Abstract
The high mortality in men with metastatic prostate cancer (PC) establishes the need for diagnostic optimization by new biomarkers. Mindful of the effect of real microgravity on metabolic pathways of carcinogenesis, we attended a parabolic flight (PF) mission to perform an experiment with the PC cell line PC-3, and submitted the resulting RNA to next generation sequencing (NGS) and quantitative real-time PCR (qPCR). After the first parabola, alterations of the F-actin cytoskeleton-like stress fibers and pseudopodia are visible. Moreover, numerous significant transcriptional changes are evident. We were able to identify a network of relevant PC cytokines and chemokines showing differential expression due to gravitational changes, particularly during the early flight phases. Together with differentially expressed regulatory lncRNAs and micro RNAs, we present a portfolio of 298 potential biomarkers. Via qPCR we identified IL6 and PIK3CB to be sensitive to vibration effects and hypergravity, respectively. Per NGS we detected five upregulated cytokines (CCL2, CXCL1, IL6, CXCL2, CCL20), one zink finger protein (TNFAIP3) and one glycoprotein (ICAM1) related to c-REL signaling and thus relevant for carcinogenesis as well as inflammatory aspects. We found regulated miR-221 and the co-localized lncRNA MIR222HG induced by PF maneuvers. miR-221 is related to the PC-3 growth rate and MIR222HG is a known risk factor for glioma susceptibility. These findings in real microgravity may further improve our understanding of PC and contribute to the development of new diagnostic tools.
Keywords:
parabolic flight; microgravity; hypergravity; prostate cancer; PC-3; RNA sequencing; qPCR; cytokines; non-coding 1. Introduction
Prostate cancer (PC) is the second leading cause of cancer death in the US male population after lung cancer. The American Cancer Society predicts 268,490 new cases of PC in the US in 2022, which is about 0.165% of the male population. The disease, which primarily occurs at an older age, has a predictable treatment prognosis. However, the 5-year survival prognosis is drastically reduced from 100% to 31% when metastases ensue (The American Cancer Society). Accordingly, early management of PC is a challenging matter in medicine. In the context of personalized medicine, transcriptional studies of factors influencing prostate carcinogenesis and progression may add new early-onset biomarkers to the diagnostic portfolio.
In general, cancer cells are characterized by uncontrolled proliferation, elevated survival and activated invasion potential. The genetic and epigenetic alterations lead to an impairment of the cell death mechanisms that are initiated upon sustained damage to the cell. As we know from previous studies, gravitational changes have a lasting effect on tumorigenesis [1,2] including cytoskeletal and focal adhesion changes and alterations in differentiation and proliferation [3,4,5,6,7,8], increased apoptosis [5,9,10], the release of cytokines [11], altered interleukin, OPN, TLN1, and CTGF signaling [9], and influenced PI3K/AKT/mTOR (PAM) signaling [7,8], to only name the most significant ones. Furthermore, a previous study by Corydon and coworkers also showed that gravitational changes have specific effects on the F-actin-cytoskeleton in non-carcinogenic epithelium cell lines [12].
In previous studies, DU-145 and PC-3 prostate carcinoma cell lines were exposed to simulated microgravity (s-µg) for several days using a high aspect rotating-wall vessel (HARV) or random positioning machine (RPM), respectively [13,14,15,16]. DU-145 prostate cancer cultures revealed a slowed growth, an increase in cytokeratin 8 and 18, together with an increase in cell–cell and cell–matrix interactions, and ultimately an organotypic 3D spheroid organization after a few hours under HARF influence compared to the static 1g control [13,14]. Furthermore, significant changes in VEGF signaling were detected in aggressive PC-3 prostate carcinoma cells, resulting in early (4 h s-µg) and late (3 days to 5 days s-µg) downregulation of VEGFA expression [15,16], indicating attenuation of PC-3 aggressiveness [15]. In contrast, a significant increase in IL6 and CXCL8 gene expression in the first 24 h of s-µg hints towards a more aggressive phenotype of PC-3 [16]. Moreover, substantial components of PAM signaling were deregulated after a 5-day RPM exposure and AKT1 was upregulated in multi-cellular spheroids, but even more in adherent cells [15]. Consequently, AKT kinase inhibitors are already being assessed in phase 3 clinical trials and seem to be particularly effective in castration-resistant metastatic prostate cancer [17,18], which supports the idea that µg-based research is relevant for increasing the current knowledge about cancer progression and metastases on Earth.
In summary, it was found in previous studies that crucial key positions of carcinogenesis are influenced by microgravity and various g sensitive key players tend to promote a milder progression. Since the s-µg influence on already known major actors of prostate carcinogenesis have been investigated, the idea of our current study was to identify transcriptome-wide early responders to gravitational changes in PC-3 using a parabolic flight scenario. Parabolic flights are a cost-effective and proven approach to briefly expose cell lines to gravitational changes. During a parabolic flight (PF), which typically consists of 31 parabolas, samples experience repeating phases of free fall and hypergravity [19]. In a PF, each parabola consists of short periods (22 s) of ~0.01g (microgravity), encased by 1.8g (hypergravity) phases. These alterations allow tracking of dynamic transcriptional responses of known biomarkers of prostate cancer development and identification of, for the first time in prostate carcinoma cells, novel µg related biomarkers by transcription-wide RNA sequencing. To distinguish microgravity from other influences occurring during parabolic flight, the expression of key genes of the extracellular matrix, focal adhesion, PAM signal transduction, cell growth and apoptosis were additionally studied under hypergravity conditions (1.8g) and vibrations as they occur during a typical parabolic flight. Focusing on early responders reduces the accumulation of secondary expression changes. Finally, both transcriptional primary effects and secondary effects are relevant to research. In a biomedical context, early responses to the gravitational stimulus are closer to the cause, i.e., genetic epigenetic constitution, and subsequent secondary effects are closer to physiological, morphological changes, e.g., progression of cancer. In particular, the study of the first parabola opens the possibility to observe the primary transcriptional response to gravitational changes before the point of divergence of related signaling pathways, mostly unaffected by later secondary regulatory responses, to unravel the origin of gravitational metabolic networks. In conclusion, the knowledge of the primary transcriptional responses to gravitational changes is essential for assessing subsequent synergistic and competitive effects of prostate carcinogenesis under µg.
2. Results
2.1. Gravitational Changes of the F-Actin Cytoskeleton
When comparing the F-actin staining under 1g, first parabola (1P) and 31st parabola (31P) conditions, it is noticeable that stress fibers appear already after the first parabola, but formation of pseudopodia can be observed only sporadically (Figure S1A,B). Nevertheless, the observed effects after 1P are more pronounced than in our previous PF experiment with human chondrocytes [20]. After 31 parabolas the stress fibers are more frequent and more pronounced; the most striking change though is an increase of pseudopoidia and lamellipodia (Figure S1C).
2.2. qPCR Results
To isolate µg-related expression changes, the main intention of the qPCR analysis was to compare RNA expression changes in PF with those under hypergravity (1.8g) and vibration. Additionally, corresponding results of the RNAseq experiment (control vs. 1P and control vs. 31P) were added to check the validity of the qPCR results. Non-concordant results between qPCR and RNAseq are significantly enriched in genes with weak regulation (<2-fold) [21]. In contrast to Section 2.3, the candidate gene approach in this section also considers the differential expression with less than two-fold difference. Accordingly, a cross-validation of qPCR results with RNAseq results seemed appropriate. First, it should be noted that under hypergravity (PIK3CB) and vibration (IL6) exposure, only 1 of the 32 investigated genes was differentially expressed (Figure 1).
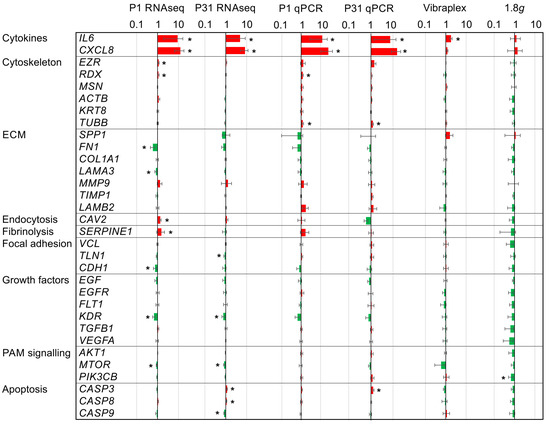
Figure 1.
