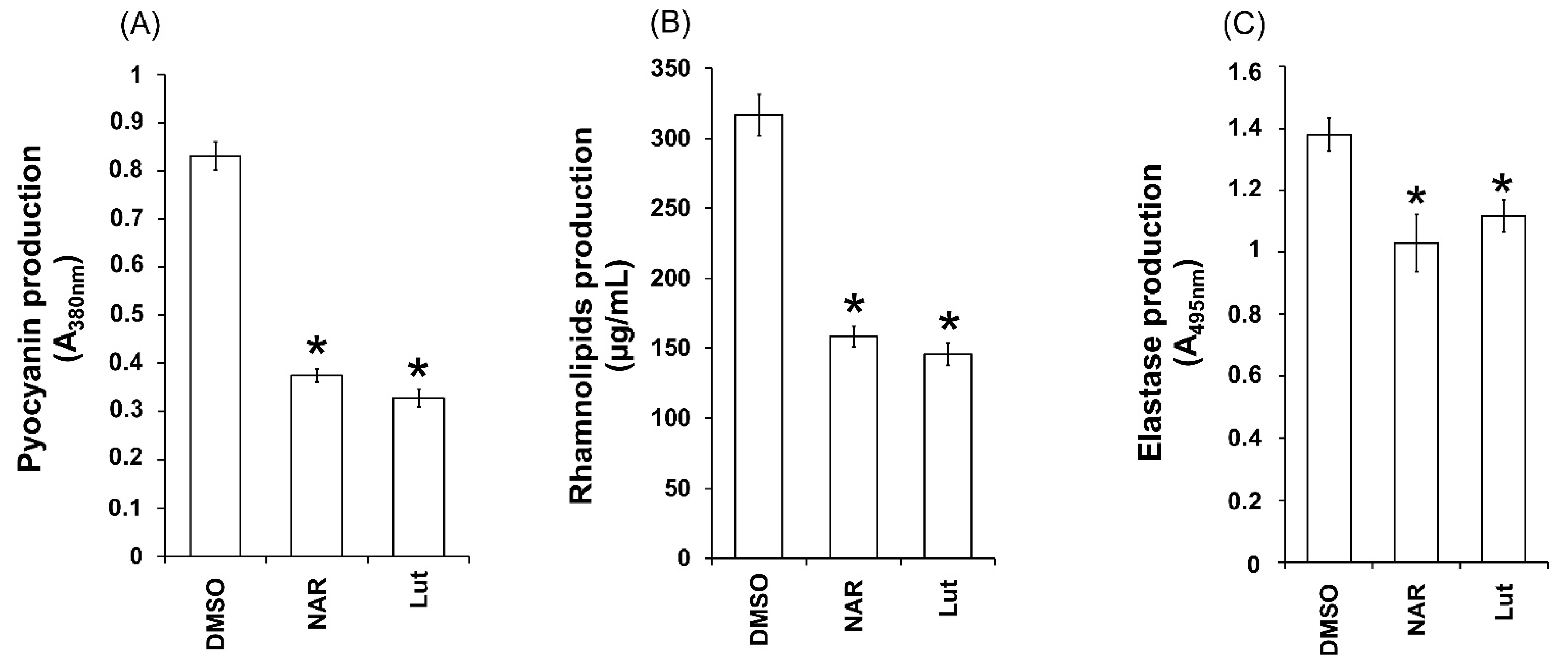
2.5. Lut Reduces Pyocyanin, Elastase and Rhamnolipids Production in PAO1
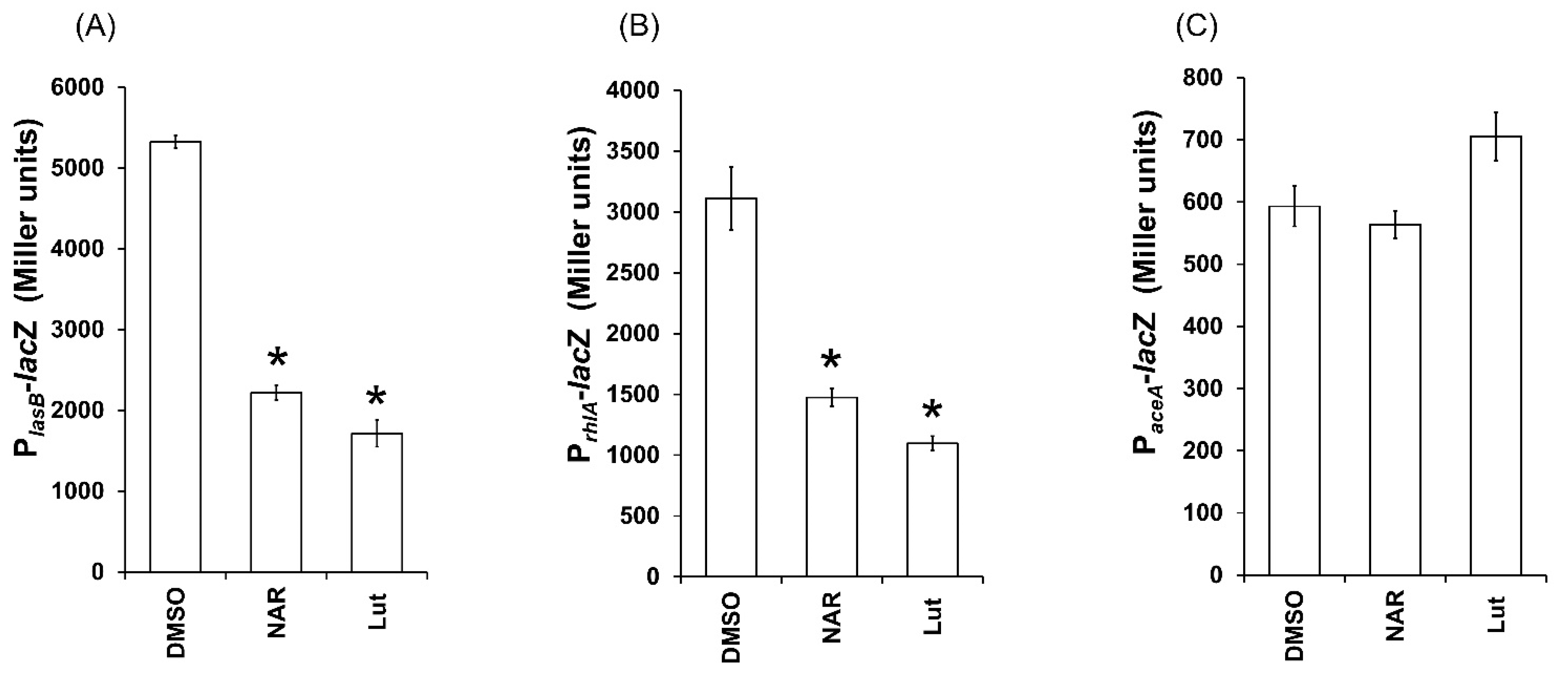
2.6. Lut Disrupts the Expression of QS-Dependent lasB and rhlA Genes in PAO1
2.7. Lut Affects the Expression of lasR/I and rhlR/I Genes in PAO1
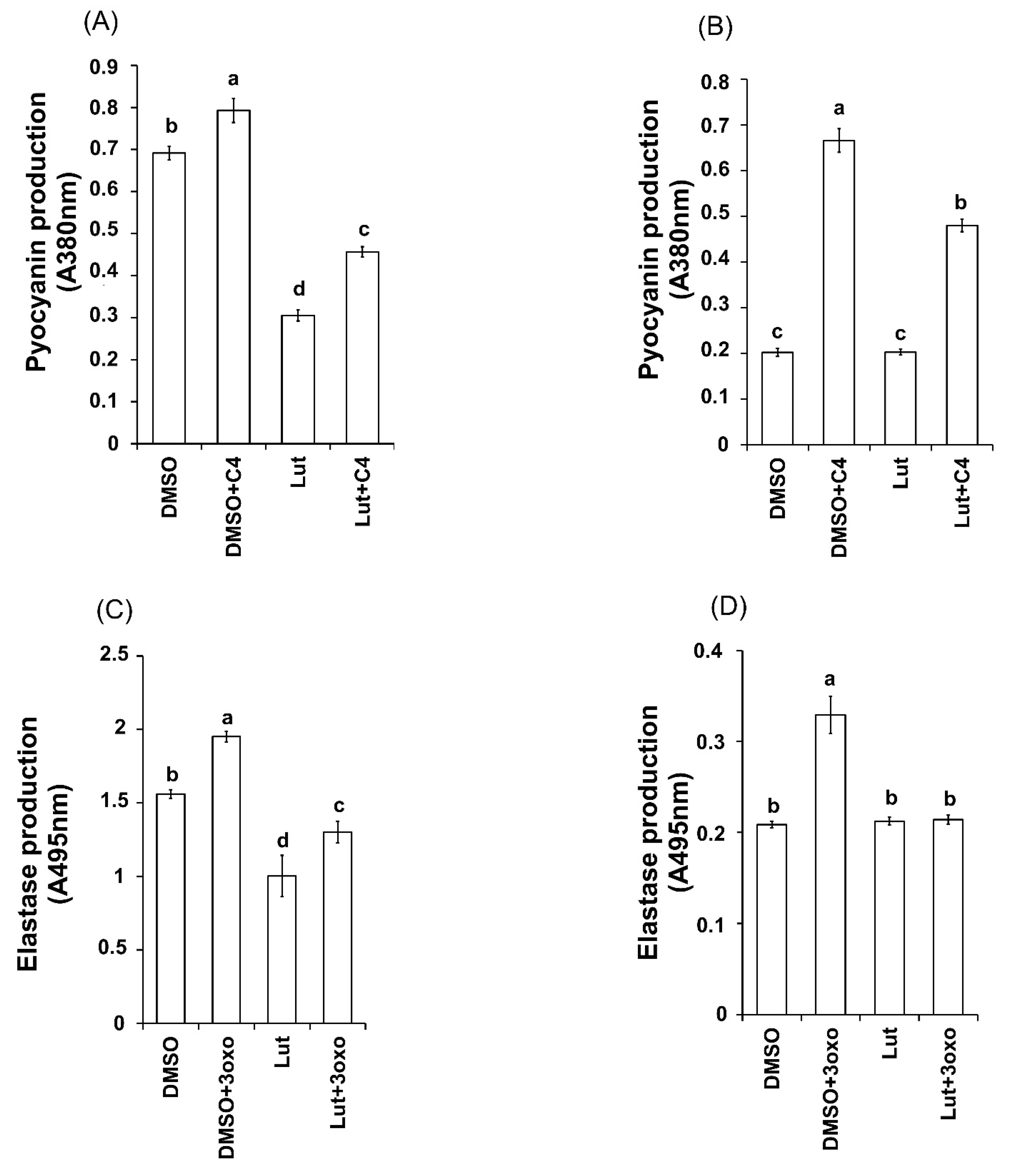
2.8. The Exogenous Addition of AHLs (3-oxo-C-12HSL and C4-HSL) Affects the Production of Pyocyanin and Elastase on Both Lut-Treated PAO1 Wild-Type and AHL-Mutant Strains
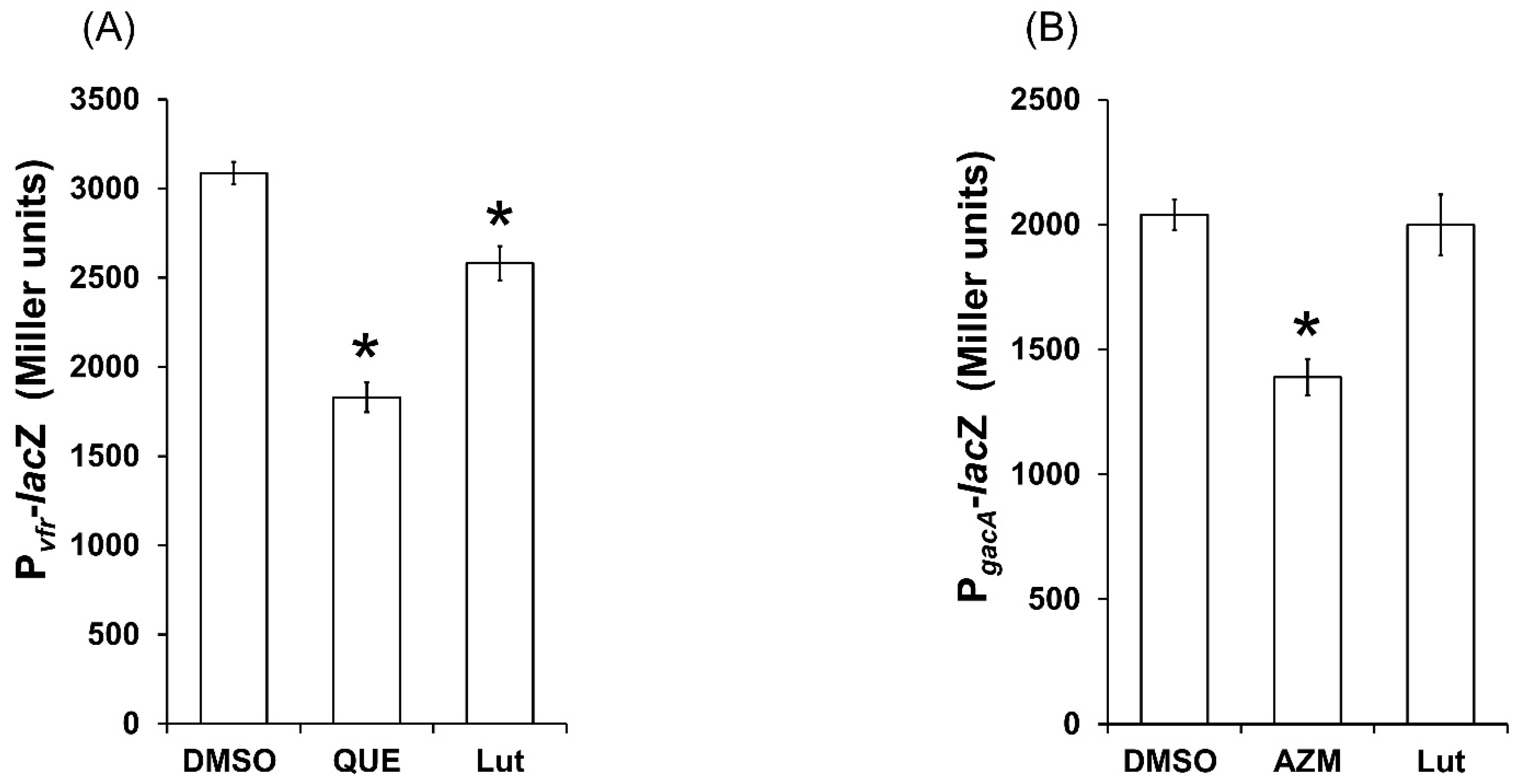
2.9. Lut Affects the Expression of Global Regulator vfr but Not gacA PAO1 Genes
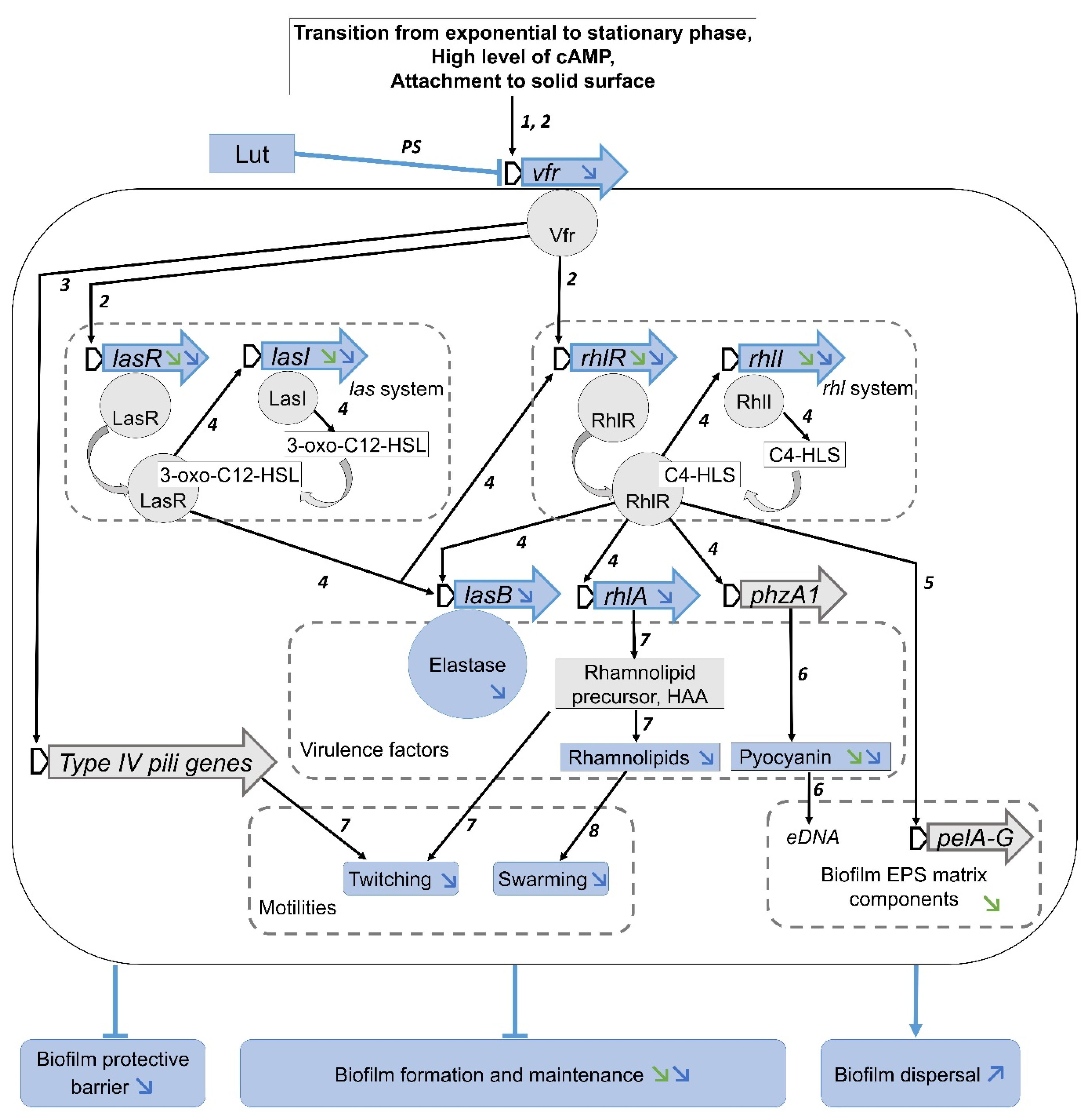
3. Discussion
4. Material and Methods
4.1. Bacterial Strains and Growth Conditions
4.2. Chemicals and Solvents
4.3. Assessment of Bacterial Growth Kinetics
4.4. Biofilm Formation and Quantification
4.5. Biofilm Phenotype and Synergistic Activity with Tobramycin in Fluorescence Microscopy
4.6. Motility Assay
4.7. Quantitative Analysis of Pyocyanin, Elastase and Rhamnolipids Production
4.8. Gene Expression and Beta-Galactosidase Measurements
4.9. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mulani, M.S.; Kamble, E.E.; Kumkar, S.N.; Tawre, M.S.; Pardesi, K.R. Emerging strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: A review. Front. Microbiol. 2019, 10, 539. [Google Scholar] [CrossRef] [PubMed]
- Perez-Rodriguez, F.; Mercanoglu, T.B. A state-of-art review on multi-drug resistant pathogens in foods of animal origin: Risk factors and mitigation strategies. Front. Microbiol. 2019, 10, 2091. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rasamiravaka, T. Antimicrobial resistance in Madagascar: A review of the current situation and challenges. Afr. J. Clin. Exp. Microbiol. 2020, 21, 165–174. [Google Scholar] [CrossRef]
- Rasamiravaka, T.; El Jaziri, M. Quorum-Sensing Mechanisms and Bacterial Response to Antibiotics in P. aeruginosa. Curr. Microbiol. 2016, 73, 747–753. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.; Thuy, D.; Pham, N.; Kim, Y. Alternative strategies for the application of aminoglycoside antibiotics against the biofilm-forming human pathogenic bacteria. Springer Nat. 2020, 104, 1955–1976. [Google Scholar] [CrossRef]
- Jamal, M.; Ahmad, W.; Andleeb, S.; Jalil, F.; Imran, M.; Asif, M.; Houssain, T.; Ali, M.; Rafiq, M.; Atif, M. Bacterial biofilm and associated infections. J. Chin. Med. Assoc. 2018, 81, 7–11. [Google Scholar] [CrossRef]
