New Insights of OLFM2 and OLFM4 in Gut-Liver Axis and Their Potential Involvement in Nonalcoholic Fatty Liver Disease
Abstract
:1. Introduction
2. Results
2.1. Baseline Characteristics of Subjects
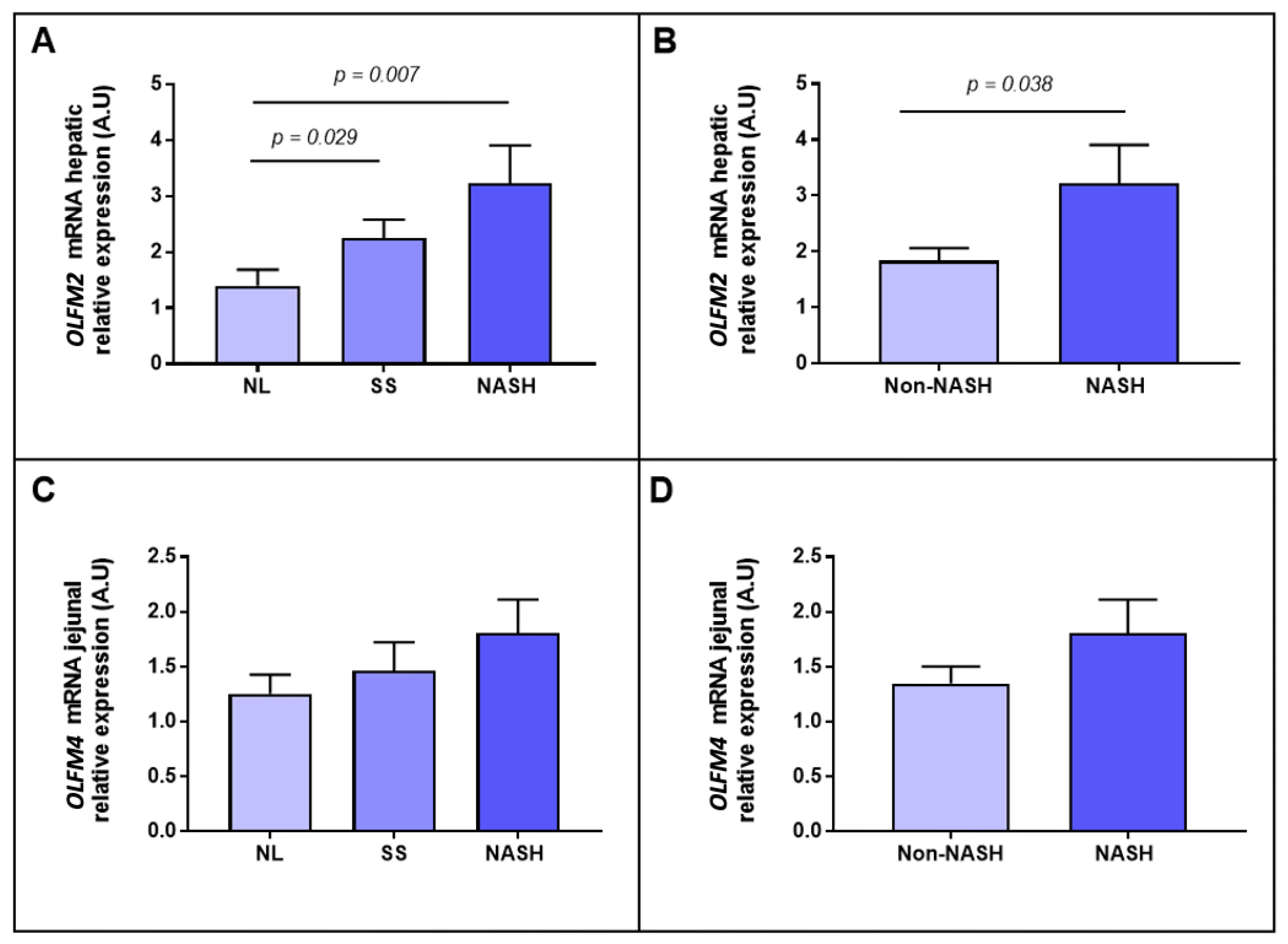
2.2. Evaluation of the Relative mRNA Abundance of OLFM2 and OLFM4 According to Hepatic Histology
2.3. Evaluation of the Relative mRNA Abundance of OLFM2 and OLFM4 According to the Severity of Steatosis
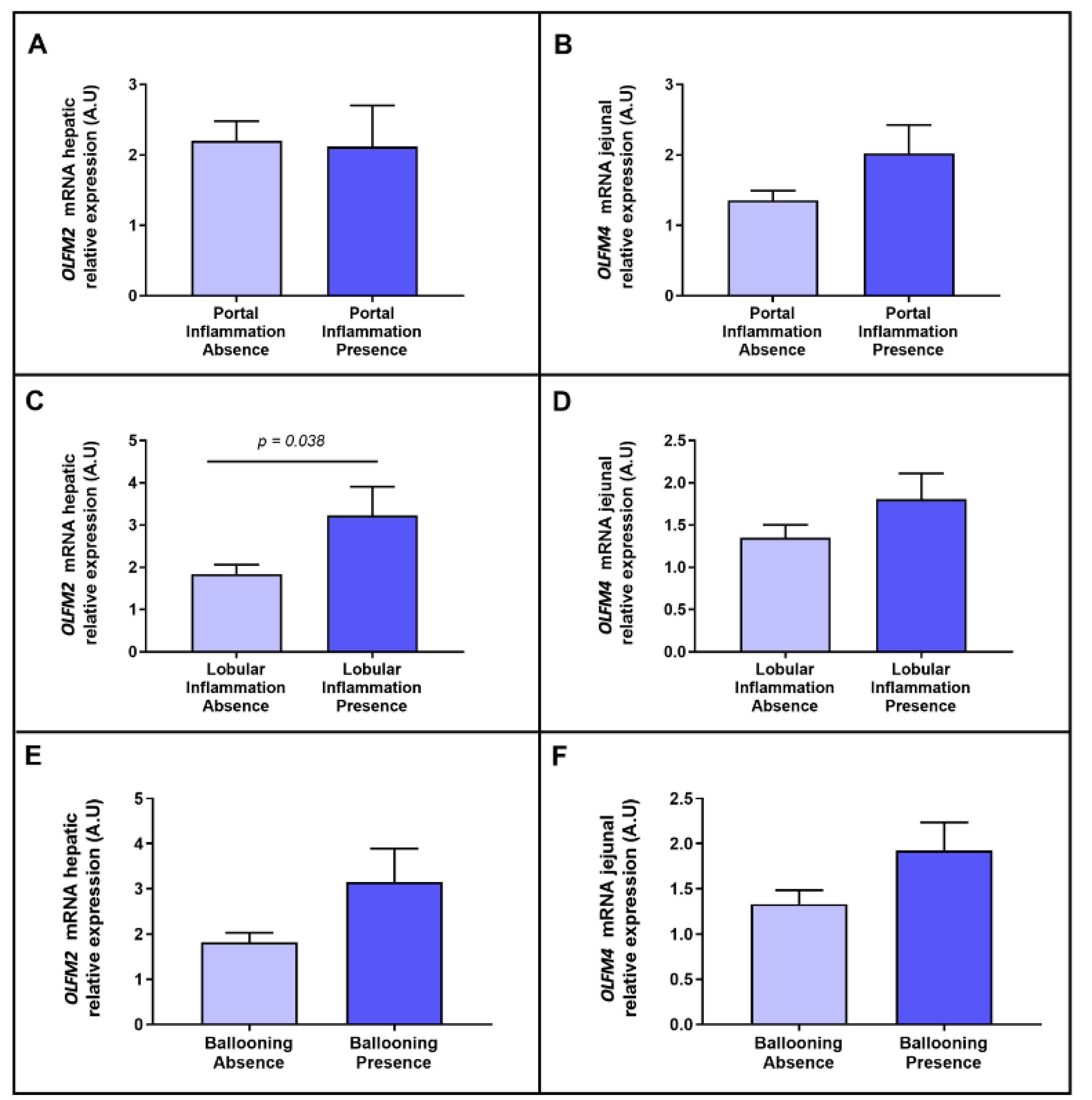
2.4. Evaluation of the Relative mRNA Abundance of OLFM2 and OLFM4 According to NASH-Related Parameters
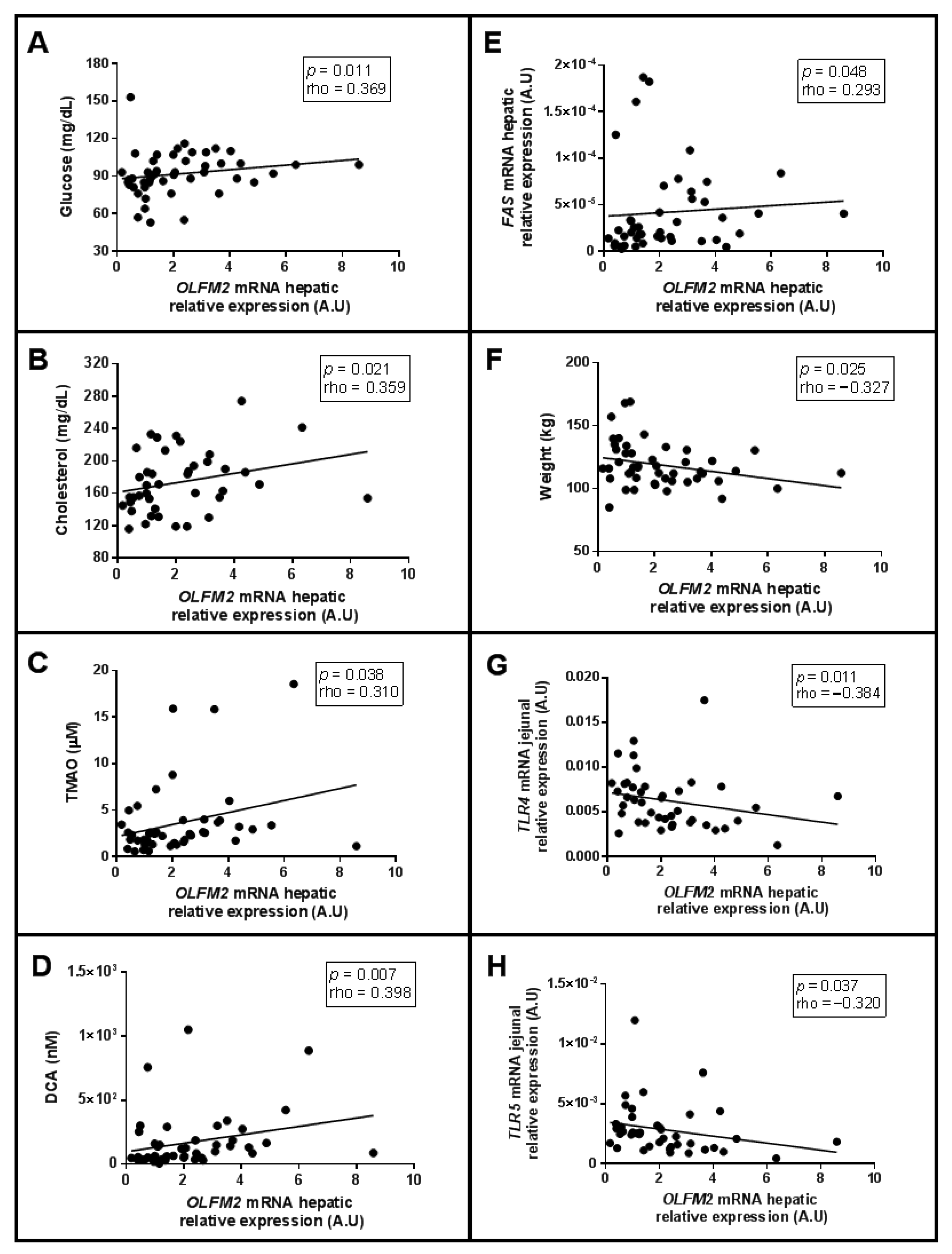
2.5. Correlations of Relative mRNA Abundance of Hepatic OLFM2 and Jejunal OLFM4, with Clinical and Biochemical-Related Parameters
3. Discussion
4. Materials and Methods
4.1. Subjects
4.2. Sample Size
4.3. Liver Pathology
4.4. Biochemical Analyses
4.5. mRNA Expression in Liver and Jejunum
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Cobbina, E.; Akhlaghi, F. Non-Alcoholic Fatty Liver Disease (NAFLD)—Pathogenesis, Classification, and Effect on Drug Metabolizing Enzymes and Transporters. Drug Metab. Rev. 2017, 49, 197–211. [Google Scholar] [CrossRef] [PubMed]
- Pappachan, J.M.; Babu, S.; Krishnan, B.; Ravindran, N.C. Non-Alcoholic Fatty Liver Disease: A Clinical Update. J. Clin. Transl. Hepatol. 2017, 5, 384–393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cataldo, I.; Sarcognato, S.; Sacchi, D.; Cacciatore, M.; Baciorri, F.; Mangia, A.; Cazzagon, N.; Guido, M. Pathology of Non-Alcoholic Fatty Liver Disease. Pathologica 2021, 113, 194–202. [Google Scholar] [CrossRef] [PubMed]
- Farrell, G.C.; Teoh, N.C.; Mccuskey, R.S. Hepatic Microcirculation in Fatty Liver Disease. Anat. Rec. Adv. Integr. Anat. Evol. Biol. 2008, 291, 684–692. [Google Scholar] [CrossRef]
- Manne, V.; Handa, P.; Kowdley, K.V. Pathophysiology of Nonalcoholic Fatty Liver Disease/Nonalcoholic Steatohepatitis. Clin. Liver Dis. 2018, 22, 23–37. [Google Scholar] [CrossRef]
- Kaufmann, B.; Reca, A.; Wang, B.; Friess, H.; Feldstein, A.E.; Hartmann, D. Mechanisms of Nonalcoholic Fatty Liver Disease and Implications for Surgery. Langenbecks Arch. Surg. 2021, 406, 1–17. [Google Scholar] [CrossRef]
- Chalasani, N.; Younossi, Z.; Lavine, J.E.; Charlton, M.; Cusi, K.; Rinella, M.; Harrison, S.A.; Brunt, E.M.; Sanyal, A.J. The Diagnosis and Management of Nonalcoholic Fatty Liver Disease. Hepatology 2018, 67, 328–357. [Google Scholar] [CrossRef]
- Kataoka, K. The Intestinal Microbiota and Its Role in Human Health and Disease. J. Med. Investig. 2016, 63, 27–37. [Google Scholar] [CrossRef] [Green Version]