Gene expression ratios of 32 key players of carcinogenesis in PC-3 prostate cancer cells under altered gravitational conditions measured by qPCR and RNAseq: Results determined after P1, after P31, after vibration-exposure and after hypergravity (1.8g)-exposure in relation to the respective 1g control samples. The red color indicates upregulated genes and the green color downregulated genes. Significant changes (nominal p < 0.05) are indicated by asterisks. The ratios are given on a logarithmic scale.
2.2.1. Cytokines
The qPCR study served to assess the influence of PF and its sub-aspects, vibration and hypergravity, on key players of carcinogenesis. Strikingly, the cytokines IL6 and CXCL8 were markedly upregulated as measured after the 1P and 31P (RNAseq and qPCR, Figure 1). However, the more moderate but significant upregulation of IL6 in the vibration experiment indicates that IL6 regulation is at least in part due to aircraft vibration.
2.2.2. Cytoskeleton
Of the six cytoskeletal genes tested via qPCR, only radixin (RDX) and tubulin, beta class I (TUBB), are differentially expressed between the 1g control and 1P. TUBB is additionally differentially expressed between control and 31P. However, TUBB qPCR results cannot be confirmed by RNAseq results, but TUBB is in RNAseq nominally significant between 1P and 31P (p = 0.009). TUBB is a structural component of microtubules and forms a dimer with alpha tubulin in this function. In RNAseq, two components of alpha tubulin TUBA4A and TUBA1B are significant differentially expressed after multiple testing corrections in comparison between 1P and 31P (Padj = 0.004 and 0.023 respectively). RDX probably possesses a role in coupling actin to the plasma membrane. Actin itself is not differentially expressed in the RNAseq experiment.
2.2.3. PI3K/AKT/mTOR (PAM) Signaling
The phosphoinositide-3-kinase, catalytic, beta polypeptide (PIK3CB) is a catalytic subunit of phosphoinositide 3-kinase (PI3K). PIK3CB is not regulated in PF, but four genes encoding catalytic subunits of PI3K are deregulated between 1g and 1P (PIK3CA, Padj = 2.8 × 10−5; PIK3R3, Padj = 2.8 × 10−5; PIK3R1, Padj = 1.9 × 10−4; PIK3C2A Padj = 2.8 × 10−3). It should be noted that the phosphoinositide 3-kinase expression is modulated by hypergravity (at least in one subunit) whereas the gene expression of other subunits is susceptible to gravitational changes in PF.
2.2.4. Apoptosis
The qPCR assessment measuring significant CASP3 upregulation after 31P is confirmed by RNAseq and complemented by a significant upregulation of CASP8 and significant downregulation of CASP9 in the RNAseq analysis. Regulations of the CASP genes are moderate and do not reach a two-fold upregulation or downregulation, respectively. Additionally, two upstream key genes of the apoptosis cascade, namely the gene coding the Tumor Necrosis Factor Receptor Type 1 (TRADD, Padj > 0.7) and the gene coding the Fas Associated Via Death Domain (FADD, Padj > 0.1), are not regulated by PF in PC-3 cells.
2.2.5. RNAseq to qPCR Comparison
As expected, additional genes could be detected in the RNAseq experiment as being regulated in PF due to the increased sensitivity (Figure 1). However, more importantly, apart from TUBB, all other qPCR PF findings could be confirmed. Apart from the cytokines IL6 and CXCL8, none of the remaining 30 qPCR-tested genes achieved an adjusted significant two-fold change in the RNAseq control to 1P comparison or in the RNAseq control to 31P comparison. The genes RDX, SERPINE1, CDH1 and MTOR are only nominally significant in the control to 1P comparison, as is KDR in the control to 31P comparison.
2.3. RNAseq Analysis
2.3.1. QC
Demultiplexed FASTQ files are used to process the 10 PF and 5 control samples individually in the secondary analysis using STAR. Fortunately, all samples exceeded a mean quality score of 35; the ratio of Q30 bases was, with one exception, >93% and the ratio of perfect barcodes >98% (Table S1). No significant difference between controls and PF samples for any of the parameters was observed.
2.3.2. General Expression Profile Reveals a Potential Habituation Effect
The general expression profile was determined using principal component analysis on the read counts. We found a high proportion of variance in the first two principal components (PCs, 65% of the variance in PC #1 and 18% of the variance in PC #2, Figure 2). This is due to the low sample number (n = 15) and to a strong effect of microgravity on the PC-3 cells. This strong effect is understandable if one considers the changes of the F-actin cytoskeleton during the PF as shown in Figure S1. The three conditions are incompletely separated in PC #1 with a tending intermediate position of the measurements after 31P. This suggests a habituation effect of the PC-3 cells to the alternating gravitational conditions during the flight.
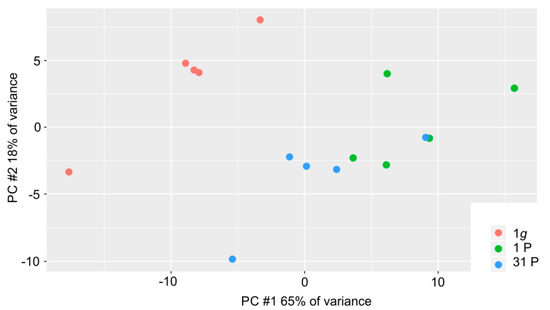
Figure 2.
Distribution of the 15 PC-3 samples in the first two principal components; 1g control samples, 1P and 31P inflight samples are color-coded in red, green and blue, respectively.
2.3.3. Differential Expression Analyses Reveal Expression Difference in Cytokines and Genes of the NFκB System Induced by Gravitational Changes
By comparing the number of genes significantly expressed until the end of 1P with the number of genes significantly expressed after 31P, the data, in terms of both stringency and fold change, reveal that the strongest changes occur in the early flight phase. DESeq2 results for the set of 298 differentially expressed genes and their dotplot visualization can be downloaded (Table S2, Figure S2). The total sum of 298 two-fold differentially expressed genes (DEGs) at Padj less than 0.05 is divided (with overlaps) into 208 DEGs between 1g and post 1P, 129 DEGs between 1g and post 31P, 47 DEGs between post 1P and post 31P. In addition to the chemokine encoding CXCL2, a significant > two-fold upregulation after P1 is followed by a significant > two-fold downregulation after P31 for another eight genes (ATOH8, AC107308.1, AC018628.2, KRTAP1-5, MIR221, CCN2, ZEB2, AC008264.2). Even though the applied RNAseq method enriches coding genes by a poly-A selection step, we found that the DEG set contains a considerable number of long non-coding RNAs (23.9% lncRNAs) and micro RNAs (2.46% miRNAs) in addition to 65.5% coding genes (Figure 3A).
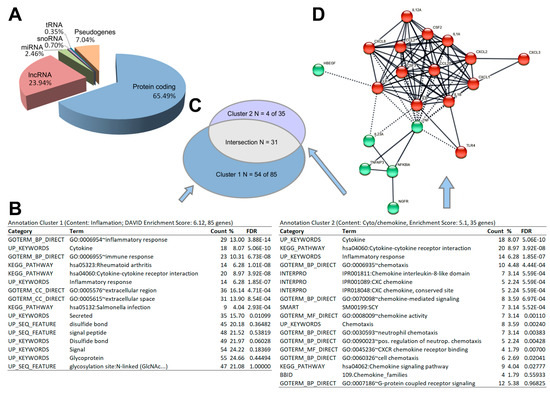
Figure 3.
