Abstract
Freezing stress is a major factor limiting production and geographical distribution of temperate crops. Elongator is a six subunit complex with histone acetyl-transferase activity and is involved in plant development and defense responses in Arabidopsis thaliana. However, it is unknown whether and how an elongator responds to freezing stress in plants. In this study, we found that wheat elongator subunit 4 (TaELP4) negatively regulates freezing tolerance through ethylene signaling. TaELP4 promoter contained cold response elements and was up-regulated in freezing stress. Subcellular localization showed that TaELP4 and AtELP4 localized in the cytoplasm and nucleus. Silencing of TaELP4 in wheat with BSMV-mediated VIGS approach significantly elevated tiller survival rate compared to control under freezing stress, but ectopic expression of TaELP4 in Arabidopsis increased leaf damage and survival rate compared with Col-0. Further results showed that TaELP4 positively regulated ACS2 and ACS6 transcripts, two main limiting enzymes in ethylene biosynthesis. The determination of ethylene content showed that TaELP4 overexpression resulted in more ethylene accumulated than Col-0 under freezing stress. Epigenetic research showed that histone H3K9/14ac levels significantly increased in coding/promoter regions of AtACS2 and AtACS6 in Arabidopsis. RT-qPCR assays showed that the EIN2/EIN3/EIL1-CBFs-COR pathway was regulated by TaELP4 under freezing stress. Taken together, our results suggest that TaELP4 negatively regulated plant responses to freezing stress via heightening histone acetylation levels of ACS2 and ACS6 and increasing their transcription and ethylene accumulation.
1. Introduction
Cold stress limits growth, reproduction, and geographical distribution of temperate crops. As for plants, cold stress is categorized into chilling stress (0–20 °C) and freezing stress (<0 °C) based on the temperatures and various physiological mechanisms that function in different temperature ranges [1,2]. Wheat (Triticum aestivum L.) is an important staple food crop worldwide that is fundamental in global food security. Cold stress often occurs in wheat-growing areas of the world, including China, the United States, Europe, and Australia [3,4,5,6]. Spring frost is wheat canopy temperature falling to 0 °C or below in the spring, causing severe damage to the micro-organelles of the cell, resulting in excessive reactive oxygen species (ROS) and the occurrence of lipid peroxidation [6,7,8]. At the vegetative phase, cold stress delays germination, poor emergence, and reduced plant density. Prolonged cold stress results in stunted growth, leaf chlorosis, and reduced root-shoot surface area, all of which significantly reduce wheat yield [9,10]. During the reproductive phase, cold stress damages flag leaves and spikes and reduces the number of effective tillers, grain number per spike, and grain filling rate, leading to grain yield loss [8,11,12,13].
To reduce the negative impact of cold stress to wheat yield, it is necessary to investigate cold-tolerant mechanisms and develop cold-tolerant wheat cultivars. The ICE-CBFs-COR pathway is a key pathway related to cold stress in plants. In the wheat genome, five ICE (inducer of CBF expression) genes, 37 CBF (C-repeat binding factor) genes, and 11 COR (cold responsive) genes have been identified [14]. TaICE41 and TaICE87 bind to the promoter of CBF gene TaCBFIVd-B9 and activate its transcription, which confers cold tolerance on TaICE41 and TaICE87 [15]. Due to a lack of systematic research, signaling pathways, gene regulatory networks, signal transduction, and molecular mechanisms related to cold stress are poorly understood in wheat.
The Elongator complex consists of six subunits (ELP1 to ELP6) and was originally isolated as an interactor of hyperphosphorylated RNA polymerase II (RNAPII) in yeast [16,17] and later identified in humans [18] and Arabidopsis [19]. Structure and function studies showed that the Elongator complex is highly conserved in plants and yeast [20]. ELP1, ELP2, and ELP3 form the core subcomplex, and ELP1 and ELP2 are WD40 proteins that serve as scaffolds for complex assembly. ELP3 is the catalytic subunit harboring a C-terminal GNAT-type histone acetyltransferase (HAT) domain and an N-terminal iron-sulfur radical S-adenosylmethionine (SAM) domain [21,22,23]. ELP4, ELP5, and ELP6 form the accessory subcomplex, and each form a RecA-ATPase-like fold and assemble into a hexameric ring-shaped structure that is important for recognizing histone H3 [24,25]. Deletion of any of the six subunits results in almost identical phenotypes, suggesting that all six subunits are required for Elongator’s cellular functions [17,19,24,26,27].
In plants, elongator participates in the development and responses to biotic stresses. Atelp mutants exhibit various developmental defects, such as narrow and elongated leaves, short primary roots, lateral root density, abnormal inflorescence phyllotaxis, and delayed seedling growth [19,26]. A mechanistic study showed that Atelp3 reduced histone H3K14 acetylation at the coding sequence and 3′-UTR of auxin-related genes SHY2/IAA3 and LAX2, which decreased expression [26]. Atelp2 and Atelp3 mediate pathogen-induced transcriptome reprogramming via altering methylation levels of defense-relate genes and increasing histone acetylation levels in coding regions of several defense genes during innate immune responses [28,29]. AtELP4 overexpression enhanced resistance to anthracnose crown rot, powdery mildew, and tomato bacterial speck in Fragaria vesca and tomato plants [30,31]. Previously, we isolated TaELP4 and demonstrated that it mediates immunity to Rhizoctonia cerealis in wheat and to Botrytis cinerea in Arabidopsis [32]. Although Elongator has been reported to regulate plant development and immunity, it is unknown whether elongator participates in cold stress.
The hormone ethylene functions directly in plant responses to freezing stress. Arabidopsis treated with the ethylene biosynthetic precursor ACC (1-aminocyclopropane-1-carboxylic acid) displayed decreased tolerance to freezing, but freezing tolerance was enhanced after the ethylene biosynthesis inhibitor AVG (aminoethoxyvinyl glycine) was applied. Mutants in the ethylene signaling pathway, including etr1-1, ein4-1, ein2-5, ein3-1, and ein3 eil1, showed enhanced freezing tolerance, while AtEIN3-overexpressing plants exhibited reduced freezing tolerance. Genetic and biochemical analyses revealed that ethylene negatively regulates cold signaling through AtEIN3 binding directly to the promoter of CBF1-3 and type-A ARR5, ARR7, and ARR15 genes, repressing its expression [33]. Type-A Arabidopsis response regulators (ARRs) act as negative regulators in cold stress signaling through the inhibition of the abscisic acid–dependent pathway [34]. In soybean, the ethylene-signaling pathway inhibits the CBF/DREB1 transcripts by EIN3, contributing to the relatively poor cold tolerance of soybean [35]. Moreover, ethylene negatively regulates the tolerance of Medicago to freezing during cold acclimation [36].
Although many studies have elucidated cold perception and response mechanisms in plants, few have been conducted in wheat. In this study, we identified a wheat elongator subunit 4 (TaELP4) that had negatively regulated freezing tolerance in wheat. Functional analysis showed that TaELP4 was induced by freezing stress; ectopic expression of TaELP4 in Arabidopsis accumulates more ethylene during freezing stress by elevating histone acetylation levels in coding/promoter regions of ACS2 and ACS6, and increasing their transcripts. The EIN2/EIN3/EIL1-CBFs-COR pathway was regulated by TaELP4. This work provides evidence that TaELP4 is involved in cold stress in wheat by histone acetylation and enriches research on the molecular mechanism of wheat response to cold stress. Moreover, TaELP4 might be an excellent gene that can be used to improve the freezing tolerance of wheat during breeding research.