- Bäckhed, F.; Ding, H.; Wang, T.; Hooper, L.V.; Koh, G.Y.; Nagy, A.; Semenkovich, C.F.; Gordon, J.I. The Gut Microbiota as an Environmental Factor That Regulates Fat Storage. Proc. Natl. Acad. Sci. USA 2004, 101, 15718–15723. [Google Scholar] [CrossRef] [Green Version]
- Imajo, K.; Yoneda, M.; Ogawa, Y.; Wada, K.; Nakajima, A. Microbiota and Nonalcoholic Steatohepatitis. Semin. Immunopathol. 2014, 36, 115–132. [Google Scholar] [CrossRef]
- Milosevic, I.; Vujovic, A.; Barac, A.; Djelic, M.; Korac, M.; Radovanovic Spurnic, A.; Gmizic, I.; Stevanovic, O.; Djordjevic, V.; Lekic, N.; et al. Gut-Liver Axis, Gut Microbiota, and Its Modulation in the Management of Liver Diseases: A Review of the Literature. Int. J. Mol. Sci. 2019, 20, 395. [Google Scholar] [CrossRef] [Green Version]
- Albillos, A.; de Gottardi, A.; Rescigno, M. The Gut-Liver Axis in Liver Disease: Pathophysiological Basis for Therapy. J. Hepatol. 2020, 72, 558–577. [Google Scholar] [CrossRef] [Green Version]
- Azzu, V.; Vacca, M.; Virtue, S.; Allison, M.; Vidal-Puig, A. Adipose Tissue-Liver Cross Talk in the Control of Whole-Body Metabolism: Implications in Nonalcoholic Fatty Liver Disease. Gastroenterology 2020, 158, 1899–1912. [Google Scholar] [CrossRef]
- Konrad, D.; Wueest, S. The Gut-Adipose-Liver Axis in the Metabolic Syndrome. Physiology 2014, 29, 304–313. [Google Scholar] [CrossRef]
- Kaya, E.; Yilmaz, Y. Metabolic-Associated Fatty Liver Disease (MAFLD): A Multi-Systemic Disease Beyond the Liver. J. Clin. Transl. Hepatol. 2022, 10, 329–338. [Google Scholar] [CrossRef]
- Anholt, R.R.H. Olfactomedin Proteins: Central Players in Development and Disease. Front. Cell Dev. Biol. 2014, 2, 6. [Google Scholar] [CrossRef] [Green Version]
- Tomarev, S.I.; Nakaya, N. Olfactomedin Domain-Containing Proteins: Possible Mechanisms of Action and Functions in Normal Development and Pathology. Mol. Neurobiol. 2009, 40, 122–138. [Google Scholar] [CrossRef] [Green Version]
- Fagerberg, L.; Hallström, B.M.; Oksvold, P.; Kampf, C.; Djureinovic, D.; Odeberg, J.; Habuka, M.; Tahmasebpoor, S.; Danielsson, A.; Edlund, K.; et al. Analysis of the Human Tissue-Specific Expression by Genome-Wide Integration of Transcriptomics and Antibody-Based Proteomics. Mol. Cell. Proteomics 2014, 13, 397–406. [Google Scholar] [CrossRef] [Green Version]
- Kutyavin, V.I.; Chawla, A. BCL6 Regulates Brown Adipocyte Dormancy to Maintain Thermogenic Reserve and Fitness. Proc. Natl. Acad. Sci. USA 2019, 116, 17071–17080. [Google Scholar] [CrossRef] [Green Version]