Results of the post hoc analyses on 298 genes. (A) Functional assembly of differentially expressed genes. (B) Result of the annotation clustering. The two most stringent clusters with an enrichment score > 5 are shown. (C) Venn diagram of the two functional enrichment clusters. (D) STRING protein–protein interactions of annotation cluster 2 genes.
The functional enrichment analyses over the annotations included in DAVID (GO, KEGG, Interpro, Smart, etc.) revealed two annotation clusters with an enrichment score of >5. The gene clusters comprise 85 genes (cluster 1) and 35 genes (cluster 2) with a strong overlap of 31 genes between the two clusters (Figure 3B,C). The annotations suggest a strong (over-)representation of cytokines (both clusters, FDR = 5.06 × 10−10) in those interactions (hsa04060; both clusters, FDR = 3.92 × 10−8), inflammatory genes (GO:0006954; cluster 1, FDR 3.88 × 10−14), chemokines and chemotaxis (IPR001089 and GO0006953; cluster 2, FDR = 5.59 × 10−4, FDR = 4.48 × 10−4 respectively). Visualization of the pairwise protein interaction of cluster 2 using the EMBL STRING database with the highest confidence (score > 0.9) reveals a core with C-X-C motif and C-C motif chemokines surrounded by interleukins and other cytokines.
3. Discussion
A PF is a long-established platform acknowledged by NASA (National Aeronautics and Space Administration), ESA (European Space Agency) and other national space agencies for studying molecular changes under the influence of gravitational changes. This platform to study the early effects of real microgravity has been used for molecular studies on, among others, Euglena gracilis [22], mouse stem cells [23] and human cardiomyocytes [24], chondrocytes [20,25], endothelial cells [26,27], but especially in cancer cells [6,8,28,29]. The parabolic flight campaign opened-up the possibility for us to generate a list of 298 gravisensitive PC-3 genes and categorize them by subsequent enrichment and interaction network analyses. However, the specific carcinogenesis reference is crucial for the usability of the study. We were able to determine an accumulation of cytokines (DAVID FDR 5.06 × 10−10, Figure 3) and, in particular, chemokines (DAVID FDR 5.59 × 10−4). These are, in turn, partially integrated into the portfolio of genes of the inflammatory response (DAVID FDR = 3.88 × 10−14, Figure 4).
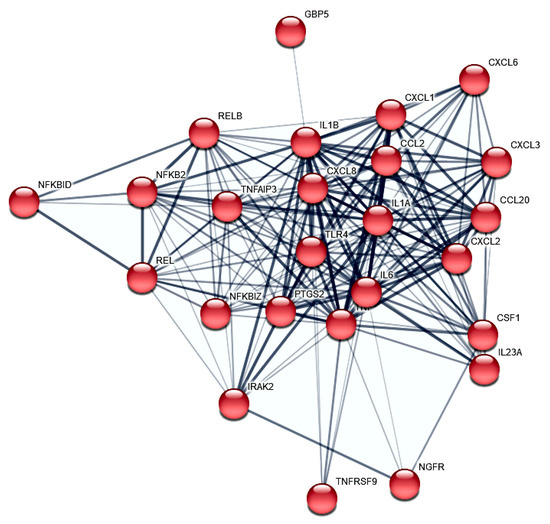
Figure 4.
STRING protein–protein interactions of PC-3 PF differential expressed genes. The 23 protein to protein interactors (confidence > 0.4) annotated in cluster 1 and involved in the inflammatory response (GO:0006954) are shown.
3.1. NF-kB Signaling and Chemokines
Because the post parabola showed one significant upregulation of the RELB proto-oncogene NF-KB subunit (Padj to 1g = 1.3 × 10−37), which is accompanied by the regulation of NF-KB-related inflammatory genes, a detailed consideration of these relationships is appropriate. de Jesús and Ramakrishnan have shown that a c-Rel knockout in mouse embryonic fibroblasts (MEF) leads to an upregulation of CCL2, CCL7, IP-10 (CXCL10), CXCL1, A20 (TNFAIP3), IL-6, CXCL2, CCL20, and ZFP36 by maintaining the expression kinetics [30]. Additionally, they found ICAM1 and VCAM1 upregulated with slowed kinetics. The gene REL encodes c-REL and was significantly upregulated after parabola one (Padj to 1g = 7.7 × 10−16) in our PF RNAseq experiment. Notably, seven of the eleven genes mentioned by de Jesús and Ramakrishnan as c-REL knock-out-dependently upregulated [30] are also highly significant and more than two-fold upregulated after P1 in our PC-3 PF (Figure 5): CCL2 (Padj to 1g = 5.0 × 10−49), CXCL1 (Padj to 1g = 2.8 × 10−50), TNFAIP3 (Padj to 1g = 9.5 × 10−23), IL6 (Padj to 1g = 4.0 × 10−6), CXCL2 (Padj to 1g = 5.2 × 10−33), CCL20 (Padj to 1g = 3.1 × 10−17), and ICAM1 (Padj to 1g = 1.7 × 10−52), whereas the joint significant upregulation of CCL2 and RELB after the first parabola is in concordance with the CCL2 expression depletion described by de Jesús and Ramakrishnan in the RELB MEF knock out experiment [30].
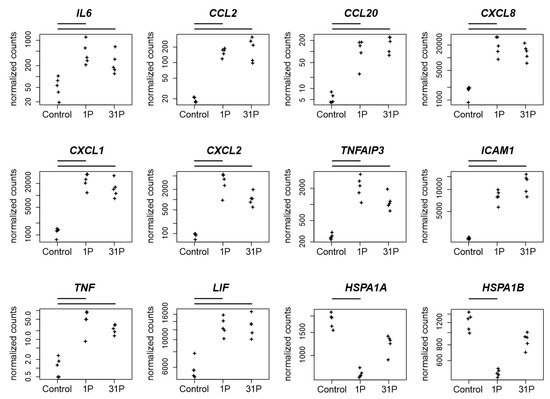
Figure 5.
Normalized PF RNAseq counts of twelve candidate genes over the conditions control, 1P and 31P. Normalized mapped RNAseq read counts to the genes encodes the cytokine IL6, two C-C motif chemokines (CCL2, CCL20), three C-X-C motif chemokines (CXCL8, CXCL1, CXCL2), the zinc finger protein TNFAIP3, the glycoprotein ICAM1, the proinflammatory cytokine TNF, the pleiotropic cytokine LIF and the heat shock 70 kDa protein (HSPA1A, HSPA1B). Bars on top of the graph indicate two-fold significant (Padj < 0.05) differential expression between the conditions.
In conclusion, although c-Rel and RELB are important factors in the regulatory cascade, the presence of other regulatory key players must be assumed. Furthermore, we were able to demonstrate the activation of genes relevant to inflammation by the gravitational change in PC-3.
3.2. Cytokines and Chemokines
The inflammatory cytokines TNF-α and LIF influence the CXCL-8 expression in pancreatic cancer progression [31]. CXCL-8 has been detected in prostate epithelium and stroma, is involved in multiple intracellular signaling [32] and upregulates the activity of Akt kinase [33]. TNF, LIF and CXCL8 are upregulated after parabola one in our PC-3 PF (Padj to 1g = 8.6 × 10−12, 1.9 × 10−15 and 2.0 × 10−22, respectively). In PC-3 cells, migration and proliferation are stimulated by CXCL3 overexpression [34]. In general, a highly increased secretion of the chemokine CXCL3 can be detected in PC-3 while an elevated expression of CXCR2 was observed in the PC cell lines DU145, LNCaP and exogenous CXCL3, which does not affect proliferation but migration [35]. The experimental autoimmune prostatitis (EAP) mice are a model for the autoimmune dysfunction of chronic prostatitis. In EAP prostate, the proinflammatory cytokine interleukin 17 and the chemokine ligands CXCL1 and CXCL2 are upregulated. These findings suggest that IL-17 promotes the production of CXCL1 and CXCL2 [36]. In support of this notion, we find CXCL1 and CXCL2 to be upregulated after P1 (Padj to 1g = 2.8 × 10−50 and 5.2 × 10−33, respectively).