- Zhou, L.; Zhang, Y.; Ge, Y.; Zhu, X.; Pan, J. Regulatory Mechanisms and Promising Applications of Quorum Sensing-Inhibiting Agents in Control of Bacterial Biofilm Formation. Front. Microbiol. 2020, 11, 589640. [Google Scholar] [CrossRef]
- Bricha, S.; Ounine, K.; Oulkheir, S.; Attarassi, B. Virulence factors and epidemiology related to Pseudomonas aeruginosa. Rev. Tunis. D’infectiol. 2009, 2, 7–14. [Google Scholar]
- Moradali, M.F.; Ghods, S.; Rehm, B.H.A. Pseudomonas aeruginosa Lifestyle: A Paradigm for Adaptation, Survival, and Persistence. Front. Cell. Infect. Microbiol. 2017, 7, 39. [Google Scholar] [CrossRef] [Green Version]
- Carette, J.; Nachtergael, A.; Duez, P.; El Jaziri, M.; Rasamiravaka, T. Natural Compounds Inhibiting Pseudomonas aeruginosa Biofilm Formation by Targeting Quorum Sensing Circuitry. In Bacterial Biofilm; Intechopen: London, UK, 2020. [Google Scholar] [CrossRef] [Green Version]
- Jimenez, P.N.; Koch, G.; Thompson, J.A.; Xavier, K.B.; Cool, R.H.; Quax, W.J. The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 2012, 76, 46–65. [Google Scholar] [CrossRef] [Green Version]
- Rasamiravaka, T.; Labtani, Q.; Duez, P.; El Jaziri, M. The Formation of Biofilms by Pseudomonas aeruginosa: A Review of the Natural and Synthetic Compounds Interfering with Control Mechanisms. BioMed Res. Int. 2015, 2015, 759348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, F.; Pham, D.T.N.; Oloketuyi, S.F.; Kim, Y.M. Regulation and controlling the motility properties of Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 2020, 104, 33–49. [Google Scholar] [CrossRef] [PubMed]
- Latifi, A.; Foglino, M.; Tanaka, K.; Williams, P.; Lazdunski, A.A. Hierarchical quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhIR (VsmR) to expression of the stationary-phase sigma factor RpoS. Mol. Microbiol. 1996, 21, 1137–1146. [Google Scholar] [CrossRef]
- Medina, G.; Juarez, K.; Diaz, R.; Soberon-Chavez, G. Transcriptional regulation of Pseudomonas aeruginosa rhlR, encoding a quorum-sensing regulatory protein. Microbiology 2003, 149, 3073–3081. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Zhang, L. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 2015, 6, 26–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albus, A.M.; Pesci, E.C.; Runyen-Janecky, L.J.; West, S.E.; Iglewski, B.H. Vfr controls quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 1997, 179, 3928–3935. [Google Scholar] [CrossRef] [Green Version]