- González-García, I.; Freire-Agulleiro, Ó.; Nakaya, N.; Ortega, F.J.; Garrido-Gil, P.; Liñares-Pose, L.; Fernø, J.; Labandeira-Garcia, J.L.; Diéguez, C.; Sultana, A.; et al. Olfactomedin 2 Deficiency Protects against Diet-Induced Obesity. Metabolism 2022, 129, 155122. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, W.-L.; Tang, D.C.; Chen, L.; Wang, M.; Pack, S.D.; Zhuang, Z.; Rodgers, G.P. Identification and Characterization of a Novel Member of Olfactomedin-Related Protein Family, HGC-1, Expressed during Myeloid Lineage Development. Gene 2002, 283, 83–93. [Google Scholar] [CrossRef]
- Liu, W.; Chen, L.; Zhu, J.; Rodgers, G.P. The Glycoprotein HGC-1 Binds to Cadherin and Lectins. Exp. Cell Res. 2006, 312, 1785–1797. [Google Scholar] [CrossRef]
- Liu, W.; Rodgers, G.P. Olfactomedin 4 Expression and Functions in Innate Immunity, Inflammation, and Cancer. Cancer Metastasis Rev. 2016, 35, 201–212. [Google Scholar] [CrossRef]
- Liu, W.; Li, H.; Hong, S.-H.; Piszczek, G.P.; Chen, W.; Rodgers, G.P. Olfactomedin 4 Deletion Induces Colon Adenocarcinoma in ApcMin/+ Mice. Oncogene 2016, 35, 5237–5247. [Google Scholar] [CrossRef] [Green Version]
- Bertran, L.; Portillo-Carrasquer, M.; Aguilar, C.; Porras, J.A.; Riesco, D.; Martínez, S.; Vives, M.; Sabench, F.; Gonzalez, E.; Del Castillo, D.; et al. Deregulation of Secreted Frizzled-Related Protein 5 in Nonalcoholic Fatty Liver Disease Associated with Obesity. Int. J. Mol. Sci. 2021, 22, 6895. [Google Scholar] [CrossRef]
- Auguet, T.; Bertran, L.; Binetti, J.; Aguilar, C.; Martínez, S.; Guiu-Jurado, E.; Sabench, F.; Adalid, L.; Porras, J.A.; Riesco, D.; et al. Hepatocyte Notch Signaling Deregulation Related to Lipid Metabolism in Women with Obesity and Nonalcoholic Fatty Liver. Obesity 2020, 28, 1487–1493. [Google Scholar] [CrossRef]
- VanDussen, K.L.; Carulli, A.J.; Keeley, T.M.; Patel, S.R.; Puthoff, B.J.; Magness, S.T.; Tran, I.T.; Maillard, I.; Siebel, C.; Kolterud, Å.; et al. Notch Signaling Modulates Proliferation and Differentiation of Intestinal Crypt Base Columnar Stem Cells. Development 2012, 139, 488–497. [Google Scholar] [CrossRef] [Green Version]
- Tian, Y.; Mok, M.; Yang, P.; Cheng, A. Epigenetic Activation of Wnt/β-Catenin Signaling in NAFLD-Associated Hepatocarcinogenesis. Cancers 2016, 8, 76. [Google Scholar] [CrossRef]
- Xu, H.; Wang, L. The Role of Notch Signaling Pathway in Non-Alcoholic Fatty Liver Disease. Front. Mol. Biosci. 2021, 8, 792667. [Google Scholar] [CrossRef]
- Daniel, P.V.; Dogra, S.; Rawat, P.; Choubey, A.; Khan, A.S.; Rajak, S.; Kamthan, M.; Mondal, P. NF-ΚB P65 Regulates Hepatic Lipogenesis by Promoting Nuclear Entry of ChREBP in Response to a High Carbohydrate Diet. J. Biol. Chem. 2021, 296, 100714. [Google Scholar] [CrossRef]