3.3. Protein Transport
The two 70 kDa heat shock protein coding genes HSPA1A and HSPA1B are both downregulated after P1 (Padj to 1g = 4.6 × 10−43 and 2.5 × 10−33 respectively, Figure 5). The proteins have chaperone and lipid-binding functions and are involved in protein stabilization and folding. The gene HspA1B is one of the most prominent biomarkers for PC [37] and HSPA1A and HSPA1B upregulation is associated with a poor survival in colon cancer [38]. Furthermore, HspA1A specifically inhibits the malignant progression of Arid2-deficient lung cancer [39]. The known translocation of HspA1A toward the plasma membrane and the resulting presentation at the cell surface [40] are possible targets of gravitational variation in our PF experiment.
3.4. Non-Coding RNA
Although the applied RNAseq method is targeted towards coding genes by poly-A filtering, 23.9% and 25% of significantly and two-fold regulated genes are lncRNAs and miRNAs, respectively (Figure 3A). miRNAs and lncRNAs are important regulators of translation and transcription, respectively, so it is interesting to investigate whether expression of the non-coding genes were altered early on by microgravity.
The role of miR-221/222 in PC remains controversial but upregulation of miR-221/222 in CRPC is associated with an androgene receptor modulation during cancer development [41]. It has been shown that elimination of miR-221 by CRISPR in PC-3 cells led to a reduced growth rate and expression of cell-cycle genes [42] through post transcriptional inhibition of CDKN1B/p27 [43]. In cells fixed after P1, we found a significantly increased miR-221 expression (q = 1.65 × 10−5) and at the same time expression depletion of CDKN1B (Padj to 1g = 5.07 × 10−5). Upstream of miR221/222 MIR222HG, a long non-coding RNA (lncRNA), is located. MIR222HG is associated with PC progression to CRPC, increases androgen-independent cell growth and represses the expression of TMPRSS2 and FKBP5 in hormone-sensitive prostate cancer [44]. We found a significant increase in MIR222HG expression in cells fixed after first parabola (Padj to 1g = 1.47 × 10−9), but no corresponding expression depletion in TMPRSS2 and FKBP5 (Figure 6).
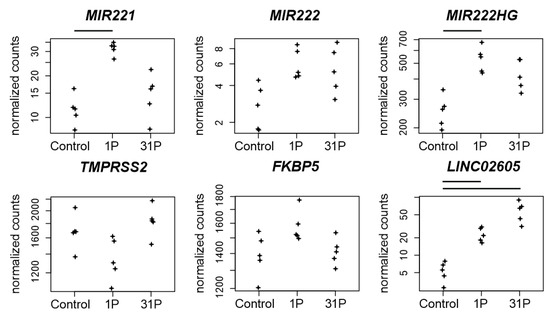
Figure 6.
Normalized PF RNAseq counts of four non-coding RNAs and two protein coding RNAs. Expressions of the micro RNAs miR-221 and miR-222, of the lncRNA MIR222HG, of the serine protease encoding TMPRSS2 gene, of the Peptidyl-Prolyl Cis-Trans Isomerase encoding gene FKBP5 and of the lncRNA LINC02605 are given. Bars on top of the graph indicate two-fold significant (Padj < 0.05) differential expression between the conditions.
MIR3142HG, a long non-coding RNA (lncRNA) associated with genomic instability, is upregulated in PC-3 cells in PF after the first parabola (Padj to 1g = 3.7 × 10−34, Figure 6). Its role in cancer progression is unclear. MIR3142HG is propagated as a risk factor for glioma susceptibility in the Chinese Han population [45], but it is also assumed to serve as a protective factor for head and neck squamous cell carcinoma (HNSCC) because the upregulated expression of MIR3142HG was associated with better survival outcomes [46].
The lncRNA LINC02605 is involved in the antiviral immune response and its knockdown leads to an increased viral replication [47]. LINC02605 is upregulated in PC-3 cells after the first parabola (Padj to 1g = 2.9 × 10−6) and even more after parabola 31 (Padj to 1g = 1.9 × 10−16). It has an effect on hsa-miR-107 regulation on the protein expression of phosphatase and tensin homolog (PTEN) [47,48]. Both miR-107 and PTEN are important players in carcinogenesis. miR-107 has a potential role in regulating NEDD9 in the invasion, migration and proliferation of breast cancer [49]. Additionally, miR-107 is hypothesized to deactivate the phosphatidylinositol 3-kinase (PI3K)/Akt pathway in a hypopharyngeal squamous cell carcinoma (HSCC) study [48]. Downregulation of miR-107 by the potential therapeutic target LINC-DUBR suppressed malignant progression of ovarian cancer [50].
3.5. qPCR: Apoptosis, Hypogravity, and Vibration
The set of genes to be validated by qPCR shows that a substantial fraction of PC key players is unaffected by the gravitational influence of PF. However, it should be noted that the cytokine interleukin 6 is at least partially upregulated due to the vibration effect. Since cytokines play a major role in early PF-induced regulation, IL6 requires further investigation. Furthermore, apoptotic processes seem to start at 31P. Although the effects on CASP3, CASP8, and CASP9 are at the limit of detection even with RNAseq, mutual validation of RNAseq and qPCR for upregulation of CASP3 indicates an albeit weak but stable finding (Figure 7A). Since more than 31P are not performed for technical reasons, it should be sufficient to keep an eye on the apoptosis problem. The downregulation of PIK3CB after 24 h of simulated microgravity obtained by random positioning published in our recent paper [16] does not technically match the downregulation measured in the present study under hypergravity (Figure 7B). PIK3CB following regulation cascades should therefore be kept in mind at least for long-term experiments with preceding hypergravity (e.g., rocket launch during ISS missions). It remains to be verified whether PIK3CB is subject to a general sensitivity to gravitational changes.
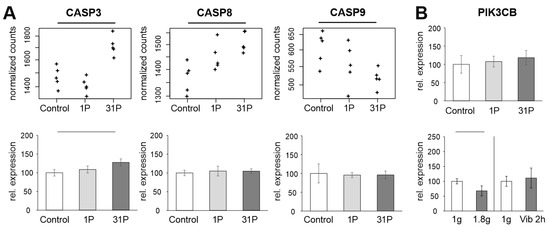
Figure 7.
Analysis of apoptosis-related genes in PF samples. (A): Normalized PF RNAseq counts and qPCR ratios of the three apoptosis-related genes CASP3, CASP8 and CASP9. (B): PIK3CB qPCR expression relative to the controls of the PF, hypergravity and Vibraplex experiment. Black and grey bars on top of the graph indicate adjusted and nominal significant (p < 0.05) differential expression, respectively.
4. Materials and Methods
4.1. Cell Culturing
The PC-3 cell line (ECACC 90112714) was purchased from the European Collection of Authenticated Cell Cultures (ECACC). The cells originated from a 62-year-old male Caucasian who suffered from a grade 4 prostate adenocarcinoma.
During cell cultivation, Ham’s F-12 Nutrient Mixture (F-12) medium (Gibco, Fisher Scientific, Schwerte, Germany) was used and was supplemented with 10% FCS (Sigma Aldrich, Steinheim, Germany) and 1% penicillin/streptomycin (Life Technologies, New York, USA). The cells were cultured in T75 cm² flasks (Sarstedt, Nümbrecht, Germany). Every 3–4 days the medium was changed. The cells were split using 0.05% trypsin-EDTA (Gibco, Life Technologies, Paisley, UK).
4.2. Parabolic Flight Campaign
The 34th PF campaign was organized by the French company Novespace at the Bordeaux-Mérignac airport on behalf of the DLR (Deutsches Zentrum für Luft- und Raumfahrt) as part of their scientific and technological research programs.