2. Results
2.1. TaELP4 Transcripts Are Induced by Freezing Stress
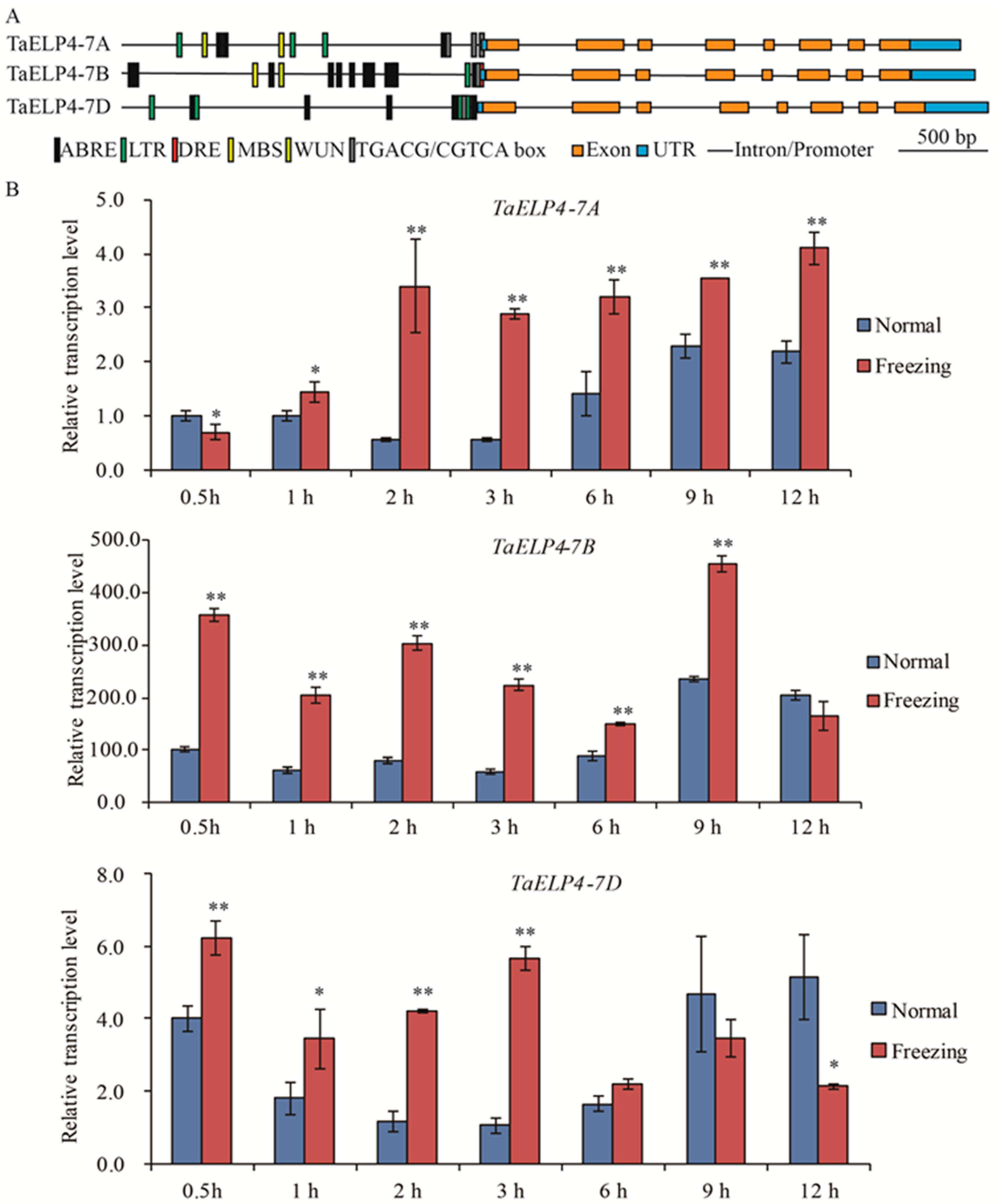
To investigate whether TaELP4 participated in abiotic stress, we first analyzed the promoter regions of TaELP4 homologs using PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/ (accessed on 10 September 2021)). Multiple regulatory elements that respond to cold stress, drought, wounding, abscisic acid (ABA), and jasmonic acid (JA) were identified, including LTR/DRE elements (CCGAAA/GCCGAC, cold stress), MBS (CAACTG, drought-induced), WUN box (AAATTTCTT, wounding), ABRE (ACGTG, ABA-induced), and TGACG box (TGACG, JA-induced) (Figure 1A). This information revealed that TaELP4 may be involved in abiotic stress responses, such as cold responses, drought, and hormones. Sequence BLAST showed that three homologs located on chromosomes 7A, 7B, and 7D (namely TaELP4-7A, TaELP4-7B, TaELP4-7D) existed in Chinese Spring RefSeq v1.0 chromosomes [37]. The analysis of coding sequence (CDS) and amino acids of TaELP4 using ClustalW software (https://www.genome.jp/tools-bin/clustalw (accessed on 1 July 2020)) showed that TaELP4-7A, TaELP4-7B, and TaELP4-7D had the same gene structure and high sequence similarity. Specifically, TaELP4-7A, TaELP4-7B, and TaELP4-7D all contained eight exons and seven introns. TaELP4-7A, TaELP4-7B, and TaELP4-7D contain a conserved PAXNEB domain (Figure S1), which is a typical domain of an RNA polymerase II Elongator protein subunit and is one part of the HAP subcomplex of elongator. The identities of CDS and amino acids ranged from 94% to 97% (Figures S2 and S3).
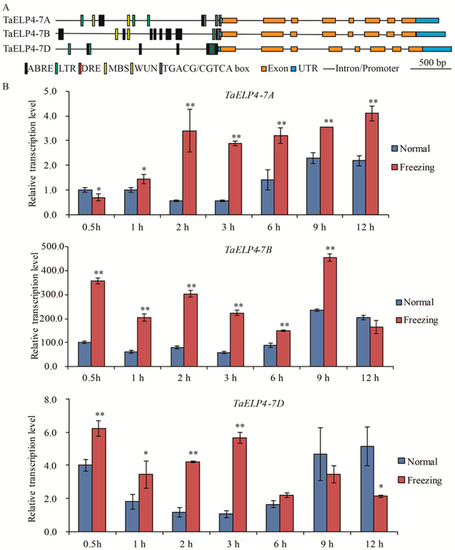
Figure 1.
Gene structure, promoter cis-acting element, and expression pattern analysis of TaELP4 homologs. (A) The gene structure of TaELP4 homologs, including 2000 bp region upstream of the ATG start codon in the promoters, untranslated region, intron, and exon. Promoter cis-elements analyzed using PLACE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/ (accessed on 10 September 2021)). (B) The relative transcript levels of TaELP4-7A, TaELP4-7B, and TaELP4-7D in shoots of Chinese Spring at 0.5, 1, 2, 3, 6, 9, and 12 h with −8 °C treatment (Freezing) or 23 °C (Normal). The transcript level of TaELP4-7A at 0.5 h of normal condition was set to 1. Asterisks (*, p < 0.05; **, p < 0.01) indicate significant differences between normal and freezing based on Student’s t-test. Bars indicate the standard deviation of the mean; TaActin was the internal control for normalization of gene expression.
To investigate the biological function of TaELP4 in detail, we first performed quantitative real-time PCR (qRT-PCR) to test the expression patterns after freezing treatment using genome-specific primers. Chinese Spring wheat was pre-treated at 4 °C for 2 h and then treated at −8 °C at the two to three leaf stage, while the control was cultured at 23 °C; the shoots were harvested at different times to test the expression pattern. Data showed that TaELP4-7A was significantly induced at 1 h, was highest at 2 h, and maintained the high level to 12 h. TaELP4-7B was induced at 0.5 h and was highest at 9 h. TaELP4-7D was significantly induced at 0.5 h and was highest at 3 h (Figure 1B). In general, TaELP4 was induced by freezing stress and TaELP4-7B might be more essential in the wheat response to freezing stress than TaELP4-7A and TaELP4-7D.
2.2. Silencing of TaELP4 Improves Wheat Tolerance to Freezing Stress
Considering that three TaELP4 homologs had high nuclear acid and amino acid identities (Figures S1–S3), they are likely to be functionally redundant. Thus, we silenced TaELP4 in wheat using the BSMV-mediated VIGS (virus induced gene silencing) approach to investigate the response of TaELP4 to freezing stress. The recombinant BSMV:TaELP4 construct was generated and inoculated into seedlings at the two-leaf stage. When seedlings grew to the four-leaf stage, we randomly selected 3 BSMV:GFP and 10 BSMV:TaELP4 wheat plants to test the silenced efficiency of VIGS. The RT-qPCR (real-time quantitative PCR) results showed that TaELP4 were all silenced and the transcripts substantially decreased 40-80% in BSMV:TaELP4-infected wheat compared to BSMV:GFP-infected plants based on primers that can simultaneously detect transcripts of TaELP4-7A, TaELP4-7B, and TaELP4-7D (Figure S4). This result implied that the three TaELP4 homologs were successfully silenced.
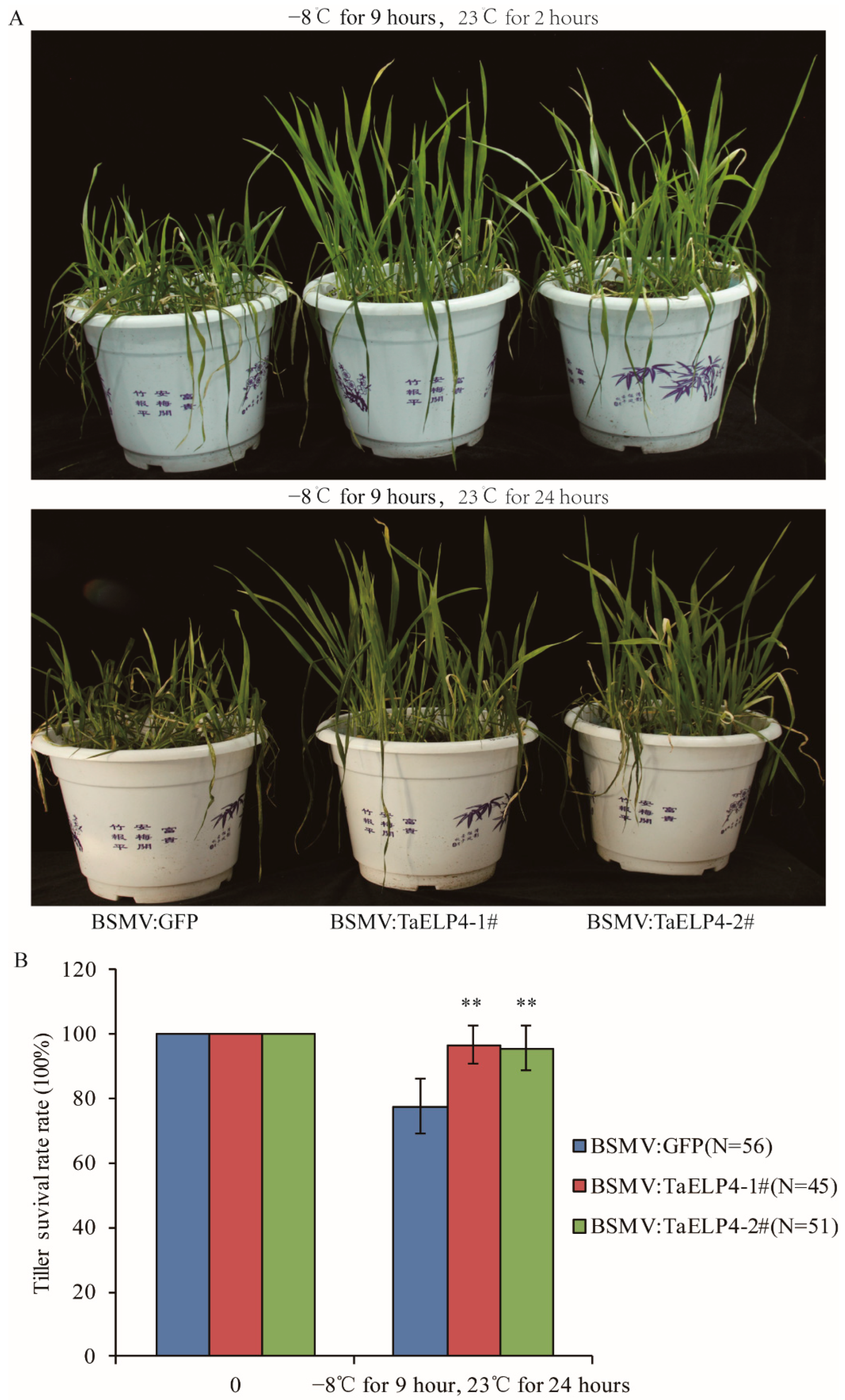
Those plants were pre-treated at 4 °C for 2 h, then treated at −8 °C for 9 h, and subsequently recovered at 23 °C. After recovery for 2 hours, most leaves of BSMV:GFP wheat were damaged and withered by freezing. Only the first or second leaves were damaged in BSMV:TaELP4 plants, while newer leaves stayed upright (Figure 2A). After recovery for 24 h, the damaged leaves and tillers turned yellow and dry in BSMV:GFP plants, while BSMV:TaELP4 wheat grew well (Figure 2A). Tiller survival rate showed 80% surviving tillers in BSMV:GFP plants, significantly lower than BSMV:TaELP4 wheat, which was nearly 100% (Figure 2B). To further study the influence of freezing on tillers, we kept the freeze-treated plants in the greenhouse. One month later, almost all BSMV:GFP plants died, and the tiller survival rate decreased to 10%, while BSMV:TaELP4 wheat grew well with normal jointing, with a tiller survival rate of 58% (Figure S5). These results imply that freezing stress severely damaged wheat growth and TaELP4-silenced wheat was more tolerant to freezing stress than control.