- Reimmann, C.; Beyeler, M.; Latifi, A.; Winteler, H.; Foglino, M.; Lazdunski, A.; Haas, D. The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol. Microbiol. 1997, 24, 309–319. [Google Scholar] [CrossRef]
- Fuchs, E.L.; Brutinel, E.D.; Jones, A.K.; Fulcher, N.B.; Urbanowski, M.L.; Yahr, T.L.; Wolfgang, M.C. The Pseudomonas aeruginosa Vfr regulator controls global virulence factor expression through cyclic AMP-dependent and independent mechanisms. J. Bacteriol. 2010, 192, 3553–3564. [Google Scholar] [CrossRef] [Green Version]
- Croda-García, G.; Grosso-Becerra, V.; Gonzalez-Valdez, A.; Servín-González, L.; Soberon-Chavez, G. Transcriptional regulation of Pseudomonas aeruginosa rhlR: Role of the CRP orthologue Vfr (virulence factor regulator) and quorum-sensing regulators LasR and RhlR. Microbiology 2011, 157, 2545–2555. [Google Scholar] [CrossRef] [Green Version]
- Luo, J.; Kong, J.; Dong, B.; Huang, H.; Wang, K.; Wu, L.; Hou, C.; Liang, Y.; Li, B.; Chen, Y. Baicalein attenuates the quorum sensing-controlled virulence factors of Pseudomonas aeruginosa and relieves the inflammatory response in P. aeruginosa-infected macrophages by downregulating the MAPK and NFκB signal-transduction pathways. Drug Des. Devel. Ther. 2016, 10, 183–203. [Google Scholar] [CrossRef] [Green Version]
- Luo, J.; Dong, B.; Wang, K.; Cai, S.; Liu, T.; Cheng, X.; Lei, D.; Chen, Y.; Li, Y.; Kong, J.; et al. Baicalin inhibits biofilm formation, attenuates the quorum sensing-controlled virulence and enhances Pseudomonas aeruginosa clearance in a mouse peritoneal implant infection model. PLoS ONE 2017, 12, e0176883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imperi, F.; Leoni, L.; Visca, P. Antivirulence activity of azithromycin in Pseudomonas aeruginosa. Front. Microbiol. 2014, 5, 178. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, J.; Feng, W.; Lai, X.; Chen, Y.; Zhang, X.; Rong, L.; Sun, F.; Chen, Y. Quercetin inhibits Pseudomonas aeruginosa biofilm formation via the vfr-mediated lasIR system. Microb. Pathog. 2020, 149, 10429. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.M.; Bu, Y.; Suga, H. Library screening for synthetic agonists and antagonists of a Pseudomonas aeruginosa autoinducer. Chem. Biol. 2003, 10, 563–571. [Google Scholar] [CrossRef] [Green Version]