The PF campaign consisted of a total of three flight days with the same experimental setup. The PF was carried out with the Airbus A310 ZERO G (Figure S3A,B) and took about two hours. When the flight altitude was reached, a total of 31 parabolas were flown. A parabola consists of a 20-s climb phase during which the acceleration due to gravity is almost twice as high (1.8g). This is followed by the actual 22-s-long µg-phase. In the nosedive, a 20-s phase of hyper-g (1,8g) follows again. During the PF, the PC-3 cells were fixed after the first and after the 31st parabola using RNAlater (Invitrogen by Thermo Fischer Scientific, Vilnius, Lithuania), thus stabilizing the RNA until isolation.
T75 cm² flasks (Sarstedt, Nümbrecht, Germany) were placed on board in an incubator preheated to 37 °C 1 h before take-off. This was secured inside the so-called flight rack (Figure S3C). The flasks were connected to syringes via a flexible tube and a three-way valve (Figure S3D). The syringes contained 50 mL RNAlater. At the time points mentioned above, the syringes were manually actuated so that the culture flasks were filled with RNAlater at a ratio of approximately 4:1 (RNAlater:medium). Post-flight, the cells were transferred into 15 mL Falcon™ tubes (Sarstedt, Nümbrecht, Germany) by scratching the cells with 25 cm cell scrapers (Sarstedt, Nümbrecht, Germany) and stored suspended in RNAlater at 4 °C until RNA isolation.
For F-actin staining, 200,000 cells were pipetted into eight slideflasks (Thermo Scientific, Waltham, MA, USA) and were incubated for 1.5 days. Of these, four slideflasks remained under 1g conditions and served as ground controls. The other four flasks were on board the aircraft in the incubator. Cells were fixed at the designated times using 4% paraformaldehyde (PFA, Sigma-Aldrich, St. Louis, MO, USA) in phosphate buffered saline (PBS; Gibco, Life Technologies, Paisley, UK). The 1g ground controls were also fixed at approximately the same times using 4% PFA. The slideflasks remained in 4% PFA until staining.
4.3. RNA Isolation
The cell suspension in the 15 mL Falcon™ tubes was diluted 1:5 with PBS. After centrifuging the dilution, the supernatant was discarded. Then, the RNA was isolated using the RNeasy Mini Kit (Qiagen, Hilden, Germany). Then, RTL lysis buffer was added, and the cell pellet was dissolved by pipetting up and down with a syringe. Afterwards, 350 µL of a 70% ethanol dilution was added, 700 µL of the solution was transferred to RNeasy Mini spin column, and the column was centrifuged for 15 s at 8000× g. Next, 700 µL RW1 buffer and 500 µL RPE buffer were added and centrifuged in between. Then, 35 µL RNAse-free water was pipetted directly to the spin column membrane and the column was put into a new Eppendorf tube. Finally, the spin column was centrifuged again to detach the RNA from the membrane.
The quality and the concentration of the RNA of each sample was evaluated by the Nanodrop technique and a spectrophotometer.
4.4. Quantitative Real-Time Polymerase Chain Reaction
A 7500 Fast Real-Time PCR System (Applied Biosystems, Darmstadt, Germany) was used both for cDNA synthesis and the following quantitative real time polymerase chain reaction (qPCR).
First, the RNA was converted into cDNA with the High-Capacity cDNA reverse Transcription Kit (Applied Biosystems, Darmstadt, Germany) on a 96-well plate. Then, 2 µL RT buffer, 0.8 µL dNTP mix, 2 µL RT random primer, 1 µL reverse transcriptase and 4.2 µL nuclease free water were added to each well. In the end, the RNA of each sample was added to get a total amount of 1 µg RNA in 20 µL solution.
The qPCR was performed with FAST SYBR™ Green Master Mix (Applied Biosystems, Foster City, CA, USA) on a 96-well plate. Every sample was measured in triplicate. Each well contained 1 µL cDNA of the corresponding sample, 7.5 µL SYBR Green Master Mix, 0.042 µL primer forward, 0.042 µL primer reverse and 6.41 µL water (Table S3). The plate was placed in the qPCR instrument and went through the following thermal cycles: 95 °C for 20 s, 95 °C for 3 s, and 60 °C for 30 s.
The samples were analyzed with the comparative threshold cycle (ΔΔCT) method, using the 18 s housekeeping gene as a reference.
4.5. F-Actin Staining
F-actin was visualized by means of rhodamine-phalloidin staining (Molecular Probes®, Eugene, OR, USA). Moreover, the nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI, Sigma-Aldrich, Merck Life Science A/S, Søborg, Denmark). The protocol was published earlier in [20].
4.6. Microscopy
The slideflasks were investigated using confocal laser scanning microscopy. The observations were made with a Leica DM 2000 microscope and an objective with a 400× magnification. The microscope was also connected to an external light source Leica EL 6000 (Leica Microsystems GmbH, Wetzlar, Germany).
4.7. Hypergravity
The hyper-g experiments were performed using the DLR Multi-Sample Incubator Centrifuge (MuSIC, DLR, Department of Gravitational Biology) as already described by Aleshcheva et al. [20]. For this purpose, four T75 cell culture flasks were placed on the MuSIC in a 37 °C incubator (Figure S3E,F). For two hours, 1.8g gravitational load was then applied to the cell cultures, which corresponds to the hyper-g-phases during the PF. At the end of the run, the samples were fixed with RNAlater for RNA isolation. The 1g control samples were cultured in parallel and also fixed using RNAlater.
4.8. Vibration Experiment
T75 flasks containing subconfluent PC-3 cells were attached to the Vibraplex device (DLR, Cologne, Germany Figure S3G) and placed on the device in a 37 °C incubator in line with the protocol published earlier by Lützenberg et al. [51]. Vibrations between 0.2 and 14 Hz were transmitted via the Vibraplex using a connected loudspeaker for about 2 h. This procedure corresponds to frequencies generated during the PF. At the end of the two hours, the samples were fixed with RNAlater for RNA isolation. Static control samples were cultured and fixed with RNAlater in parallel.
4.9. RNA Sequencing
Extracted RNA was checked for quantity and quality as described in Section 4.3. Sufficient quality RNA was converted to libraries using the TruSeq™ RNA Library Prep Kit v2 (Illumina, San Diego, CA, USA) as directed by standard protocols. Libraries were sequenced on Illumina’s novaseq 6000 with paired-end 100 base pair reads and a sequencing depth of >20 milion pairs. For all sequencing data, reads were quality checked for read counts and quality values. The unaligned reads were mapped on the hg38 genome using STAR 2.6.1a [52] and the ENSEMBL v.99 [53] annotation. A maximum of ten mismatches and a multimapping to up to 10 different positions was allowed. Secondary alignments were filtered out.
4.10. Transcriptomic Analyses
We used the Bioconductor R package DESeq2 to capture differentially expressed genes (DEG). DESeq2 relies on a negative binomial generalized linear model (GLM) and uses the uniquely mapped RNAseq counts to an exon [54]. For integrated post hoc analysis for enrichments and networks of differentially expressed genes, we used the Database for Annotation, Visualization and Integrated Discovery (DAVID v. 6.8) [55] and the protein-to-protein interaction database STRING v. 11.5 [56], respectively.
4.11. Statistical Analysis
The statistics were performed using the GraphPad Prism 7.01 software (GraphPad Software, Inc., San Diego, CA, USA) and the stats package of R 3.4.3. The sample and the related control were analyzed with the Mann–Whitney U-test at a significance level of p * < 0.05.
5. Conclusions
The objective behind our PF experiment was to identify early responses to the alternating gravitational stimulus and to put them into context with carcinogenesis and cancer progression in order to finally obtain new and fast expression-altering biomarkers for PC diagnosis. In general, we observed that the highly differential genes, which include some cytokines and especially chemokines, on the one hand provide a stable expression and are therefore robust for a diagnostic approach, and on the other hand are already related to the disease. Notably, the NGS analysis identified five cytokines (CCL2, CXCL1, IL6, CXCL2, CCL20), one zink finger protein (TNFAIP3), one glycoprotein (ICAM1) related to c-REL signaling, and the microRNA (miR-221) to be differentially regulated during PFs. Since these factors are relevant in cancer as well as inflammatory aspects, the presented findings may thus contribute to the development of improved diagnostic tools relevant to PC. CCL2/CCR2 have been associated with PC advancement, metastasis and relapse [57]. A similar action is known for IL6, which plays a key role in cancer progression [58]. In particular, non-coding RNAs and micro-RNAs have recently been increasingly used in cancer medicine as diagnostic, prognostic and therapeutic biomarkers [59,60,61,62] and cytokines are well established in cancer immunotherapy [63,64,65,66].