Figure 2.
Phenotypes and tiller survival rates of BSMV-TaELP4 and BSMV-GFP with freezing stress. (A) BSMV-TaELP4 and BSMV-GFP wheat seedlings at the four-leaf stage were pre-treated at 4 °C for 2 h, then treated at −8 °C for 9 h, and subsequently recovered at 23 °C for 2 and 24 h; the phenotypes are shown. (B) The surviving tillers from the heart leaf without freezing injury were calculated. N means numbers of tillers. Double asterisks (**, p < 0.01) indicate significant differences between BSMV-TaELP4 and BSMV-GFP wheat based on Student’s t-test. Bars indicate the standard deviation of the mean.
2.3. Ectopic-Expression of TaELP4 Decreased Tolerance to Freezing Stress in Arabidopsis
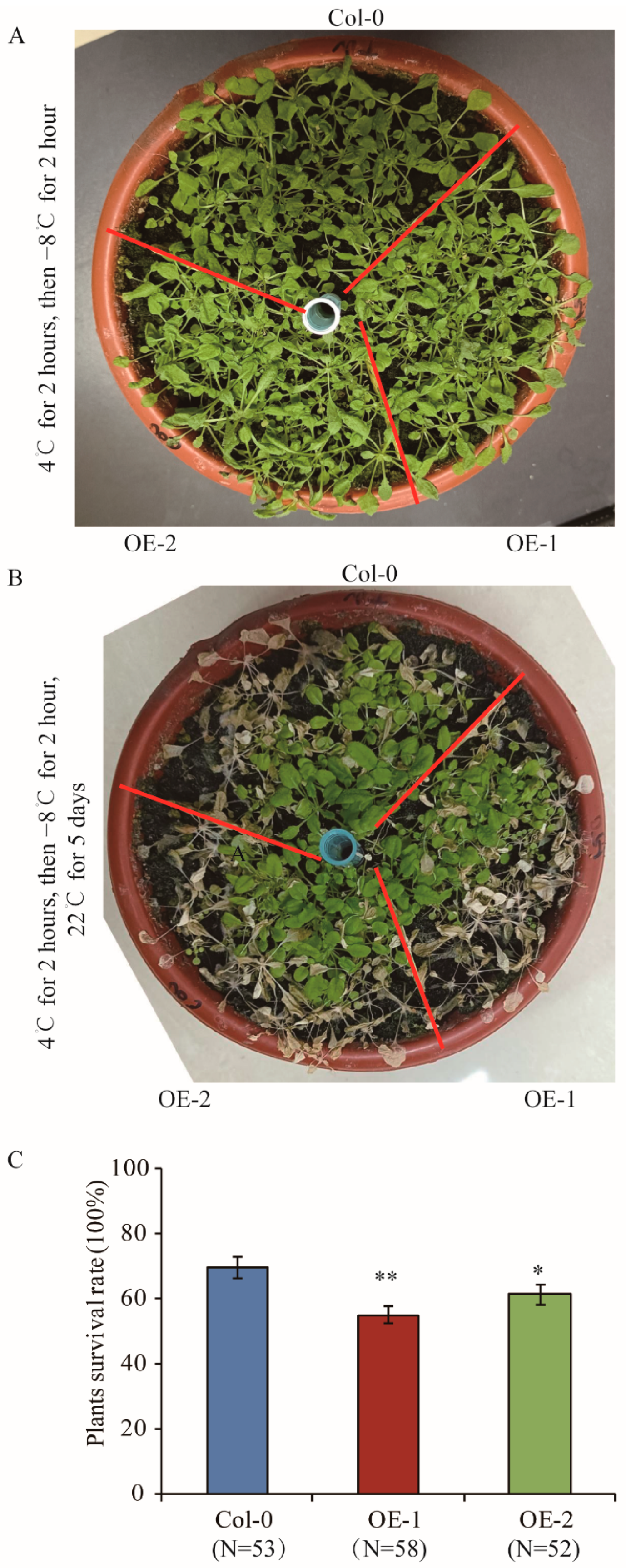
To investigate whether TaELP4 overexpressing lines were more sensitive to freezing stress, we over-expressed TaELP4-7B in Arabidopsis thaliana. Two representative TaELP4-overexpressing transgenic lines (OE-1 and OE-2) in the T3 generation were selected in this study. We first tested the expression of TaELP4 in Arabidopsis using confocal microscopy. GFP fluorescence signal showed that TaELP4 localized at the cytoplasm and nucleus (Figure S6). The Col-0 and transgenic lines were pre-treated at 4 °C for 2 h, then treated with −8 °C for 2 h, and finally recovered at 22 °C. After recovery for 2 h, there was no difference between TaELP4-overexpressing lines and Col-0 (Figure 3A). After recovery for 5 days, some plants withered and died, and OE-1 and OE-2 had more leaves with severe freezing injury than Col-0 (Figure 3B). Statistical analysis showed that plant survival rates of OE-1 and OE-2 were 52% and 59%, while Col-0 had a survival rate of 70% (Figure 3C). This demonstrated that TaELP4 overexpression decreased freezing tolerance in Arabidopsis.

Figure 3.
Phenotypes and plant survival rates of TaELP4-overexpression transgenic Arabidopsis and Col-0 with freezing stress. (A,B) Plants of TaELP4-overexpression transgenic Arabidopsis and Col-0 before flowering were pre-treated at 4 °C for 2 h, then treated at −8 °C for 2 h, and subsequently recovered at 22 °C for 2 h and 5 days; the phenotypes are shown. (C) The surviving plants without withering were calculated. N means numbers of plants. Asterisks (*, p < 0.05; **, p < 0.01) indicate significant differences between TaELP4-overexpression transgenic Arabidopsis and Col-0 based on Student’s t-test. Bars indicate the standard deviation of the mean.
Subcellular localization showed that TaELP4 localized at both the cytoplasm and nucleus in Arabidopsis, so we wanted to confirm that this localization was correct. We cloned TaELP4 and AtELP4 genes and separately constructed them into p16318h-GFP vector, then transformed them into wheat mesophyll protoplast cells. Subcellular localization assay showed that both TaELP4 and AtELP4 localized at the cytoplasm and nucleus and co-localized with a nuclear marker OsMADS3 (Figure S7A). We then extracted the total proteins to detect whether TaELP4 and AtELP4 were intact. Western blot showed that TaELP4-GFP and AtELP4-GFP were intact proteins with molecular weights of 68 kDa (Figure S7B). This result implied that TaELP4 might have the same function as AtELP4, and TaELP4 had normal function in Arabidopsis.