- Liu, W.; Yan, M.; Liu, Y.; Wang, R.; Li, C.; Deng, C.; Singh, A.; Coleman, W.G.; Rodgers, G.P. Olfactomedin 4 Down-Regulates Innate Immunity against Helicobacter Pylori Infection. Proc. Natl. Acad. Sci. USA 2010, 107, 11056–11061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.-Y.; Chen, S.-H.; Zhang, Y.-N.; Xu, C.-F. Olfactomedin-4 in Digestive Diseases: A Mini-Review. World J. Gastroenterol. 2018, 24, 1881–1887. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zhu, J.; Cao, L.; Rodgers, G.P. Expression of HGC-1 Is Correlated with Differentiation of Gastric Carcinoma. Histopathology 2007, 51, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Grover, P.K.; Hardingham, J.E.; Cummins, A.G. Stem Cell Marker Olfactomedin 4: Critical Appraisal of Its Characteristics and Role in Tumorigenesis. Cancer Metastasis Rev. 2010, 29, 761–775. [Google Scholar] [CrossRef]
- Schuster, S.; Cabrera, D.; Arrese, M.; Feldstein, A.E. Triggering and Resolution of Inflammation in NASH. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 349–364. [Google Scholar] [CrossRef]
- Inczefi, O.; Bacsur, P.; Resál, T.; Keresztes, C.; Molnár, T. The Influence of Nutrition on Intestinal Permeability and the Microbiome in Health and Disease. Front. Nutr. 2022, 9, 718710. [Google Scholar] [CrossRef]
- Sala, P.; Torrinhas, R.S.M.d.M.; Fonseca, D.C.; Machado, N.M.; Singer, J.; Singer, P.; Ravacci, G.R.; Belarmino, G.; Ferreira, B.A.M.; Marques, M.; et al. Intestinal Expression of Toll-like Receptor Gene Changes Early after Gastric Bypass Surgery and Association with Type 2 Diabetes Remission. Nutrition 2020, 79–80, 110885. [Google Scholar] [CrossRef]
- Enjoji Impact of Cholesterol Metabolism and the LXRα-SREBP-1c Pathway on Nonalcoholic Fatty Liver Disease. Int. J. Mol. Med. 2009, 23, 603–608. [CrossRef] [Green Version]
- Donnelly, K.L.; Smith, C.I.; Schwarzenberg, S.J.; Jessurun, J.; Boldt, M.D.; Parks, E.J. Sources of Fatty Acids Stored in Liver and Secreted via Lipoproteins in Patients with Nonalcoholic Fatty Liver Disease. J. Clin. Investig. 2005, 115, 1343–1351. [Google Scholar] [CrossRef] [Green Version]
- Parlati, L.; Régnier, M.; Guillou, H.; Postic, C. New Targets for NAFLD. JHEP Rep. 2021, 3, 100346. [Google Scholar] [CrossRef]
- Auguet, T.; Berlanga, A.; Guiu-Jurado, E.; Martinez, S.; Porras, J.; Aragonès, G.; Sabench, F.; Hernandez, M.; Aguilar, C.; Sirvent, J.; et al. Altered Fatty Acid Metabolism-Related Gene Expression in Liver from Morbidly Obese Women with Non-Alcoholic Fatty Liver Disease. Int. J. Mol. Sci. 2014, 15, 22173–22187. [Google Scholar] [CrossRef] [Green Version]