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms23147876/s1.
Author Contributions
Conceptualization H.S., D.G.; methodology, H.S. and D.D.; software H.S. and M.W.; validation D.G., H.S., T.J.C. and M.W.; formal analysis H.S. and D.D.; investigation H.S., D.D., M.W., T.J.C., R.H., C.L., K.S., N.H. and D.M.; resources M.I., R.H. and D.G.; writing—original draft preparation, H.S.; writing—review and editing, D.G., T.J.C., R.H. and M.W.; visualization, H.S., T.J.C. and D.G.; supervision, D.G.; project administration, D.G.; funding acquisition, D.G. and M.I. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Deutsches Zentrum für Luft- und Raumfahrt (DLR) projects 50WB2219 and 50WB1924 (D.G.).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors express their thanks to Yale T. Kozinski for his thorough language review of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| PF | parabolic flight, |
| PC | prostate cancer |
| HNSCC | head and neck squamous cell carcinoma |
| HSCC | hypopharyngeal squamous cell carcinoma |
| PCA | principal component analysis |
| µg | Microgravity |
| s-µg | simulated microgravity |
| p | probability-value |
| Padj | adjusted probability-value |
| 1P | one parabola |
| 31P | 31 parabolas |
| NGS | next generation sequencing |
| qPCR | quantitative real-time PCR |
| RPM | random positioning machine |
| HARV | high aspect rotating-wall vessel |
| PAM | PI3K/AKT/mTOR |
References
- Grimm, D.; Schulz, H.; Krüger, M.; Cortés-Sánchez, J.L.; Egli, M.; Kraus, A.; Sahana, J.; Corydon, T.J.; Hemmersbach, R.; Wise, P.M.; et al. The Fight against Cancer by Microgravity: The Multicellular Spheroid as a Metastasis Model. Int. J. Mol. Sci. 2022, 23, 3073. [Google Scholar] [CrossRef] [PubMed]
- Pietsch, J.; Kussian, R.; Sickmann, A.; Bauer, J.; Weber, G.; Nissum, M.; Westphal, K.; Egli, M.; Grosse, J.; Schönberger, J.; et al. Application of free-flow IEF to identify protein candidates changing under microgravity conditions. Proteomics 2010, 10, 904–913. [Google Scholar] [CrossRef] [PubMed]
- Nassef, M.Z.; Kopp, S.; Wehland, M.; Melnik, D.; Sahana, J.; Krüger, M.; Corydon, T.J.; Oltmann, H.; Schmitz, B.; Schütte, A.; et al. Real Microgravity Influences the Cytoskeleton and Focal Adhesions in Human Breast Cancer Cells. Int. J. Mol. Sci. 2019, 20, 3156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Häder, D.P.; Braun, M.; Grimm, D.; Hemmersbach, R. Gravireceptors in eukaryotes-a comparison of case studies on the cellular level. NPJ Microgravity 2017, 3, 13. [Google Scholar] [CrossRef] [Green Version]
- Grimm, D.; Bauer, J.; Kossmehl, P.; Shakibaei, M.; Schöberger, J.; Pickenhahn, H.; Schulze-Tanzil, G.; Vetter, R.; Eilles, C.; Paul, M.; et al. Simulated microgravity alters differentiation and increases apoptosis in human follicular thyroid carcinoma cells. FASEB J. 2002, 16, 604–606. [Google Scholar] [CrossRef]
- Corydon, T.J.; Kopp, S.; Wehland, M.; Braun, M.; Schütte, A.; Mayer, T.; Hülsing, T.; Oltmann, H.; Schmitz, B.; Hemmersbach, R.; et al. Alterations of the cytoskeleton in human cells in space proved by life-cell imaging. Sci. Rep. 2016, 6, 20043. [Google Scholar] [CrossRef] [Green Version]
- Tan, X.; Xu, A.; Zhao, T.; Zhao, Q.; Zhang, J.; Fan, C.; Deng, Y.; Freywald, A.; Genth, H.; Xiang, J. Simulated microgravity inhibits cell focal adhesions leading to reduced melanoma cell proliferation and metastasis via FAK/RhoA-regulated mTORC1 and AMPK pathways. Sci. Rep. 2018, 8, 3769. [Google Scholar] [CrossRef] [Green Version]
- Nassef, M.Z.; Kopp, S.; Melnik, D.; Corydon, T.J.; Sahana, J.; Krüger, M.; Wehland, M.; Bauer, T.J.; Liemersdorf, C.; Hemmersbach, R.; et al. Short-Term Microgravity Influences Cell Adhesion in Human Breast Cancer Cells. Int. J. Mol. Sci. 2019, 20, 5730. [Google Scholar] [CrossRef] [Green Version]
- Grosse, J.; Wehland, M.; Pietsch, J.; Schulz, H.; Saar, K.; Hübner, N.; Eilles, C.; Bauer, J.; Abou-El-Ardat, K.; Baatout, S.; et al. Gravity-sensitive signaling drives 3-dimensional formation of multicellular thyroid cancer spheroids. FASEB J. 2012, 26, 5124–5140. [Google Scholar] [CrossRef] [Green Version]
- Kossmehl, P.; Shakibaei, M.; Cogoli, A.; Infanger, M.; Curcio, F.; Schönberger, J.; Eilles, C.; Bauer, J.; Pickenhahn, H.; Schulze-Tanzil, G.; et al. Weightlessness induced apoptosis in normal thyroid cells and papillary thyroid carcinoma cells via extrinsic and intrinsic pathways. Endocrinology 2003, 144, 4172–4179. [Google Scholar] [CrossRef] [Green Version]
- Warnke, E.; Pietsch, J.; Kopp, S.; Bauer, J.; Sahana, J.; Wehland, M.; Krüger, M.; Hemmersbach, R.; Infanger, M.; Lützenberg, R.; et al. Cytokine Release and Focal Adhesion Proteins in Normal Thyroid Cells Cultured on the Random Positioning Machine. Cell Physiol. Biochem. 2017, 43, 257–270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corydon, T.J.; Mann, V.; Slumstrup, L.; Kopp, S.; Sahana, J.; Askou, A.L.; Magnusson, N.E.; Echegoyen, D.; Bek, T.; Sundaresan, A.; et al. Reduced Expression of Cytoskeletal and Extracellular Matrix Genes in Human Adult Retinal Pigment Epithelium Cells Exposed to Simulated Microgravity. Cell Physiol. Biochem. 2016, 40, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Clejan, S.; O’Connor, K.C.; Cowger, N.L.; Cheles, M.K.; Haque, S.; Primavera, A.C. Effects of simulated microgravity on DU 145 human prostate carcinoma cells. Biotechnol. Bioeng. 1996, 50, 587–597. [Google Scholar] [CrossRef]
- Clejan, S.; O’Connor, K.; Rosensweig, N. Tri-dimensional prostate cell cultures in simulated microgravity and induced changes in lipid second messengers and signal transduction. J. Cell Mol. Med. 2001, 5, 60–73. [Google Scholar] [CrossRef]
- Hybel, T.E.; Dietrichs, D.; Sahana, J.; Corydon, T.J.; Nassef, M.Z.; Wehland, M.; Krüger, M.; Magnusson, N.E.; Bauer, J.; Utpatel, K.; et al. Simulated Microgravity Influences VEGF, MAPK, and PAM Signaling in Prostate Cancer Cells. Int. J. Mol. Sci. 2020, 21, 1263. [Google Scholar] [CrossRef] [Green Version]