2.4. TaELP4 Regulated Ethylene Biosynthesis
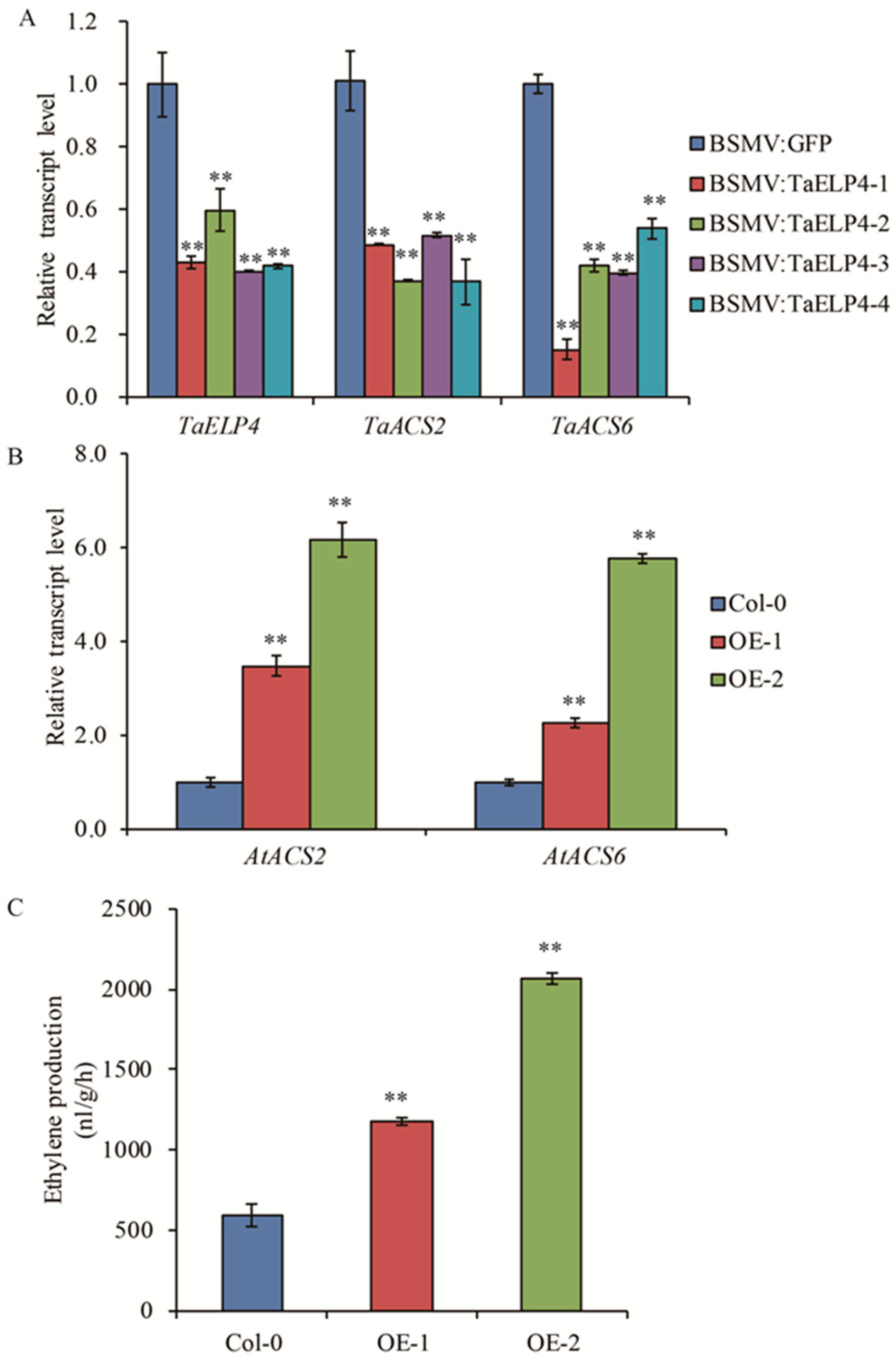
The hormone ethylene negatively regulates tolerance to freezing in Arabidopsis, soybean, and Medicago [33,35,36]. To investigate whether TaELP4 negatively regulates freezing-tolerance through the ethylene pathway, we tested the expression of ACS2 and ACS6, two key rate-limiting enzymes that catalyze the committing reaction in ethylene biosynthesis [38]. First, we tested the transcript levels of TaACS2 and TaACS6 in TaELP4-silenced and BSMV:GFP-infected plants after freezing treatment for 3 h. The RT-qPCR results showed that the transcripts of TaACS2 and TaACS6 were significantly decreased in TaELP4-silenced plants compared to BSMV:GFP plants (Figure 4A), suggesting that TaELP4 might regulate ethylene biosynthesis by regulating the transcripts of TaACS2 and TaACS6. We then tested the transcripts of AtACS2 and AtACS6 in OE-1 and OE-2 as well as in Col-0 after freezing treatment for 2 h. The RT-qPCR results showed that the transcripts of AtACS2 and AtACS6 were increased in OE-1 and OE-2 lines compared with Col-0 (Figure 4B), suggesting that TaELP4 positively regulates the expression of AtACS2 and AtACS6 in Arabidopsis.

Figure 4.
TaELP4 positively regulates expression of ACS2 and ACS6 and enhances ethylene content. (A,B) The relative transcript levels of ACS2 and ACS6 in TaELP4-silenced and -overexpressed plants and their control after freezing treatment. TaActin or AtActin was the internal controls for normalization of gene expression. (C) Ethylene production in TaELP4-overexpressed and Col-0 after freezing treatment. Double asterisks (**, p < 0.01) indicate significant differences based on Student’s t-test. Bars indicate the standard deviation of the mean.
The result that TaELP4 positively regulated AtACS2 and AtACS6 gene transcription in Arabidopsis prompted us to test the production of ethylene in OE-1, OE-2, and Col-0. We examined the ethylene production in transgenic plants and Col-0 plants after freezing treatment. We found that ethylene production in Col-0 was 637.8 nl/g/h, while the values in OE-1 and OE-2 were 1181.5 and 2067.8 nl/g/h, respectively (Figure 4C), which were 1.85 and 3.24-fold higher than Col-0. This result showed that TaELP4 transgenic plants accumulated high levels of ethylene in Arabidopsis during freezing stress.
2.5. TaELP4 Increases Histone H3K9/14ac Levels of AtACS2 and AtACS6 in Arabidopsis
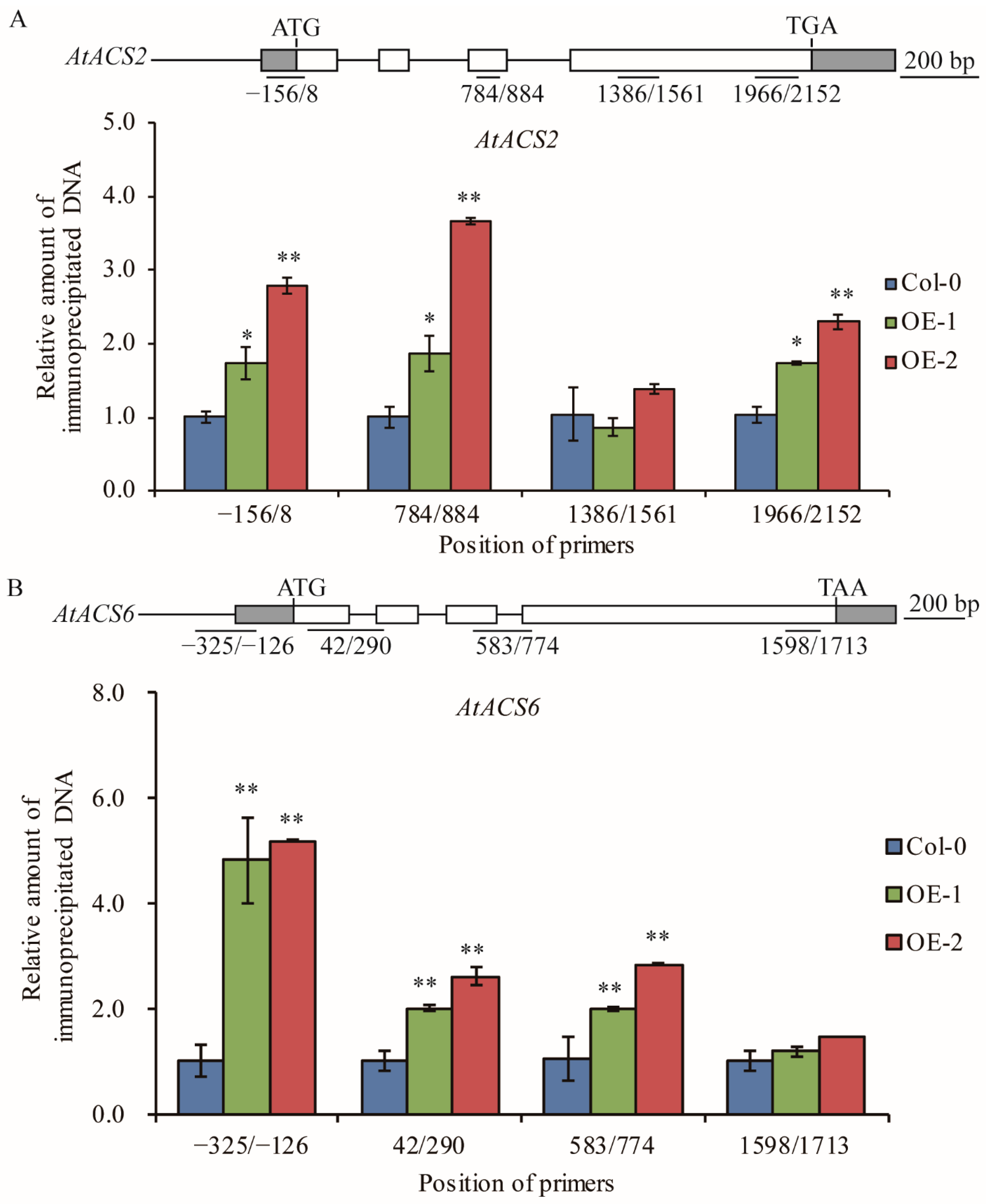
Previous papers reported that the acetylation level of histone H3 is generally associated with active transcription and TaELP4 epigenetically regulates immune responses to R. cerealis and B. cinerea in wheat and Arabidopsis through facilitating chromatin histone H3K9/14 acetylation of defense genes [32,39,40]. To test whether TaELP4 regulates histone acetylation levels in ACS2 and ACS6, ChIP assay via histone H3K9/14ac (Histone H3 acetylated at lysines 9 and 14) antibody was deployed to examine histone H3 acetylation level in regions of AtACS2 and AtACS6 in OE-1 and OE-2 as well as in Col-0. ChIP-qPCR results showed that histone H3K9/14ac levels in the promoter and coding regions of AtACS2 and AtACS6 were significantly higher in OE-1 and OE-2 compared to Col-0 (Figure 5). These data indicated that TaELP4 overexpression boosted histone H3K9/14ac levels in the promoter and coding regions of AtACS2 and AtACS6, which were consistent with higher transcript levels of AtACS2 and AtACS6 in TaELP4-transgenic plants than in Col-0.