- Mahavy, C.M.; Duez, P.; El Jaziri, M.; Rasamiravaka, T. African plant-based natural products with antivirulence activities to the rescue of antibiotics. Antibiotics 2020, 9, 830. [Google Scholar] [CrossRef]
- Vandeputte, O.M.; Kiendrebeogo, M.; Rasamiravaka, T.; Stévigny, C.; Duez, P.; Rajaonson, S.; Diallo, B.; Mol, A.; Baucher, M.; El Jaziri, M. The flavanone naringenin reduces the production of quorum sensing-controlled virulence factors in Pseudomonas aeruginosa PAO1. Microbiology 2011, 157, 2120–2132. [Google Scholar] [CrossRef] [Green Version]
- Rasamiravaka, T.; Vandeputte, O.M.; Pottier, L.; Huet, J.; Rabemanantsoa, C.; Kiendrebeogo, M.; Andriantsimahavandy, A.; Rasamindrakotroka, A.; Stévigny, C.; Duez, P.; et al. Pseudomonas aeruginosa biofilm formation and persistence, along with the production of quorum sensing-dependent virulence factors, are disrupted by a triterpenoid coumarate ester isolated from Dalbergia trichocarpa, a tropical legume. PLoS ONE 2015, 10, e0132791. [Google Scholar] [CrossRef]
- Rasamiravaka, T.; Ngezahayo, J.; Pottier, L.; Oliveira Ribeiro, S.; Souard, F.; Hari, L.; Stévigny, C.; El Jaziri, M.; Duez, P. Terpenoids from Platostoma rotundifolium (Briq.) A. J. Paton Alter the Expression of Quorum Sensing-Related Virulence Factors and the Formation of Biofilm in Pseudomonas aeruginosa PAO1. Int. J. Mol. Sci. 2017, 18, 1270. [Google Scholar] [CrossRef] [Green Version]
- Sampathkumar, S.J.; Srivastava, P.; Ramachandran, S.; Sivashanmugam, K.; Gothandam, K.M. Lutein: A potential antibiofilm and antiquorum sensing molecule from green microalga Chlorella pyrenoidosa. Microb. Pathog. 2019, 135, 103658. [Google Scholar] [CrossRef]
- Gökalsın, B.; Aksoydan, B.; Erman, B.; Sesal, N.C. Reducing Virulence and Biofilm of Pseudomonas aeruginosa by Potential Quorum Sensing Inhibitor Carotenoid: Zeaxanthin. Microb. Ecol. 2017, 74, 466–473. [Google Scholar] [CrossRef]
- Mogayzel, P.J., Jr.; Naureckas, E.T.; Robinson, K.A.; Mueller, G.; Hadjiliadis, D.; Hoag, J.B.; Lubsch, L.; Hazle, L.; Sabadosa, K.; Marshall, B.; et al. Pulmonary Clinical Practice Guidelines Committee. Cystic for maintenance of lung health. Am. J. Respir. Crit. Care Med. 2013, 187, 680–689. [Google Scholar] [CrossRef] [PubMed]
- Drenkard, E. Antimicrobial resistance of Pseudomonas aeruginosa biofilms. Microb. Infect. 2003, 5, 1213–1219. [Google Scholar] [CrossRef] [PubMed]