- Aragonès, G.; Colom-Pellicer, M.; Aguilar, C.; Guiu-Jurado, E.; Martínez, S.; Sabench, F.; Antonio Porras, J.; Riesco, D.; Del Castillo, D.; Richart, C.; et al. Circulating Microbiota-Derived Metabolites: A “liquid Biopsy? Int. J. Obes. 2020, 44, 875–885. [Google Scholar] [CrossRef] [Green Version]
- Paul, B.; Lewinska, M.; Andersen, J.B. Lipid Alterations in Chronic Liver Disease and Liver Cancer. JHEP Rep. 2022, 4, 100479. [Google Scholar] [CrossRef]
- Friedman, S.L.; Neuschwander-Tetri, B.A.; Rinella, M.; Sanyal, A.J. Mechanisms of NAFLD Development and Therapeutic Strategies. Nat. Med. 2018, 24, 908–922. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, Y.; Zhou, R.; Chen, X.; Wang, C.; Tan, X.; Wang, L.; Zheng, R.; Zhang, H.; Ling, W.; et al. Associations of Gut-Flora-Dependent Metabolite Trimethylamine-N-Oxide, Betaine and Choline with Non-Alcoholic Fatty Liver Disease in Adults. Sci. Rep. 2016, 6, 19076. [Google Scholar] [CrossRef]
- Gao, X.; Liu, X.; Xu, J.; Xue, C.; Xue, Y.; Wang, Y. Dietary Trimethylamine N-Oxide Exacerbates Impaired Glucose Tolerance in Mice Fed a High Fat Diet. J. Biosci. Bioeng. 2014, 118, 476–481. [Google Scholar] [CrossRef]
- Tan, X.; Liu, Y.; Long, J.; Chen, S.; Liao, G.; Wu, S.; Li, C.; Wang, L.; Ling, W.; Zhu, H. Trimethylamine N-Oxide Aggravates Liver Steatosis through Modulation of Bile Acid Metabolism and Inhibition of Farnesoid X Receptor Signaling in Nonalcoholic Fatty Liver Disease. Mol. Nutr. Food Res. 2019, 63, 1900257. [Google Scholar] [CrossRef]
- Jaillier-Ramírez, A.M.; Waitzberg, D.; Becerra Romero, A. Relación Entre Los Ácidos Biliares y La Microbiota Intestinal ¿es Posible Considerarlo Como Un Factor Etiológico En Diversas Colangiopatías? Una Revisión Narrativa. Rev. Nutr. Clínica Metab. 2021, 4, 40–55. [Google Scholar] [CrossRef]
- Grzych, G.; Chávez-Talavera, O.; Descat, A.; Thuillier, D.; Verrijken, A.; Kouach, M.; Legry, V.; Verkindt, H.; Raverdy, V.; Legendre, B.; et al. NASH-Related Increases in Plasma Bile Acid Levels Depend on Insulin Resistance. JHEP Rep. 2021, 3, 100222. [Google Scholar] [CrossRef]
- Miura, K. Role of Gut Microbiota and Toll-like Receptors in Nonalcoholic Fatty Liver Disease. World J. Gastroenterol. 2014, 20, 7381. [Google Scholar] [CrossRef]
- Bettini, S.; Belligoli, A.; Fabris, R.; Busetto, L. Diet Approach before and after Bariatric Surgery. Rev. Endocr. Metab. Disord. 2020, 21, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Muscogiuri, G.; Barrea, L.; Laudisio, D.; Pugliese, G.; Salzano, C.; Savastano, S.; Colao, A. The Management of Very Low-Calorie Ketogenic Diet in Obesity Outpatient Clinic: A Practical Guide. J. Transl. Med. 2019, 17, 356. [Google Scholar] [CrossRef] [PubMed]