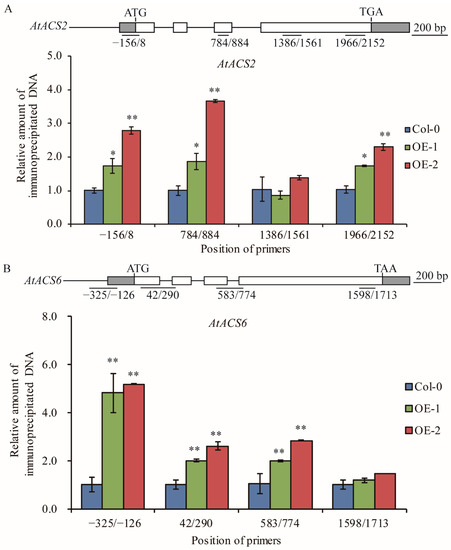
Figure 5.
Histone H3K9/14 acetylation levels of AtACS2 (A) and AtACS6 (B) in TaELP4-overexpressed transgenic Arabidopsis and Col-0. The position of the primers is relative to the initiation ATG codon. The relative amount of immunoprecipitated chromatin fragments (as determined by qPCR) from TaELP4-overexpressing lines were compared with that from Col-0. AtActin was the internal control. Asterisks (*, p < 0.05; **, p < 0.01) indicate significant differences based on Student’s t-test. Bars indicate standard error of the mean.
2.6. TaELP4 Regulates the EIN3/EIL1-CBFs-CORs Pathway
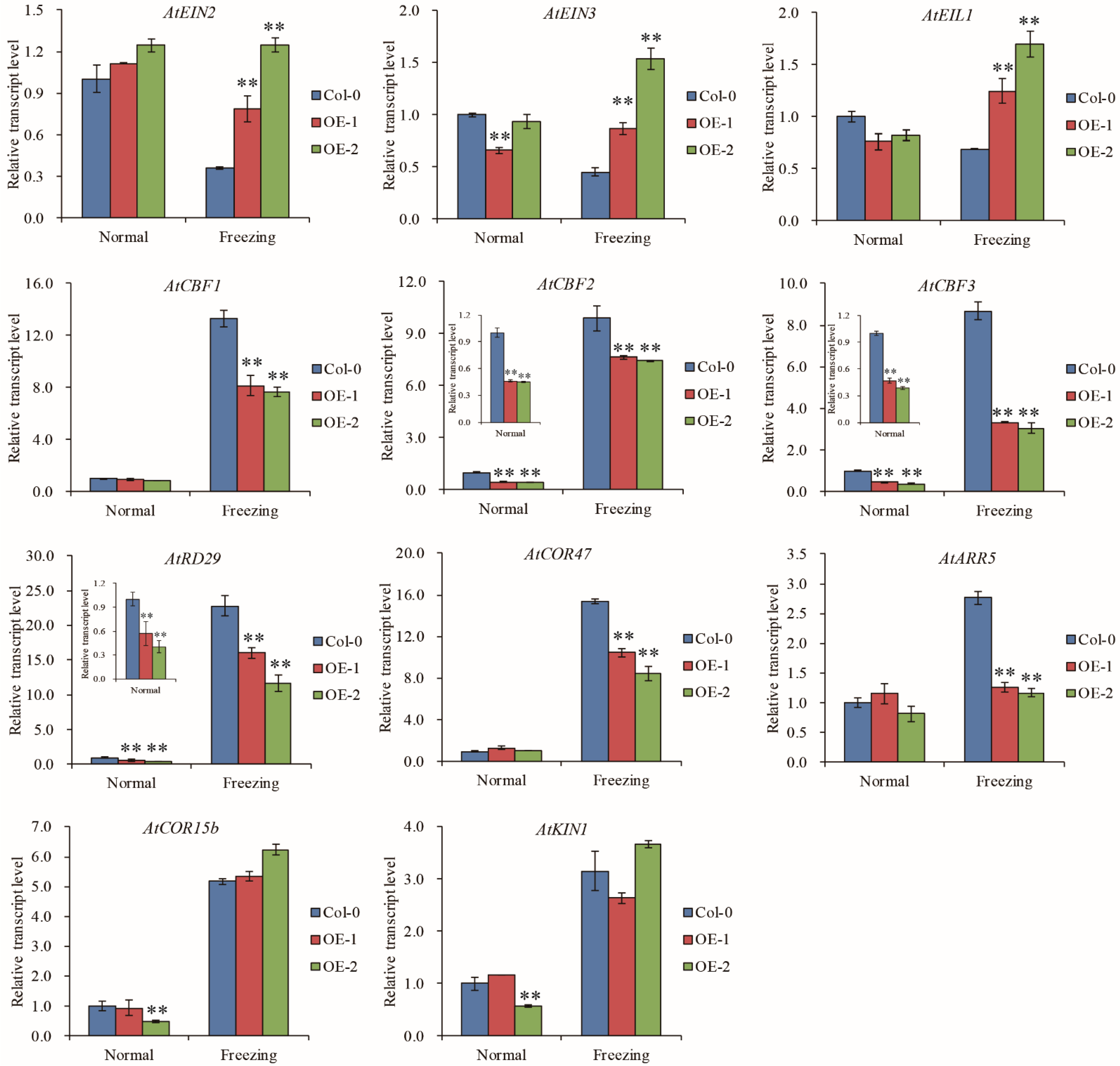
In Arabidopsis, ethylene negatively regulates cold signaling partially through direct transcriptional control of cold-regulated CBFs-COR genes by EIN3 [33]. We tested whether TaELP4 regulated the EIN3/EIL1-CBFs-CORs pathway during responses to freezing stress. RT-qPCR showed that AtEIN2, AtEIN3, and AtEIL1 had no differences between TaELP4-overexpressing lines and Col-0 under normal conditions. After freezing stress, AtEIN2, AtEIN3, and AtEIL1 transcripts in Col-0 were decreased, and the transcripts were significantly higher in OE-1 and OE-2 than in Col-0, implying that TaELP4 boosted AtEIN2, AtEIN3, and AtEIL1 transcription under freezing stress. We next tested the expression levels of AtCBF1, AtCBF2, and AtCBF3, and showed that AtCBF2 and AtCBF3 were down-regulated in OE-1 and OE-2 compared with Col-0 under normal conditions and freezing stress, while AtCBF1 showed no differences under normal conditions and was significantly lower in OE-1 and OE-2 than in Col-0 under freezing stress. This demonstrated that TaELP4 down regulated AtCBFs expression. In Arabidopsis, three CBFs (CBF1, CBF2, and CBF3) bind to the promoters of COR genes and then activate expression in a large subset of COR genes under cold stress, including AtRD29, AtCOR15b, AtCOR47, and AtKIN1 [33,41,42]. We further detected the expression levels of selected COR genes and showed that AtRD29 and AtCOR47 were down regulated in OE-1 and OE-2 compared with Col-0 during freezing stress, while AtCOR15b and AtKIN1 showed no difference, which implied that TaELP4 regulated AtRD29 and AtCOR47 expression. Moreover, we found that TaELP4 overexpression decreased type-A ARR genes AtARR5 under freezing stress (Figure 6). Those results showed that TaELP4 regulated the EIN3/EIL1-CBFs-COR pathway to respond to freezing.
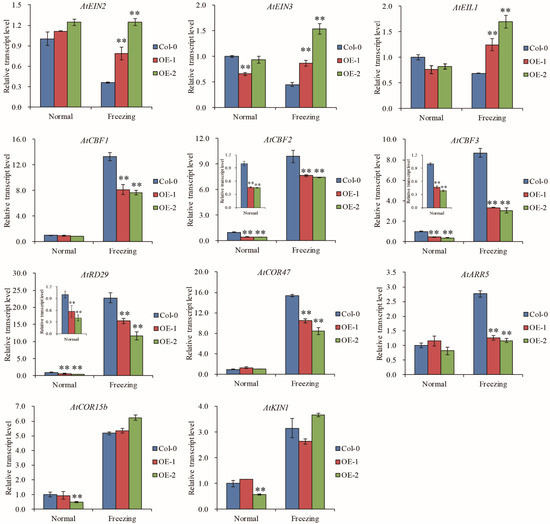
Figure 6.
The relative expression levels of ethylene-signaling components, CBFs, and selected COR genes in TaELP4-overexpressing lines and Col-0. The relative expression levels of AtEIN2, AtEIN3, AtEIL1, AtCBF1, AtCBF2, AtCBF3, AtRD29, AtCOR47, AtARR5, AtCOR15b, and AtKIN1 in TaELP4-overexpressing lines and Col-0 with or without freezing treatment. The transcription level in Col-0 was set to 1, AtActin was the internal control for normalization of gene expression. Asterisks (**, p < 0.01) indicate significant differences based on Student’s t-test. Bars indicate the standard deviation of the mean.
3. Discussion
Cold stress often occurs in the growth season of crops, including wheat and rice [2,10]. The genetic and molecular mechanisms of plant responses to cold stress have been identified in rice and Arabidopsis, but are much less known in wheat. In this study, we found that TaELP4 was a negative regulator in wheat responses to cold stress. Several LTR/DRE elements that responded to cold stress existed in the promoter of TaELP4, and the expression of TaELP4 homologs were induced by freezing at different times (Figure 1), implying that TaELP4 participates in freezing stress. Further evidence showed that TaELP4 is a negative regulator of freezing tolerance. When we silenced three TaELP4 homologs simultaneously in wheat, BSMV:TaELP4 wheat significantly improved resistance to freezing stress compared to BSMV:GFP (Figure 2 and Figure S5). When we overexpressed TaELP4 in Arabidopsis, TaELP4 transgenic lines showed severe injury and a higher death rate than Col-0 after freezing treatment (Figure 3). Those results further showed that TaELP4 negatively regulates freezing tolerance. We know that the soil is a buffer preventing rapid effects of temperature, however, in this freezing experiment, we can see the Arabidopsis that lived at the edge of the pot that died first; we speculated that −8 °C may cause the soil to become frozen and that may have caused severe damage to plants with underdeveloped roots. Once thawed, the roots cannot quickly recover to meet normal growth, resulting in the death of plants. Previous papers documented that AtELP1-6 complex regulates plants growth, development, and immunity [19]. In wheat, TaELP4, acting as a functional regulator, is required for immune the response of wheat to R. cerealis infection as with the response of Arabidopsis to B. cinerea [32]. Thus, we deduced that ELP4 has multiple functions.