- Otton, L.M.; Campos, M.D.S.; Meneghetti, K.L.; Corção, G. Influence of twitching and swarming motilities on biofilm formation in Pseudomonas strains. Arch. Microbiol. 2017, 199, 677–682. [Google Scholar] [CrossRef] [PubMed]
- Buscemi, S.; Corleo, D.; Di Pace, F.; Petroni, M.L.; Satriano, A.; Marchesini, G. The effect of lutein on eye and extra-eye health. Nutrients 2018, 10, 1321. [Google Scholar] [CrossRef] [Green Version]
- Różanowska, M.; Czuba-Pelech, B.; Landrum, J.T.; Rozanowski, B. Comparison of Antioxidant Properties of Dehydrolutein with Lutein and Zeaxanthin, and their Effects on Retinal Pigment Epithelial Cells. Antioxidants 2021, 10, 753. [Google Scholar] [CrossRef]
- Songca, S.P.; Sebothoma, C.; Samuel, B.B.; Eloff, J.N. A biflavonoid and a carotenoid from Rhus leptodictya: Isolation, characterization and antibacterial properties. Afr. J. Biochem. Res. 2012, 6, 172–178. [Google Scholar]
- Liu, C.; Sun, D.; Zhu, J.; Liu, J.; Liu, W. The regulation of bacterial biofilm formation by cAMP-CRP: A mini-review. Front. Microbiol. 2020, 11, 802. [Google Scholar] [CrossRef]
- Coggan, K.A.; Wolfgang, M.C. Global regulatory pathways and cross-talk control Pseudomonas aeruginosa environmental lifestyle and virulence phenotype. Curr. Issues Mol. Biol. 2012, 14, 47–70. [Google Scholar]
- Kohler, T.; Curty, L.K.; Barja, F.; Van Delden, C.; Pechère, J.C. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J. Bacteriol. 2000, 182, 5990–5996. [Google Scholar] [CrossRef] [Green Version]
- Melville, S.; Craig, L. Type IV pili in Gram-positive bacteria. Microbiol. Mol. Biol. Rev. 2013, 77, 323–341. [Google Scholar] [CrossRef] [Green Version]
- Beatson, S.A.; Whitchurch, C.B.; Sargent, J.L.; Levesque, R.C.; Mattick, J.S. Differential regulation of twitching motility and elastase by Vfr in Pseudomonas aeruginosa. J. Bacteriol. 2002, 184, 3605–3613. [Google Scholar] [CrossRef] [Green Version]
- Haley, C.L.; Kruczek, C.; Qaisar, U.; Colmer-Hamood, J.A.; Hamood, A.N. Mucin inhibits Pseudomonas aeruginosa biofilm formation by significantly enhancing twitching motility. Can. J. Microbiol. 2014, 60, 155–166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakuragi, Y.; Kolter, R. Quorum-sensing regulation of the biofilm matrix genes (pel) of Pseudomonas aeruginosa. J. Bacteriol. 2007, 189, 5383–5386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, T.; Ibugo, A.I.; Klare, W.; Manefield, M. Role of pyocyanin and extracellular DNA in facilitating Pseudomonas aeruginosa biofilm formation. In Microbial Biofilms—Importance Applications; IntechOpen: London, UK, 2016; Volume 13, pp. 23–42. [Google Scholar]