- Dietrichs, D.; Grimm, D.; Sahana, J.; Melnik, D.; Corydon, T.J.; Wehland, M.; Krüger, M.; Vermeesen, R.; Baselet, B.; Baatout, S.; et al. Three-Dimensional Growth of Prostate Cancer Cells Exposed to Simulated Microgravity. Front. Cell Dev. Biol. 2022, 10, 841017. [Google Scholar] [CrossRef]
- Gasmi, A.; Roubaud, G.; Dariane, C.; Barret, E.; Beauval, J.B.; Brureau, L.; Créhange, G.; Fiard, G.; Fromont, G.; Gauthé, M.; et al. Overview of the Development and Use of Akt Inhibitors in Prostate Cancer. J. Clin. Med. 2021, 11, 160. [Google Scholar] [CrossRef]
- Bachmann, S.; Iding, H.; Lautz, C.; Thomé-Pfeiffer, I.; Maierhofer, C.; Mondière, R.; Schmidt, P.; Strasser, C.; Bär, T.; Aebi, A.; et al. Development of the Commercial Manufacturing Process for Ipatasertib. Chimia 2021, 75, 605–613. [Google Scholar] [CrossRef]
- Riwaldt, S.; Corydon, T.J.; Pantalone, D.; Sahana, J.; Wise, P.; Wehland, M.; Krüger, M.; Melnik, D.; Kopp, S.; Infanger, M.; et al. Role of Apoptosis in Wound Healing and Apoptosis Alterations in Microgravity. Front. Bioeng. Biotechnol. 2021, 9, 679650. [Google Scholar] [CrossRef]
- Aleshcheva, G.; Wehland, M.; Sahana, J.; Bauer, J.; Corydon, T.J.; Hemmersbach, R.; Frett, T.; Egli, M.; Infanger, M.; Grosse, J.; et al. Moderate alterations of the cytoskeleton in human chondrocytes after short-term microgravity produced by parabolic flight maneuvers could be prevented by up-regulation of BMP-2 and SOX-9. FASEB J. 2015, 29, 2303–2314. [Google Scholar] [CrossRef]
- Everaert, C.; Luypaert, M.; Maag, J.L.V.; Cheng, Q.X.; Dinger, M.E.; Hellemans, J.; Mestdagh, P. Benchmarking of RNA-sequencing analysis workflows using whole-transcriptome RT-qPCR expression data. Sci. Rep. 2017, 7, 1559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kruger, J.; Richter, P.; Stoltze, J.; Strauch, S.M.; Kruger, M.; Daiker, V.; Prasad, B.; Sonnewald, S.; Reid, S.; Lebert, M. Changes of Gene Expression in Euglena gracilis Obtained During the 29(th) DLR Parabolic Flight Campaign. Sci. Rep. 2019, 9, 14260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Acharya, A.; Brungs, S.; Henry, M.; Rotshteyn, T.; Singh Yaduvanshi, N.; Wegener, L.; Jentzsch, S.; Hescheler, J.; Hemmersbach, R.; Boeuf, H.; et al. Modulation of Differentiation Processes in Murine Embryonic Stem Cells Exposed to Parabolic Flight-Induced Acute Hypergravity and Microgravity. Stem. Cells Dev. 2018, 27, 838–847. [Google Scholar] [CrossRef] [PubMed]
- Acharya, A.; Brungs, S.; Lichterfeld, Y.; Hescheler, J.; Hemmersbach, R.; Boeuf, H.; Sachinidis, A. Parabolic, Flight-Induced, Acute Hypergravity and Microgravity Effects on the Beating Rate of Human Cardiomyocytes. Cells 2019, 8, 352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wehland, M.; Aleshcheva, G.; Schulz, H.; Saar, K.; Hubner, N.; Hemmersbach, R.; Braun, M.; Ma, X.; Frett, T.; Warnke, E.; et al. Differential gene expression of human chondrocytes cultured under short-term altered gravity conditions during parabolic flight maneuvers. Cell Commun. Signal. 2015, 13, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grosse, J.; Wehland, M.; Pietsch, J.; Ma, X.; Ulbrich, C.; Schulz, H.; Saar, K.; Hubner, N.; Hauslage, J.; Hemmersbach, R.; et al. Short-term weightlessness produced by parabolic flight maneuvers altered gene expression patterns in human endothelial cells. FASEB J. 2012, 26, 639–655. [Google Scholar] [CrossRef]
- Wehland, M.; Ma, X.; Braun, M.; Hauslage, J.; Hemmersbach, R.; Bauer, J.; Grosse, J.; Infanger, M.; Grimm, D. The impact of altered gravity and vibration on endothelial cells during a parabolic flight. Cell Physiol. Biochem. 2013, 31, 432–451. [Google Scholar] [CrossRef]
- Ulbrich, C.; Pietsch, J.; Grosse, J.; Wehland, M.; Schulz, H.; Saar, K.; Hubner, N.; Hauslage, J.; Hemmersbach, R.; Braun, M.; et al. Differential gene regulation under altered gravity conditions in follicular thyroid cancer cells: Relationship between the extracellular matrix and the cytoskeleton. Cell Physiol. Biochem. 2011, 28, 185–198. [Google Scholar] [CrossRef]
- Ma, X.; Pietsch, J.; Wehland, M.; Schulz, H.; Saar, K.; Hubner, N.; Bauer, J.; Braun, M.; Schwarzwalder, A.; Segerer, J.; et al. Differential gene expression profile and altered cytokine secretion of thyroid cancer cells in space. FASEB J. 2014, 28, 813–835. [Google Scholar] [CrossRef]
- de Jesús, T.J.; Ramakrishnan, P. NF-κB c-Rel Dictates the Inflammatory Threshold by Acting as a Transcriptional Repressor. iScience 2020, 23, 100876. [Google Scholar] [CrossRef] [Green Version]
- Kamohara, H.; Takahashi, M.; Ishiko, T.; Ogawa, M.; Baba, H. Induction of interleukin-8 (CXCL-8) by tumor necrosis factor-alpha and leukemia inhibitory factor in pancreatic carcinoma cells: Impact of CXCL-8 as an autocrine growth factor. Int. J. Oncol. 2007, 31, 627–632. [Google Scholar] [PubMed]
- Liu, Q.; Li, A.; Tian, Y.; Wu, J.D.; Liu, Y.; Li, T.; Chen, Y.; Han, X.; Wu, K. The CXCL8-CXCR1/2 pathways in cancer. Cytokine Growth Factor Rev. 2016, 31, 61–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Culig, Z. CXCL8, an underestimated “bad guy” in prostate cancer. Eur. Urol. 2013, 64, 189–190. [Google Scholar] [CrossRef] [PubMed]
- Xin, H.; Cao, Y.; Shao, M.L.; Zhang, W.; Zhang, C.B.; Wang, J.T.; Liang, L.C.; Shao, W.W.; Qi, Y.L.; Li, Y.; et al. Chemokine CXCL3 mediates prostate cancer cells proliferation, migration and gene expression changes in an autocrine/paracrine fashion. Int. Urol. Nephrol. 2018, 50, 861–868. [Google Scholar] [CrossRef]
- Gui, S.L.; Teng, L.C.; Wang, S.Q.; Liu, S.; Lin, Y.L.; Zhao, X.L.; Liu, L.; Sui, H.Y.; Yang, Y.; Liang, L.C.; et al. Overexpression of CXCL3 can enhance the oncogenic potential of prostate cancer. Int. Urol. Nephrol. 2016, 48, 701–709. [Google Scholar] [CrossRef]
- Zhang, C.; Chen, J.; Wang, H.; Chen, J.; Zheng, M.J.; Chen, X.G.; Zhang, L.; Liang, C.Z.; Zhan, C.S. IL-17 exacerbates experimental autoimmune prostatitis via CXCL1/CXCL2-mediated neutrophil infiltration. Andrologia 2022, e14455. [Google Scholar] [CrossRef]
- Zhou, K.; Arslanturk, S.; Craig, D.B.; Heath, E.; Draghici, S. Discovery of primary prostate cancer biomarkers using cross cancer learning. Sci. Rep. 2021, 11, 10433. [Google Scholar] [CrossRef]