Ethylene is a versatile phytohormone, and ethylene-signaling participates in cold stress [43]. In Arabidopsis, ethylene-insensitive mutants, including ein2 and ein3 eil1, displayed enhanced tolerance to freezing stress by increasing the expression of cold-inducible CBF1/2/3 and type-A ARR genes [33]. Cold stress induces CBFs expression, which can activate expression in a series of COR genes [41,42]. Here, we found that TaELP4 positively regulated ACS2 and ACS6 expression, two key rate-limiting enzymes that catalyze the committing reaction in ethylene biosynthesis, and elevated ethylene accumulation after freezing stress (Figure 4). These results implied that TaELP4 might regulate freezing tolerance through ethylene signaling. Next, we tested the expression of ethylene signaling transduction components and downstream genes and showed that TaELP4 overexpression increased the expression of AtEIN2, AtEIN3, and AtEIL1, and reduced the expression of AtCBF1, AtCBF2, ACBF3, AtRD29, AtCOR47, and AtARR5 (Figure 6). These results demonstrated that TaELP4 overexpression elevated ethylene content and regulated the EIN2/EIN3/EIL1-CBFs-COR pathway after freezing stress.
Elongator complex was originally identified in yeast as a histone acetyltransferase complex that activates RNAPII [17]. The acetylation level of histone H3 is generally associated with active transcription [28,29,39,40]. A recent study showed that TaELP4 up-regulates transcription of a subset of defense-related genes through elevating the histone H3K9/14ac levels of their chromatin, in turn boosting plant innate immune responses [32]. In this study, we demonstrated that TaELP4 overexpression in Arabidopsis elevated the histone H3K9/14ac levels of AtACS2 and AtACS6 in the promoter and gene body region (Figure 5). This result implies that TaELP4 overexpression increases H3K9/14ac levels of AtACS2 and AtACS6, and then elevates their expression during freezing stress. This result links TaELP4 overexpression to enhanced ethylene content during freezing. However, how TaELP4 directly affects histone acetylation level is not clear, and whether there are expression or sequence differences of TaELP4 between winter wheat and spring wheat is not clear, and needs to be studied in a further study.
4. Materials and Methods
4.1. Plant Materials
Chinese Spring wheat (Triticum aestivum L.) was used to investigate TaELP4 transcription profiles and in VIGS experiments. Arabidopsis thaliana Columbia-0 (Col-0) ecotype was used to ectopically-express TaELP4-7B.
4.2. Freezing Tolerance Assay and Phenotype Analyses
Wheat plants were grown in a greenhouse at 23 °C/14 h light (250–300 μmol m−2 s−1) and 10 °C/10 h dark. Seedlings at the four-leaf stage were pre-treated at 4 °C for 2 h, then treated at −8 °C for 9 h, and subsequently recovered at 23 °C. The wheat growth state was photographed and the tiller survival rate was calculated. The tiller survival rate (%) was calculated as: 100% × dead tiller number/total tiller number. Arabidopsis were grown in a greenhouse at 22 °C/14 h light (100–150 μmol m−2 s−1) and 22 °C/10 h dark. Plants before flowering (about 20 days after germination) were cultured at 4 °C for 2 h, then treated at −8 °C for 2 h, and subsequently recovered at 22 °C. After recovery for 5 days, the plant survival rate (%) was calculated as 100% × dead plant number/total plant number. The plants with all dead rosette leaves were defined as death after recovery for 5 days. The freezing experiments were repeated at least twice and processed in the dark.
4.3. RNA Extraction and RT-qPCR
Total RNA was extracted from wheat or Arabidopsis samples using Trizol reagent (Invitrogen, Carlsbad, CA, USA). RNA was first purified and then checked for integrity. For RT-qPCR, total RNA was reverse-transcribed to cDNA using the FastQuant RT Kit (Tiangen, Beijing, China). RT-qPCR was carried out in a Roche LightCycler480 system using SYBR Premix Ex Taq kit (TaKaRa, Shiga, Japan). Reactions were set up using the following thermal cycling profile: 95 °C for 30 s followed by 40 cycles of 95 °C for 5 s, 58 °C for 30 s, and 72 °C for 34 s. Each experiment was replicated three times. The relative expression of the target genes was calculated using the 2−ΔΔCT method [44], with the wheat Actin gene (TaActin) or Arabidopsis actin gene (AtActin) used as the internal reference genes for the analysis.
4.4. Plasmid Construction and Plant Transformation
To generate BSMV-TaELP4 vector for barley stripe mosaic virus (BSMV)-mediated VIGS experiment, a 272 bp fragment matching the CDS of TaELP4-7A, TaELP4-7B, and TaELP4-7D (from 414 to 686 nucleotides in TaELP4-7B sequence) was selected based on SiFi software and then subcloned in antisense orientation into the Nhe I restriction site of the RNA γ of BSMV. The inoculation method followed the protocols described by Holzberg et al. [45] and Wang et al. [32].
To generate the 35S:TaELP4-7B-GFP vector for ectopic expression, the CDS of TaELP4-7B was amplified and cloned into pCAMBIA1300-GFP (Hind III) using in-fusion PCR cloning systems. The resulting 35S:TaELP4-7B-GFP vector was introduced into Arabidopsis Col-0 plants by Agrobacterium-mediated transformation [46]. Transgenic plants were selected by hygromycin B, and T3 homozygous transgenic plants were used in this study.
To generate 35S:TaELP4-7B-GFP, 35S:AtELP4-GFP vector for subcellular localization and the CDS of TaELP4-7B or AtELP4 were respectively amplified and cloned into p16318h-GFP (BamH I) using in-fusion PCR cloning systems.
4.5. Subcellular Localization and Western Blot
To detect the subcellular localization of TaELP4 and AtELP4 proteins, 35S:TaELP4-7B-GFP and 35S:AtELP4-GFP vectors were transformed into wheat protoplasts via PEG-calcium transfection method, following the protocol of Yoo et al. [47]. Nuclear marker OsMADS3 was used as the co-localization marker. GFP fluorescence was observed by confocal microscopy (ZEISS710; Carl Zeiss, Jena, Germany) with excitation light at 488 nm.
For Western blot, the transformed wheat protoplasts were collected by centrifugation at 100× g for 2 min, and then total proteins were extracted using plant protein extraction kit (CW0885M, CWBIO, Taizhou, China). Total soluble proteins (~20 μg) were separated on 12% SDS-PAGE and transferred to polyvinyl difluoride membranes (Amersham). The blotting membranes were incubated with 2500-fold diluted Anti-GFP Mouse Monoclonal Antibody (TransGen Biotech, Beijing, China) at 4 °C overnight and then incubated with 4000-fold diluted Goat Anti-Mouse IgG (H + L), HPR conjugated secondary antibody (TransGen Biotech, China) at 22–23 °C for 1 h. The fusion protein was visualized using the Pro-light HRP Chemiluminescent Kit (TransGen Biotech, China).
4.6. Measurement of Ethylene Content
Ethylene emission was measured with a gas chromatograph as described previously [48]. In brief, seedlings (20–25) of Col-0 and TaELP4-overexpression transgenic plants were grown in 15 mL gas chromatography vials containing 5 mL 1/2 Murashige & Skoog medium. Seedlings were cultured under a 16 h: 8 h, light: dark cycle at 22 °C for 2 weeks. Seedlings were pre-treated at 4 °C for 2 h, and then treated at −8 °C for 2 h, and finally the vials were sealed for 12 h at 22 °C. In total, 1 mL of gas from each vial was used to analyze the ethylene emissions with a gas chromatograph (Hitachi, Tokyo, Japan).
4.7. Chromatin Immunoprecipitation with Acetylated-Histone3 Lysine 9/14 and qPCR
Chromatin immunoprecipitation (ChIP) was performed as described by Gendrel et al. [49]. Briefly, approximately 2.0 g of Arabidopsis plants after freezing treatment were cross-linked in 1% formaldehyde under a vacuum. Chromatin was extracted and fragmented via ultrasound treatment to a size of 200–500 bp. The fragments were incubated with acetylated Histone H3 K9/K14ac (Histone H3 acetylated at lysines 9 and 14) antibody (Santa Cruz Biotechnology, Heidelberg, Germany), and the immune complex was collected by Dynabeads protein G (Invitrogen). After extensive washing, the immunoprecipitated chromatin was recovered using the phenol-chloroform method. The amount of precipitated DNA corresponding to a specific gene region was determined by qPCR and normalized by both input DNA and a constitutively expressed gene AtActin.
4.8. Statistical Analyses
All experimental results are means ± standard deviations of at least three independent replicates. Student’s t-test was used for comparing two data sets in the SPSS System.
4.9. Accession Numbers
Sequence data from this article can be found in the EnsemblPlants (http://plants.ensembl.org/index.html (accessed on 2 June 2021)) or TAIR (https://www.arabidopsis.org/ (accessed on 2 June 2021)) databases under accession numbers: TaELP4-7A (TraesCS7A02G522900), TaELP4-7B (TraesCS7D02G512100), TaELP4-7D (TraesCS7B02G439900), TaACS2 (TraesCS2B02G414800), TaACS6 (TraesCS2A02G396400), TaActin (TraesCS5D02G516200), AtACS2 (AT1G01480), AtACS6 (AT4G11280), AtEIN2 (At5g03280), AtEIN3 (At3g20770), AtEIL1 (At2g27050), AtCBF1 (At4g25490), AtCBF2 (At4g25470), AtCBF3 (At4g25480), AtRD29 (At5g52310), AtCOR15b (At2g42530), AtCOR47 (At1g20440), AtKIN1 (At5g15960), AtARR5 (At3g48100), and AtActin (At3g18780).