- Rogero, M.; Calder, P. Obesity, Inflammation, Toll-Like Receptor 4 and Fatty Acids. Nutrients 2018, 10, 432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferreira, D.F.; Fiamoncini, J.; Prist, I.H.; Ariga, S.K.; de Souza, H.P.; de Lima, T.M. Novel Role of TLR4 in NAFLD Development: Modulation of Metabolic Enzymes Expression. Biochim. Biophys. Acta BBA Mol. Cell Biol. Lipids 2015, 1851, 1353–1359. [Google Scholar] [CrossRef]
- Zhang, W.; Hartmann, R.; Tun, H.M.; Elson, C.O.; Khafipour, E.; Garvey, W.T. Deletion of the Toll-Like Receptor 5 Gene Per Se Does Not Determine the Gut Microbiome Profile That Induces Metabolic Syndrome: Environment Trumps Genotype. PLoS ONE 2016, 11, e0150943. [Google Scholar] [CrossRef]
- Etienne-Mesmin, L.; Vijay-Kumar, M.; Gewirtz, A.T.; Chassaing, B. Hepatocyte Toll-Like Receptor 5 Promotes Bacterial Clearance and Protects Mice Against High-Fat Diet–Induced Liver Disease. Cell. Mol. Gastroenterol. Hepatol. 2016, 2, 584–604. [Google Scholar] [CrossRef] [Green Version]
- Chassaing, B.; Ley, R.E.; Gewirtz, A.T. Intestinal Epithelial Cell Toll-like Receptor 5 Regulates the Intestinal Microbiota to Prevent Low-Grade Inflammation and Metabolic Syndrome in Mice. Gastroenterology 2014, 147, 1363–1377.e17. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Wang, L.; Sun, X.-H.; Liu, X.; Xiao, Y.; Zhang, J.; Wang, T.; Chen, H.; Zhan, Y.-Q.; Yu, M.; et al. Toll-like Receptor 5-Mediated Signaling Enhances Liver Regeneration in Mice. Mil. Med. Res. 2021, 8, 16. [Google Scholar] [CrossRef]
- Vijay-Kumar, M.; Aitken, J.D.; Carvalho, F.A.; Cullender, T.C.; Mwangi, S.; Srinivasan, S.; Sitaraman, S.V.; Knight, R.; Ley, R.E.; Gewirtz, A.T. Metabolic Syndrome and Altered Gut Microbiota in Mice Lacking Toll-Like Receptor 5. Science 2010, 328, 228–231. [Google Scholar] [CrossRef] [Green Version]
- Glass, O.; Henao, R.; Patel, K.; Guy, C.D.; Gruss, H.J.; Syn, W.; Moylan, C.A.; Streilein, R.; Hall, R.; Mae Diehl, A.; et al. Serum Interleukin-8, Osteopontin, and Monocyte Chemoattractant Protein 1 Are Associated With Hepatic Fibrosis in Patients With Nonalcoholic Fatty Liver Disease. Hepatol. Commun. 2018, 2, 1344–1355. [Google Scholar] [CrossRef]
- Ruiz de Morales, J.M.G.; Puig, L.; Daudén, E.; Cañete, J.D.; Pablos, J.L.; Martín, A.O.; Juanatey, C.G.; Adán, A.; Montalbán, X.; Borruel, N.; et al. Critical Role of Interleukin (IL)-17 in Inflammatory and Immune Disorders: An Updated Review of the Evidence Focusing in Controversies. Autoimmun. Rev. 2020, 19, 102429. [Google Scholar] [CrossRef]