- Pamp, S.J.; Tolker-Nielsen, T. Multiple roles of biosurfactants in structural biofilm development by Pseudomonas aeruginosa. J. Bacteriol. 2007, 189, 2531–2539. [Google Scholar] [CrossRef] [Green Version]
- Caiazza, N.C.; Shanks, R.M.; O’toole, G.A. Rhamnolipids modulate swarming motility patterns of Pseudomonas aeruginosa. J. Bacteriol. 2005, 187, 7351–7361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brindhadevi, K.; LewisOscar, F.; Mylonakis, E.; Shanmugam, S.; Verma, T.N.; Pugazhendhi, A. Biofilm and Quorum sensing mediated pathogenicity in Pseudomonas aeruginosa. Process Biochem. 2020, 96, 49–57. [Google Scholar] [CrossRef]
- Deziel, E.; Le’pine, F.; Milot, S.; Villemur, R. rhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3-(3-hydroxyalkanoyloxy)alkanoic acids (HAAs), the precursors of rhamnolipids. Microbiology 2003, 149, 2005–2013. [Google Scholar] [CrossRef] [Green Version]
- Kaplan, J.Á. Biofilm dispersal: Mechanisms, clinical implications, and potential therapeutic uses. J. Dent. Res. 2010, 89, 205–218. [Google Scholar] [CrossRef] [Green Version]
- Yin, W.; Wang, Y.; Liu, L.; He, J. Biofilms: The microbial “protective clothing” in extreme environments. Int. J. Mol. Sci. 2019, 20, 3423. [Google Scholar] [CrossRef] [Green Version]
- Gloyne, L.S.; Grant, G.D.; Perkins, A.V.; Powell, K.L.; McDermott, C.M.; Johnson, P.V.; Anderson, G.J.; Kiefel, M.; Anoopkumar-Dukie, S. Pyocyanin-induced toxicity in A549 respiratory cells is causally linked to oxidative stress. Toxicol. In Vitro 2011, 25, 1353–1358. [Google Scholar] [CrossRef]
- Muller, M. Premature cellular senescence induced by pyocyanin, a redox-active Pseudomonas aeruginosa toxin. Free Radic. Biol. Med. 2006, 41, 1670–1677. [Google Scholar] [CrossRef] [PubMed]
- Klare, W.; Das, T.; Ibugo, A.; Buckle, E.; Manefield, M.; Manos, J. Glutathione-disrupted biofilms of clinical Pseudomonas aeruginosa strains exhibit an enhanced antibiotic effect and a novel biofilm transcriptome. Antimicrob. Agents Chemother. 2016, 60, 4539–4551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okshevsky, M.; Meyer, R.L. The role of extracellular DNA in the establishment, maintenance and perpetuation of bacterial biofilms. Crit. Rev. Microbiol. 2015, 41, 341–352. [Google Scholar] [CrossRef]