5. Conclusions
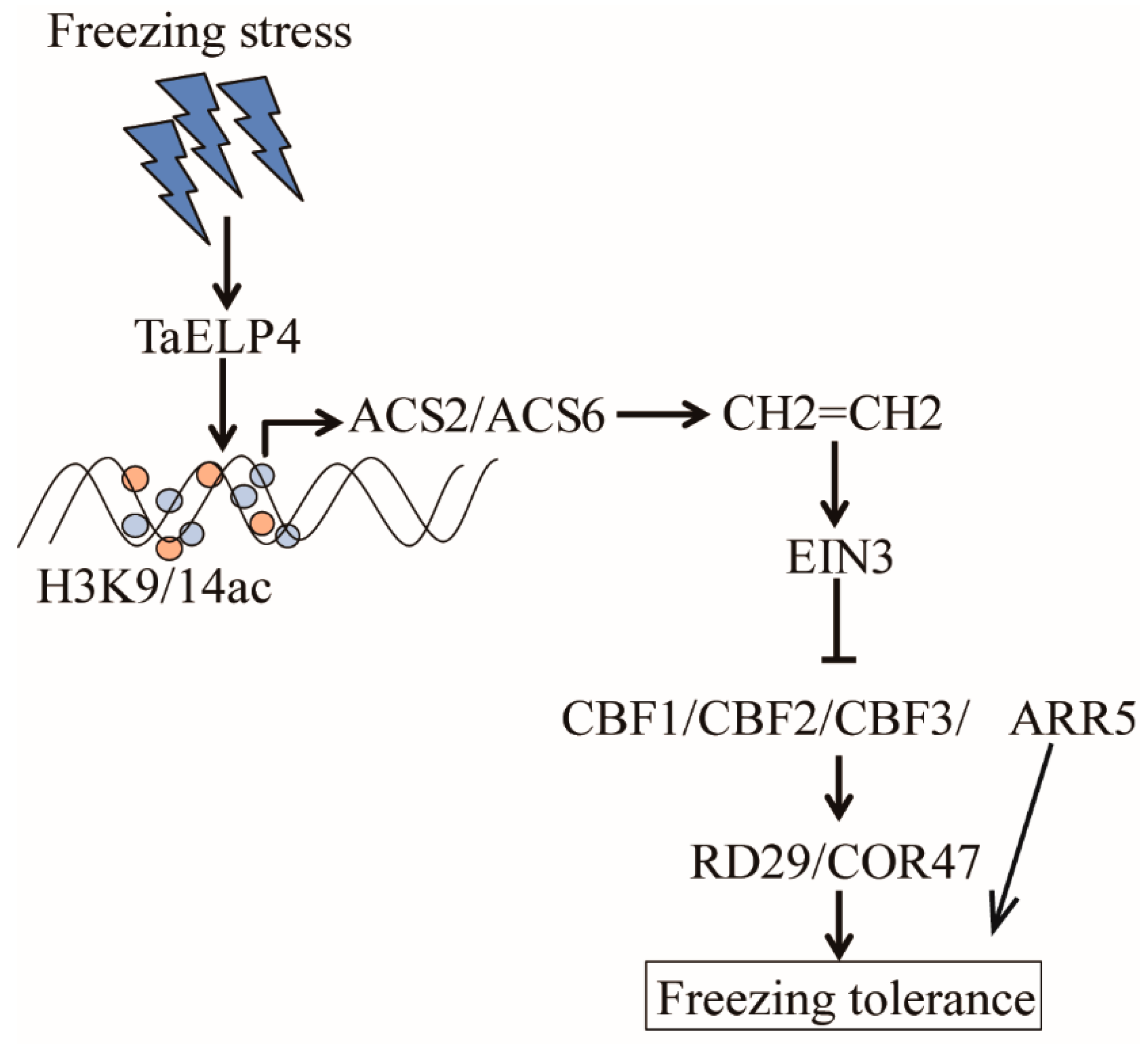
TaELP4 negatively regulates freezing tolerance by regulating ethylene signaling. TaELP4 was induced by freezing stress, which elevates the histone H3K9/14ac levels of AtACS2 and AtACS6, and increases the expression of AtACS2 and AtACS6, resulting in more ethylene content being accumulated. Ethylene signaling negatively regulates freezing tolerance through CBFs-COR pathway (Figure 7). This study provides insights on how TaELP4 negatively regulates freezing tolerance in monocot plant species.

Figure 7.
Proposed model of TaELP4 negatively regulates freezing tolerance. Freezing stress induces TaELP4 expression, which elevates the histone H3K9/14ac levels of AtACS2 and AtACS6, and increases expression of AtACS2 and AtACS6, resulting in more ethylene content accumulated. Ethylene signaling negatively regulates CBFs-COR pathway to reduce freezing tolerance.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms23147634/s1.
Author Contributions
C.L. conceived and supervised the research plan and designed the experiments; K.W. and M.Z. performed the majority of these experiments; R.H., X.W., W.X., X.Z. and G.Q. provided technical assistance; C.L. and K.W. wrote the paper; T.K. provided suggestions. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (31971874), Taishan Scholars Project (tsqn201812123), the Shandong Agricultural Seed Improvement Project (2019LZGC016), and the Research Start-up Foundation for Young Talent of Shandong Academy of Agricultural Sciences to Kai Wang (CXGC2022E01), Double Hundred Talent Program of Shandong Province (WST2020012).
Informed Consent Statement
Not applicable.
Acknowledgments
The authors are very grateful to Zengyan Zhang (Institute of Crop Science, Chinese Academy of Agricultural Sciences) for providing seeds of TaELP4 overexpression transgenic Arabidopsis.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Tchagang, A.B.; Fauteux, F.; Tulpan, D.; Pan, Y.L. Bioinformatics identification of new targets for improving low temperature stress tolerance in spring and winter wheat. BMC Bioinform. 2017, 18, 174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, X.Y.; Liu, D.F.; Chong, K. Cold signaling in plants: Insights into mechanisms and regulation. J. Integr. Plant Biol. 2018, 60, 745–756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, L.J.; Liu, L.L.; Asseng, S.; Xia, Y.M.; Tang, L.; Liu, B.; Cao, W.X.; Zhu, Y. Estimating spring frost and its impact on yield across winter wheat in China. Agr. Forest Meteorol. 2018, 260, 154–164. [Google Scholar] [CrossRef]
- Holman, J.D.; Schlegel, A.J.; Thompson, C.R.; Lingenfelser, J.E. Influence of precipitation, temperature, and 56 years on winter wheat yields in western Kansas. Crop Manag. 2011, 10, 1–10. [Google Scholar] [CrossRef]
- Trnka, M.; Rötter, R.P.; Ruiz-Ramos, M.; Kersebaum, K.C.; Olesen, J.E.; Žalud, Z.; Semenov, M.A. Adverse weather conditions for European wheat production will become more frequent with climate change. Nat. Clim. Chang. 2014, 4, 637–643. [Google Scholar] [CrossRef]
- Zheng, B.Y.; Chapman, S.C.; Christopher, J.T.; Frederiks, T.M.; Chenu, K. Frost trends and their estimated impact on yield in the Australian wheatbelt. J. Exp. Bot. 2015, 66, 3611–3623. [Google Scholar] [CrossRef] [Green Version]
- Frederiks, T.M.; Christopher, J.T.; Sutherland, M.W.; Borrell, A.K. Post-head-emergence frost in wheat and barley: Defining the problem, assessing the damage, and identifying resistance. J. Exp. Bot. 2015, 66, 3487–3498. [Google Scholar] [CrossRef] [Green Version]
- Thakur, P.; Kumar, S.; Malik, J.A.; Berger, J.D.; Nayyar, H. Cold stress effects on reproductive development in grain crops: An overview. Environ. Exp. Bot. 2010, 67, 429–443. [Google Scholar] [CrossRef]
- Jame, Y.W.; Cutforth, H.W. Simulating the effects of temperature and seeding depth on germination and emergence of spring wheat. Agr. Forest Meteorol. 2004, 124, 207–218. [Google Scholar] [CrossRef]
- Hussain, H.A.; Hussain, S.; Khaliq, A.; Ashraf, U.; Anjum, S.A.; Men, S.; Wang, L. Chilling and drought stresses in crop plants: Implications, cross talk, and potential management opportunities. Front. Plant Sci. 2018, 9, 393. [Google Scholar] [CrossRef]
- Fuller, M.P.; Fuller, A.M.; Kaniouras, S.; Christophers, J.; Fredericks, T. The freezing characteristics of wheat at ear emergence. Eur. J. Agron. 2007, 26, 435–441. [Google Scholar] [CrossRef]
- Valluru, R.; Link, J.; Claupein, W. Consequences of early chilling stress in two Triticum species: Plastic responses and adaptive significance. Plant Biol. 2012, 14, 641–651. [Google Scholar] [CrossRef] [PubMed]
- Li, X.N.; Cai, J.; Liu, F.L.; Zhou, Q.; Dai, T.B.; Cao, W.X.; Jiang, D. Wheat plants exposed to winter warming are more susceptible to low temperature stress in the spring. Plant Growth Regul. 2015, 77, 11–19. [Google Scholar] [CrossRef]
- Guo, J.; Ren, Y.K.; Tang, Z.H.; Shi, W.P.; Zhou, M.X. Characterization and expression profiling of the ICE-CBF-COR genes in wheat. PeerJ 2019, 7, e8190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Badawi, M.; Reddy, Y.V.; Agharbaoui, Z.; Tominaga, Y.; Danyluk, J.; Sarhan, F.; Houde, M. Structure and functional analysis of wheat ICE (inducer of CBF expression) genes. Plant Cell Physiol. 2008, 49, 1237–1249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Otero, G.; Fellows, J.; Li, Y.; de Bizemont, T.; Dirac, A.M.; Gustafsson, C.M.; Erdjument-Bromage, H.; Tempst, P.; Svejstrup, J.Q. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol. Cell. 1999, 3, 109–118. [Google Scholar] [CrossRef]
- Krogan, N.J.; Greenblatt, J.F. Characterization of a six-subunit holo-elongator complex required for the regulated expression of a group of genes in Saccharomyces cerevisiae. Mol. Cell Biol. 2001, 21, 8203–8212. [Google Scholar] [CrossRef] [Green Version]
- Hawkes, N.A.; Otero, G.; Winkler, G.S.; Marshall, N.; Dahmus, M.E.; Krappmann, D.; Scheidereit, C.; Thomas, C.L.; Schiavo, G.; Erdjument-Bromage, H.; et al. Purification and characterization of the human elongator complex. J. Biol. Chem. 2002, 277, 3047–3052. [Google Scholar] [CrossRef] [Green Version]
- Nelissen, H.; Fleury, D.; Bruno, L.; Robles, P.; De Veylder, L.; Traas, J.; Micol, J.L.; Van Montagu, M.; Inzé, D.; Van Lijsebettens, M. The elongata mutants identify a functional Elongator complex in plants with a role in cell proliferation during organ growth. Proc. Natl. Acad. Sci. USA 2005, 102, 7754–7759. [Google Scholar] [CrossRef] [Green Version]
- Ding, Y.Z.; Mou, Z.L. Elongator and its epigenetic role in plant development and responses to abiotic and biotic stresses. Front. Plant Sci. 2015, 6, 296. [Google Scholar] [CrossRef] [Green Version]