- Gomes, A.L.; Teijeiro, A.; Burén, S.; Tummala, K.S.; Yilmaz, M.; Waisman, A.; Theurillat, J.-P.; Perna, C.; Djouder, N. Metabolic Inflammation-Associated IL-17A Causes Non-Alcoholic Steatohepatitis and Hepatocellular Carcinoma. Cancer Cell 2016, 30, 161–175. [Google Scholar] [CrossRef]
- Chin, K.L.; Aerbajinai, W.; Zhu, J.; Drew, L.; Chen, L.; Liu, W.; Rodgers, G.P. The Regulation of OLFM4 Expression in Myeloid Precursor Cells Relies on NF-ΚB Transcription Factor. Br. J. Haematol. 2008, 143, 421–432. [Google Scholar] [CrossRef]
- Engin, A. The Pathogenesis of Obesity-Associated Adipose Tissue Inflammation. In Obesity and Lipotoxicity; Engin, A.B., Engin, A., Eds.; Advances in Experimental Medicine and Biology; Springer International Publishing: Cham, Switzerland, 2017; Volume 960, pp. 221–245. ISBN 978-3-319-48380-1. [Google Scholar]
- Couper, K.N.; Blount, D.G.; Riley, E.M. IL-10: The Master Regulator of Immunity to Infection. J. Immunol. 2008, 180, 5771–5777. [Google Scholar] [CrossRef]
- Cintra, D.E.; Pauli, J.R.; Araújo, E.P.; Moraes, J.C.; de Souza, C.T.; Milanski, M.; Morari, J.; Gambero, A.; Saad, M.J.; Velloso, L.A. Interleukin-10 Is a Protective Factor against Diet-Induced Insulin Resistance in Liver. J. Hepatol. 2008, 48, 628–637. [Google Scholar] [CrossRef]
- Esposito, K.; Pontillo, A.; Giugliano, F.; Giugliano, G.; Marfella, R.; Nicoletti, G.; Giugliano, D. Association of Low Interleukin-10 Levels with the Metabolic Syndrome in Obese Women. J. Clin. Endocrinol. Metab. 2003, 88, 1055–1058. [Google Scholar] [CrossRef]
- Tedgui, A.; Mallat, Z. Anti-Inflammatory Mechanisms in the Vascular Wall. Circ. Res. 2001, 88, 877–887. [Google Scholar] [CrossRef] [Green Version]
- Shepard, C.R. TLR9 in MAFLD and NASH: At the Intersection of Inflammation and Metabolism. Front. Endocrinol. 2021, 11, 613639. [Google Scholar] [CrossRef]
- He, Y.; Hwang, S.; Cai, Y.; Kim, S.; Xu, M.; Yang, D.; Guillot, A.; Feng, D.; Seo, W.; Hou, X.; et al. MicroRNA-223 Ameliorates Nonalcoholic Steatohepatitis and Cancer by Targeting Multiple Inflammatory and Oncogenic Genes in Hepatocytes. Hepatology 2019, 70, 1150–1167. [Google Scholar] [CrossRef]
- Kleiner, D.E.; Brunt, E.M.; Van Natta, M.; Behling, C.; Contos, M.J.; Cummings, O.W.; Ferrell, L.D.; Liu, Y.-C.; Torbenson, M.S.; Unalp-Arida, A.; et al. Design and Validation of a Histological Scoring System for Nonalcoholic Fatty Liver Disease. Hepatology 2005, 41, 1313–1321. [Google Scholar] [CrossRef]
- Brunt, E.M.; Janney, C.G.; Di Bisceglie, A.M.; Neuschwander-Tetri, B.A.; Bacon, B.R. Nonalcoholic Steatohepatitis: A Proposal for Grading and Staging The Histological Lesions. Am. J. Gastroenterol. 1999, 94, 2467–2474. [Google Scholar] [CrossRef]