- Guan, Y.; Zhu, X.; Liang, J.; Wei, M.; Huang, S.; Pan, X.X. Upregulation of HSPA1A/HSPA1B/HSPA7 and Downregulation of HSPA9 Were Related to Poor Survival in Colon Cancer. Front. Oncol. 2021, 11, 749673. [Google Scholar] [CrossRef]
- Wang, X.; Wang, Y.; Fang, Z.; Wang, H.; Zhang, J.; Zhang, L.; Huang, H.; Jiang, Z.; Jin, Y.; Han, X.; et al. Targeting HSPA1A in ARID2-deficient lung adenocarcinoma. Natl. Sci. Rev. 2021, 8, nwab014. [Google Scholar] [CrossRef]
- Smulders, L.; Daniels, A.J.; Plescia, C.B.; Berger, D.; Stahelin, R.V.; Nikolaidis, N. Characterization of the Relationship between the Chaperone and Lipid-Binding Functions of the 70-kDa Heat-Shock Protein, HspA1A. Int. J. Mol. Sci. 2020, 21, 5995. [Google Scholar] [CrossRef]
- Gui, B.; Hsieh, C.L.; Kantoff, P.W.; Kibel, A.S.; Jia, L. Androgen receptor-mediated downregulation of microRNA-221 and -222 in castration-resistant prostate cancer. PLoS ONE 2017, 12, e0184166. [Google Scholar] [CrossRef] [PubMed]
- Dart, D.A.; Koushyar, S.; Lanning, B.E.; Jiang, W. MiR-221 Is Specifically Elevated in PC3 Cells and its Deletion Reduces Adhesion, Motility and Growth. Anticancer Res. 2019, 39, 5311–5327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fornari, F.; Gramantieri, L.; Ferracin, M.; Veronese, A.; Sabbioni, S.; Calin, G.A.; Grazi, G.L.; Giovannini, C.; Croce, C.M.; Bolondi, L.; et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene 2008, 27, 5651–5661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, T.; Du, S.Y.; Armenia, J.; Qu, F.; Fan, J.; Wang, X.; Fei, T.; Komura, K.; Liu, S.X.; Lee, G.M.; et al. Expression of lncRNA MIR222HG co-transcribed from the miR-221/222 gene promoter facilitates the development of castration-resistant prostate cancer. Oncogenesis 2018, 7, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, J.; Ouyang, Y.; Xu, D.; He, Q.; Liu, D.; Fan, X.; Xu, P.; Mo, Y. Genetic variants in MIR17HG affect the susceptibility and prognosis of glioma in a Chinese Han population. BMC Cancer 2020, 20, 976. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhao, Y.; Lu, R.; Zhao, H.; Guo, Y. Identification and Validation of a Novel Genomic Instability-Associated Long Non-Coding RNA Prognostic Signature in Head and Neck Squamous Cell Carcinoma. Front. Cell Dev. Biol. 2021, 9, 787766. [Google Scholar] [CrossRef]
- Xu, R.; Yu, S.S.; Yao, R.R.; Tang, R.C.; Liang, J.W.; Pang, X.; Zhang, J. Interferon-Inducible LINC02605 Promotes Antiviral Innate Responses by Strengthening IRF3 Nuclear Translocation. Front. Immunol. 2021, 12, 755512. [Google Scholar] [CrossRef]
- Gao, X.; Fan, X.; Zeng, W.; Liang, J.; Guo, N.; Yang, X.; Zhao, Y. Overexpression of microRNA-107 suppressed proliferation, migration, invasion, and the PI3K/Akt signaling pathway and induced apoptosis by targeting Nin one binding (NOB1) protein in a hypopharyngeal squamous cell carcinoma cell line (FaDu). Bioengineered 2022, 13, 7881–7893. [Google Scholar] [CrossRef]
- Zhou, J.; Sun, X.; Zhang, X.; Yang, H.; Jiang, Z.; Luo, Q.; Liu, Y.; Wang, G. miR-107 is involved in the regulation of NEDD9-mediated invasion and metastasis in breast cancer. BMC Cancer 2022, 22, 533. [Google Scholar] [CrossRef]
- Han, Z.; Li, D.; Yang, Y.; Zhang, H. LINC-DUBR Suppresses Malignant Progression of Ovarian Cancer by Downregulating miR-107 to Induce SMAC Expression. J. Healthc. Eng. 2022, 2022, 4535655. [Google Scholar] [CrossRef]
- Lützenberg, R.; Wehland, M.; Solano, K.; Nassef, M.Z.; Buken, C.; Melnik, D.; Bauer, J.; Kopp, S.; Krüger, M.; Riwaldt, S.; et al. Beneficial Effects of Low Frequency Vibration on Human Chondrocytes in Vitro. Cell Physiol. Biochem. 2019, 53, 623–637. [Google Scholar] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Yates, A.D.; Achuthan, P.; Akanni, W.; Allen, J.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R.; et al. Ensembl 2020. Nucleic Acids Res. 2020, 48, D682–D688. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dennis, G.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003, 4, P3. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids. Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Hao, Q.; Vadgama, J.V.; Wang, P. CCL2/CCR2 signaling in cancer pathogenesis. Cell Commun. Signal. 2020, 18, 82. [Google Scholar] [CrossRef]
- Smith, P.C.; Hobisch, A.; Lin, D.L.; Culig, Z.; Keller, E.T. Interleukin-6 and prostate cancer progression. Cytokine Growth Factor Rev. 2001, 12, 33–40. [Google Scholar] [CrossRef]
- Tornesello, M.L.; Faraonio, R.; Buonaguro, L.; Annunziata, C.; Starita, N.; Cerasuolo, A.; Pezzuto, F.; Tornesello, A.L.; Buonaguro, F.M. The Role of microRNAs, Long Non-coding RNAs, and Circular RNAs in Cervical Cancer. Front. Oncol. 2020, 10, 150. [Google Scholar] [CrossRef] [Green Version]
- Casarotto, M.; Fanetti, G.; Guerrieri, R.; Palazzari, E.; Lupato, V.; Steffan, A.; Polesel, J.; Boscolo-Rizzo, P.; Fratta, E. Beyond MicroRNAs: Emerging Role of Other Non-Coding RNAs in HPV-Driven Cancers. Cancers 2020, 12, 1246. [Google Scholar] [CrossRef]
- Zheng, J.; Chen, L. Non-coding RNAs-EZH2 regulatory mechanisms in cervical cancer: The current state of knowledge. Biomed. Pharmacother. 2022, 146, 112123. [Google Scholar] [CrossRef] [PubMed]
- Razavi, Z.S.; Tajiknia, V.; Majidi, S.; Ghandali, M.; Mirzaei, H.R.; Rahimian, N.; Hamblin, M.R.; Mirzaei, H. Gynecologic cancers and non-coding RNAs: Epigenetic regulators with emerging roles. Crit. Rev. Oncol. Hematol. 2021, 157, 103192. [Google Scholar] [CrossRef] [PubMed]
- Lippitz, B.E. Cytokine patterns in patients with cancer: A systematic review. Lancet Oncol. 2013, 14, e218–e228. [Google Scholar] [CrossRef]
- Rallis, K.S.; Corrigan, A.E.; Dadah, H.; George, A.M.; Keshwara, S.M.; Sideris, M.; Szabados, B. Cytokine-based Cancer Immunotherapy: Challenges and Opportunities for IL-10. Anticancer Res. 2021, 41, 3247–3252. [Google Scholar] [CrossRef]
- Bonati, L.; Tang, L. Cytokine engineering for targeted cancer immunotherapy. Curr. Opin. Chem. Biol. 2021, 62, 43–52. [Google Scholar] [CrossRef]
- Conlon, K.C.; Miljkovic, M.D.; Waldmann, T.A. Cytokines in the Treatment of Cancer. J. Interferon Cytokine Res. 2019, 39, 6–21. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).