- Winkler, G.S.; Petrakis, T.G.; Ethelberg, S.; Tokunaga, M.; Erdjument-Bromage, H.; Tempst, P.; Svejstrup, J.Q. RNA polymerase II elongator holoenzyme is composed of two discrete subcomplexes. J. Biol. Chem. 2001, 276, 32743–32749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chinenov, Y. A second catalytic domain in the Elp3 histone acetyltransferases: A candidate for histone demethylase activity? Trends Biochem. Sci. 2002, 27, 115–117. [Google Scholar] [CrossRef]
- DeFraia, C.T.; Zhang, X.D.; Mou, Z.L. Elongator subunit 2 is an accelerator of immune responses in Arabidopsis thaliana. Plant J. 2010, 64, 511–523. [Google Scholar] [CrossRef] [PubMed]
- Glatt, S.; Létoquart, J.; Faux, C.; Taylor, N.M.; Séraphin, B.; Müller, C.W. The Elongator subcomplex Elp456 is a hexameric RecA-like ATPase. Nat. Struct. Mol. Biol. 2012, 19, 314–320. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.J.; Zhao, W.J.; Diao, W.T.; Xie, X.Q.; Wang, Z.; Zhang, J.X.; Shen, Y.Q.; Long, J.F. Crystal structure of elongator subcomplex Elp4-6. J. Biol. Chem. 2012, 287, 21501–21508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelissen, H.; De Groeve, S.; Fleury, D.; Neyt, P.; Bruno, L.; Bitonti, M.B.; Vandenbussche, F.; Van der Straeten, D.; Yamaguchi, T.; Tsukaya, H.; et al. Plant Elongator regulates auxin-related genes during RNA polymerase II transcription elongation. Proc. Natl. Acad. Sci. USA 2010, 107, 1678–1683. [Google Scholar] [CrossRef] [Green Version]
- Mehlgarten, C.; Jablonowski, D.; Wrackmeyer, U.; Tschitschmann, S.; Sondermann, D.; Jäger, G.; Gong, Z.; Byström, A.S.; Schaffrath, R.; Breunig, K.D. Elongator function in tRNA wobble uridine modification is conserved between yeast and plants. Mol. Microbiol. 2010, 76, 1082–1094. [Google Scholar] [CrossRef]
- Wang, C.G.; Ding, Y.Z.; Yao, J.; Zhang, Y.P.; Sun, Y.J.; Colee, J.; Mou, Z.L. Arabidopsis Elongator subunit 2 positively contributes to resistance to the necrotrophic fungal pathogens Botrytis cinerea and Alternaria brassicicola. Plant J. 2015, 83, 1019–1033. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.S.; An, C.F.; Zhang, X.D.; Yao, J.Q.; Zhang, Y.P.; Sun, Y.J.; Yu, F.H.; Amador, D.M.; Mou, Z.L. The Arabidopsis elongator complex subunit2 epigenetically regulates plant immune responses. Plant Cell 2016, 25, 762–776. [Google Scholar] [CrossRef] [Green Version]
- Silva, K.J.P.; Brunings, A.M.; Pereira, J.A.; Peres, N.A.; Folta, K.M.; Mou, Z. The Arabidopsis ELP3/ELO3 and ELP4/ELO1 genes enhance disease resistance in Fragaria vesca L. BMC Plant Biol. 2017, 17, 230. [Google Scholar] [CrossRef] [Green Version]
- Pereira, J.A.; Yu, F.H.; Zhang, Y.P.; Jones, J.B.; Mou, Z.L. The Arabidopsis Elongator subunit ELP3 and ELP4 confer resistance to bacterial speck in tomato. Front. Plant Sci. 2018, 9, 1066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, K.; Rong, W.; Liu, Y.P.; Li, H.; Zhang, Z.Y. Wheat Elongator subunit 4 is required for epigenetic regulation of host immune response to Rhizoctonia cerealis. Crop J. 2020, 8, 565–576. [Google Scholar] [CrossRef]
- Shi, Y.T.; Tian, S.W.; Hou, L.Y.; Huang, X.Z.; Zhang, X.Y.; Guo, H.W.; Yang, S.H. Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-A ARR genes in Arabidopsis. Plant Cell 2012, 24, 2578–2595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeon, J.; Kim, N.Y.; Kim, S.; Kang, N.Y.; Novák, O.; Ku, S.J.; Cho, C.; Lee, D.J.; Lee, E.J.; Strnad, M.; et al. A subset of cytokinin two-component signaling system plays a role in cold temperature stress response in Arabidopsis. J. Biol. Chem. 2010, 285, 23371–23386. [Google Scholar] [CrossRef] [Green Version]
- Robison, J.D.; Yamasaki, Y.; Randall, S.K. The ethylene signaling pathway negatively impacts CBF/DREB-regulated cold response in Soybean (Glycine max). Front. Plant Sci. 2019, 10, 121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, M.G.; Liu, W.J.; Xia, X.Z.; Wang, T.Z.; Zhang, W.H. Cold acclimation-induced freezing tolerance of Medicago truncatula seedlings is negatively regulated by ethylene. Physiol. Plant 2014, 152, 115–129. [Google Scholar] [CrossRef]
- Ma, S.W.; Wang, M.; Wu, J.H.; Guo, W.L.; Chen, Y.M.; Li, G.W.; Wang, Y.P.; Shi, W.M.; Xia, G.M.; Fu, D.L.; et al. WheatOmics: A platform combining multiple omics data to accelerate functional genomics studies in wheat. Mol. Plant 2021, 14, 1965–1968. [Google Scholar] [CrossRef]
- Han, L.; Li, G.J.; Yang, K.Y.; Mao, G.H.; Wang, R.G.; Liu, Y.D.; Zhang, S.Q. Mitogen-activated protein kinase 3 and 6 regulate Botrytis cinerea-induced ethylene production in Arabidopsis. Plant J. 2010, 64, 114–127. [Google Scholar] [CrossRef]
- Winkler, G.S.; Kristjuhan, A.; Erdjument-Bromage, H.; Tempst, P.; Svejstrup, J.Q. Elongator is a histone H3 and H4 acetyltransferase important for normal histone acetylation levels in vivo. Proc. Natl. Acad. Sci. USA 2002, 99, 3517–3522. [Google Scholar] [CrossRef] [Green Version]
- Svejstrup, J.Q. Elongator complex: How many roles does it play? Curr. Opin. Cell Biol. 2007, 19, 331–336. [Google Scholar] [CrossRef]
- Maruyama, K.; Sakuma, Y.; Kasuga, M.; Ito, Y.; Seki, M.; Goda, H.; Shimada, Y.; Yoshida, S.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J. 2004, 38, 982–993. [Google Scholar] [CrossRef] [PubMed]
- Vogel, J.T.; Zarka, D.G.; Van Buskirk, H.A.; Fowler, S.G.; Thomashow, M.F. Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J. 2005, 41, 195–211. [Google Scholar] [CrossRef]
- Zhang, Z.J.; Huang, R.F. Enhanced tolerance to freezing in tobacco and tomato overexpressing transcription factor TERF2/LeERF2 is modulated by ethylene biosynthesis. Plant Mol. Biol. 2010, 73, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔCT) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Holzberg, S.; Brosio, P.; Gross, C.; Pogue, G.P. Barley stripe mosaic virus-induced gene silencing in a monocot plant. Plant J. 2002, 30, 315–327. [Google Scholar] [CrossRef]
- Bechtold, N.; Pelletier, G. In planta agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 1998, 82, 259–266. [Google Scholar] [PubMed]
- Yoo, S.D.; Cho, Y.H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef] [Green Version]
- Dong, Z.J.; Yu, Y.W.; Li, S.H.; Wang, J.; Tang, S.J.; Huang, R.F. Abscisic acid antagonizes ethylene production through the ABI4-mediated transcriptional repression of ACS4 and ACS8 in Arabidopsis. Mol. Plant 2016, 9, 126–135. [Google Scholar] [CrossRef] [Green Version]
- Gendrel, A.V.; Lippman, Z.; Martienssen, R.; Colot, V. Profiling histone modification patterns in plants using genomic tiling microarrays. Nat. Methods 2005, 2, 213–218. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).