| Variables | NL (n = 27) | SS (n = 26) | NASH (n = 16) |
|---|---|---|---|
| Weight (kg) | 117.00 (107.00–131.00) | 114.00 (108.98–128.60) | 110.50 (104.33–120.75) |
| BMI (kg/m2) | 43.50 (40.89–46.88) | 44.35 (40.87–46.80) | 44.19 (40.69–45.80) |
| SBP (mmHg) | 120.00 (100.00–132.50) | 117.50 (108.50–127.00) | 115.00 (102.00–127.00) |
| DBP (mmHg) | 63.00 (57.50–73.00) | 62.00 (59.50–73.75) | 64.00 (55.00–70.00) |
| HOMA1-IR | 2.05 (1.03–3.45) | 2.52 (1.38–3.68) | 1.63 (1.26–4.23) |
| Glucose (mg/dL) | 85.00 (76.00–93.00) | 93.00 (87.25–107.00) * | 91.50 (82.25–101.75) |
| Insulin (mUI/L) | 9.57 (5.55–16.82) | 10.17 (7.23–13.93) | 7.19 (5.14–26.02) |
| HbA1c (%) | 5.50 (5.30–5.70) | 5.55 (5.30–5.95) | 5.55 (5.15–6.13) |
| TG (mg/dL) | 106.50 (94.00–136.00) | 117.50 (82.25–172.50) | 153.00 (116.50–256.50) * |
| Cholesterol (mg/dL) | 170.00 (148.25–209.50) | 171.15 (136.25–194.25) | 183.90 (152.75–229.50) |
| HDL-C (mg/dL) | 40.60 (32.05–48.50) | 43.50 (33.75–47.00) | 37.80 (33.50–48.50) |
| LDL-C (mg/dL) | 107.90 (86.00–134.20) | 104.10 (77.20–126.25) | 94.00 (79.30–128.03) |
| AST (UI/L) | 20.00 (15.50–36.50) | 23.00 (17.00–35.00) | 27.00 (17.25–43.50) |
| ALT (UI/L) | 22.50 (16.00–37.50) | 31.00 (22.00–32.25) | 32.00 (16.25–41.00) |
| GGT (UI/L) | 18.00 (15.25–26.25) | 21.00 (16.00–32.25) | 25.50 (18.00–28.75) |
| ALP (Ul/L) | 58.50 (49.25–71.25) | 74.00 (64.00–86.25) * | 63.00 (55.00–74.50) $ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bertran, L.; Jorba-Martin, R.; Barrientos-Riosalido, A.; Portillo-Carrasquer, M.; Aguilar, C.; Riesco, D.; Martínez, S.; Vives, M.; Sabench, F.; Castillo, D.D.; et al. New Insights of OLFM2 and OLFM4 in Gut-Liver Axis and Their Potential Involvement in Nonalcoholic Fatty Liver Disease. Int. J. Mol. Sci. 2022, 23, 7442. https://doi.org/10.3390/ijms23137442
Bertran L, Jorba-Martin R, Barrientos-Riosalido A, Portillo-Carrasquer M, Aguilar C, Riesco D, Martínez S, Vives M, Sabench F, Castillo DD, et al. New Insights of OLFM2 and OLFM4 in Gut-Liver Axis and Their Potential Involvement in Nonalcoholic Fatty Liver Disease. International Journal of Molecular Sciences. 2022; 23(13):7442. https://doi.org/10.3390/ijms23137442
Chicago/Turabian StyleBertran, Laia, Rosa Jorba-Martin, Andrea Barrientos-Riosalido, Marta Portillo-Carrasquer, Carmen Aguilar, David Riesco, Salomé Martínez, Margarita Vives, Fàtima Sabench, Daniel Del Castillo, and et al. 2022. "New Insights of OLFM2 and OLFM4 in Gut-Liver Axis and Their Potential Involvement in Nonalcoholic Fatty Liver Disease" International Journal of Molecular Sciences 23, no. 13: 7442. https://doi.org/10.3390/ijms23137442