- Dasgupta, N.; Ferrell, E.P.; Kanack, K.J.; West, S.E.; Ramphal, R. fleQ, the gene encoding the major flagellar regulator of Pseudomonas aeruginosa, is σ70 dependent and is downregulated by Vfr, a homolog of Escherichia coli cyclic AMP receptor protein. J. Bacteriol. 2002, 184, 5240–5250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, L.Z.W.; Hong, Z.; Yam, J.K.H.; Salido, M.M.S.; Woo, B.Y.; Li, S.F.Y.; Yang, L.; Givskov, M.; Chng, S.S. Auranofin inhibits virulence in Pseudomonas aeruginosa. BioRxiv 2017, 198820. [Google Scholar] [CrossRef] [Green Version]
- Calvo, M.M. Lutein: A valuable ingredient of fruit and vegetables. Crit. Rev. Food Sci. Nutr. 2005, 45, 671–696. [Google Scholar] [CrossRef]
- Vandeputte, O.M.; Kiendrebeogo, M.; Rajaonson, S.; Diallo, B.; Mol, A.; El Jaziri, M.; Baucher, M. Identification of catechin as one of the flavonoids from Combretum albiflorum bark extract that reduces the production of quorum-sensing-controlled virulence factors in Pseudomonas aeruginosa PAO1. Appl. Environ. Microbiol. 2010, 76, 243–253. [Google Scholar] [CrossRef] [Green Version]
- Rasamiravaka, T.; Jedrzejowski, A.; Kiendrebeogo, M.; Rajaonson, S.; Randriamampionona, D.; Rabemanantsoa, C.; Andriantsimahavandy, A.; Rasamindrakotroka, A.; Duez, P.; El Jaziri, M.; et al. Endemic Malagasy Dalbergia species inhibit quorum sensing in Pseudomonas aeruginosa PAO1. Microbiology 2013, 159, 924–938. [Google Scholar] [CrossRef] [Green Version]
- Jacobs, M.A.; Alwood, A.; Thaipisuttikul, I.; Spencer, D.; Haugen, E.; Ernst, S.; Will, O.; Kaul, R.; Raymond, C.; Levy, R.; et al. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2003, 100, 14339–14344. [Google Scholar] [CrossRef] [Green Version]
- Kiplimo, J.J.; Koorbanally, N.A.; Chenia, H.Y. Triterpenoids from Vernonia auriculifera Hiern exhibit antimicrobial activity. Afr. J. Pharm. Pharmacol. 2011, 5, 1150–1156. [Google Scholar]
- Rasamiravaka, T.; Quentin, L.; Mol, A.; Megalizzi, V.; Rabemanantsoa, C.; Duez, P.; Jaziri, M.E. An Active Fraction from DalbergiaTrichocarpa Baker Disrupts the Formation and Maintenance of Biofilms in Pseudomonas aeruginosa PAO1. Iarjset 2016, 3, 124–133. [Google Scholar] [CrossRef]
- Darzins, A. The pilG gene product, required for Pseudomonas aeruginosa pilus production and twitching motility, is homologous to the enteric, single-domain response regulator CheY. J. Bacteriol. 1993, 175, 5934–5944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rasamiravaka, T.; Vandeputte, O.M.; El Jaziri, M. Procedure for Rhamnolipids Quantification Using Methylene-blue. Bio-Protocol 2016, 6, e1783. [Google Scholar] [CrossRef]
- Müh, U.; Schuster, M.; Heim, R.; Singh, A.; Olson, E.R.; Greenberg, E.P. Novel Pseudomonas aeruginosa quorum-sensing inhibitors identified in an ultra-high-throughput screen. Antimicrob. Agents Chemother. 2006, 50, 3674–3679. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Martínez, I.; Haas, D. Azithromycin inhibits expression of the GacA-dependent small RNAs RsmY and RsmZ in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2011, 55, 3399–3405. [Google Scholar] [CrossRef] [Green Version]
- Bisson, J.; McAlpine, J.B.; Friesen, J.B.; Chen, S.N.; Graham, J.; Pauli, G.F. Can Invalid Bioactives Undermine Natural Product-Based Drug Discovery? J. Med. Chem. 2016, 59, 1671–1690. [Google Scholar] [CrossRef]
- Donovan, G.T.; Norton, J.P.; Bower, J.M.; Mulvey, M.A. Adenylate Cyclase and the Cyclic AMP Receptor Protein Modulate Stress Resistance and Virulence Capacity of Uropathogenic Escherichia coli. Infect. Immun. 2012, 81, 249–258. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mahavy, C.E.; Mol, A.; Andrianarisoa, B.; Duez, P.; Jaziri, M.E.; Baucher, M.; Rasamiravaka, T. The Xanthophyll Carotenoid Lutein Reduces the Invasive Potential of Pseudomonas aeruginosa and Increases Its Susceptibility to Tobramycin. Int. J. Mol. Sci. 2022, 23, 7199. https://doi.org/10.3390/ijms23137199
Mahavy CE, Mol A, Andrianarisoa B, Duez P, Jaziri ME, Baucher M, Rasamiravaka T. The Xanthophyll Carotenoid Lutein Reduces the Invasive Potential of Pseudomonas aeruginosa and Increases Its Susceptibility to Tobramycin. International Journal of Molecular Sciences. 2022; 23(13):7199. https://doi.org/10.3390/ijms23137199
Chicago/Turabian StyleMahavy, Christian Emmanuel, Adeline Mol, Blandine Andrianarisoa, Pierre Duez, Mondher El Jaziri, Marie Baucher, and Tsiry Rasamiravaka. 2022. "The Xanthophyll Carotenoid Lutein Reduces the Invasive Potential of Pseudomonas aeruginosa and Increases Its Susceptibility to Tobramycin" International Journal of Molecular Sciences 23, no. 13: 7199. https://doi.org/10.3390/ijms23137199






