Impact of Exogenous Application of Potato Virus Y-Specific dsRNA on RNA Interference, Pattern-Triggered Immunity and Poly(ADP-ribose) Metabolism
Abstract
1. Introduction
2. Results
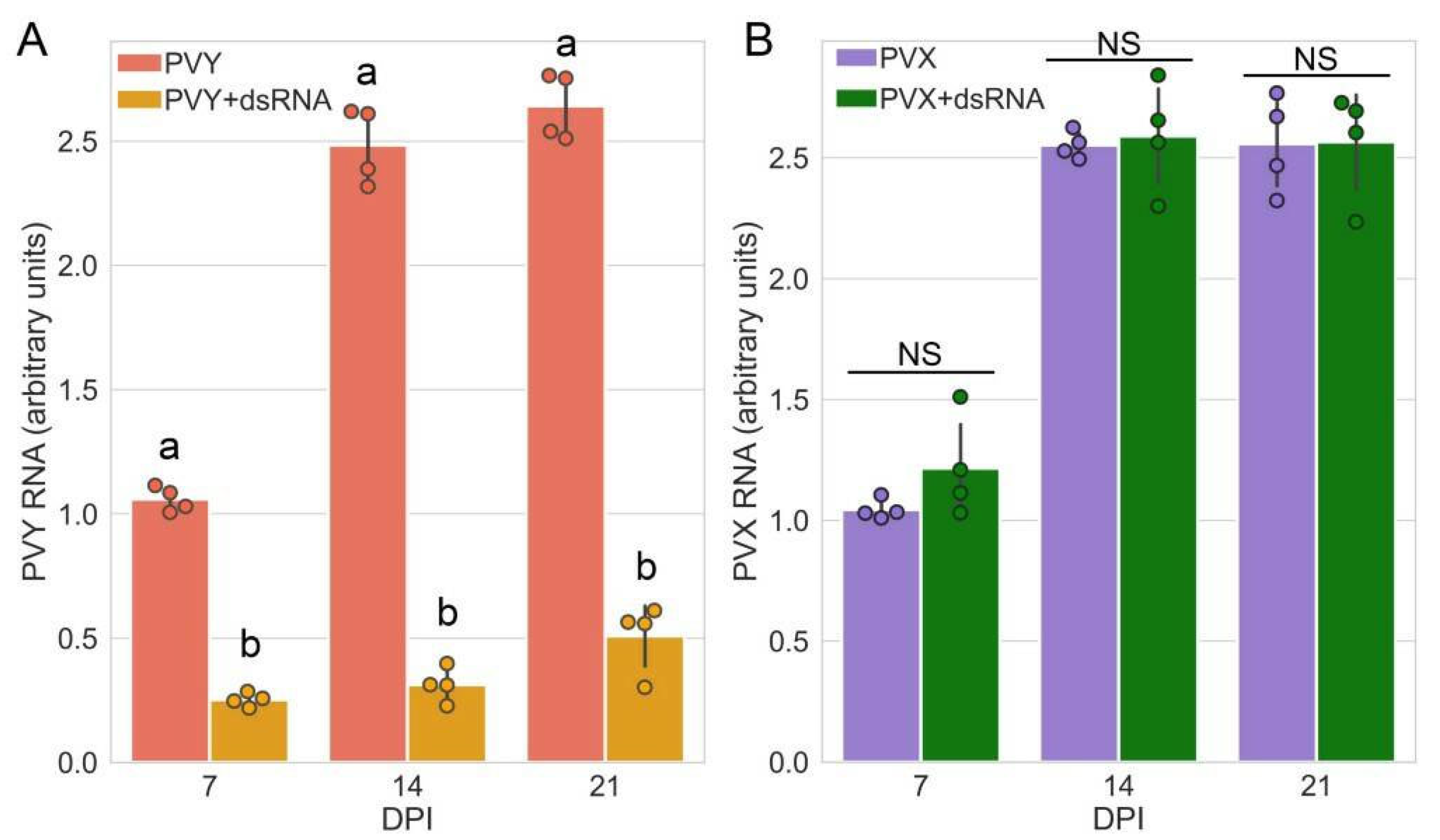
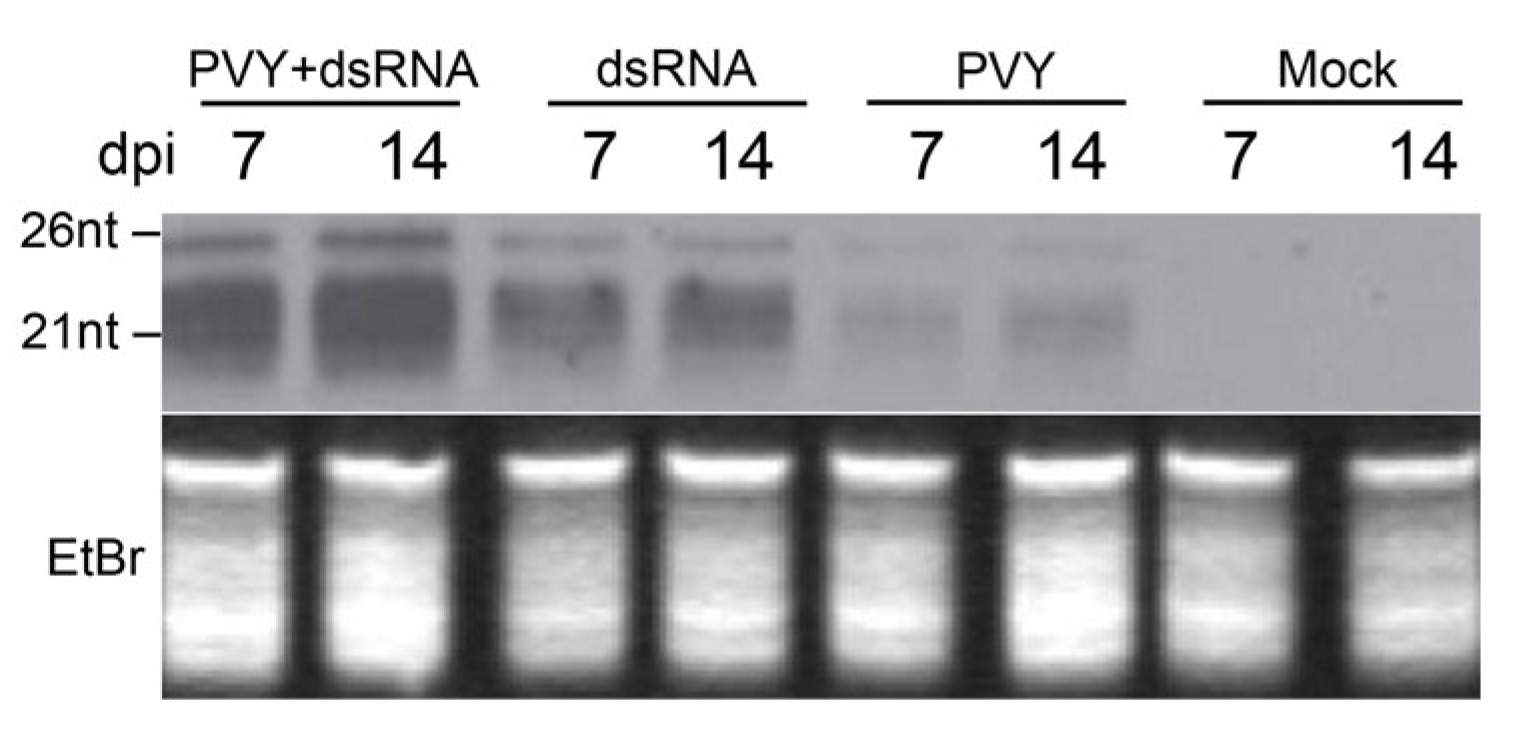
2.1. Specific Protection against PVY Infection by External dsRNA Application
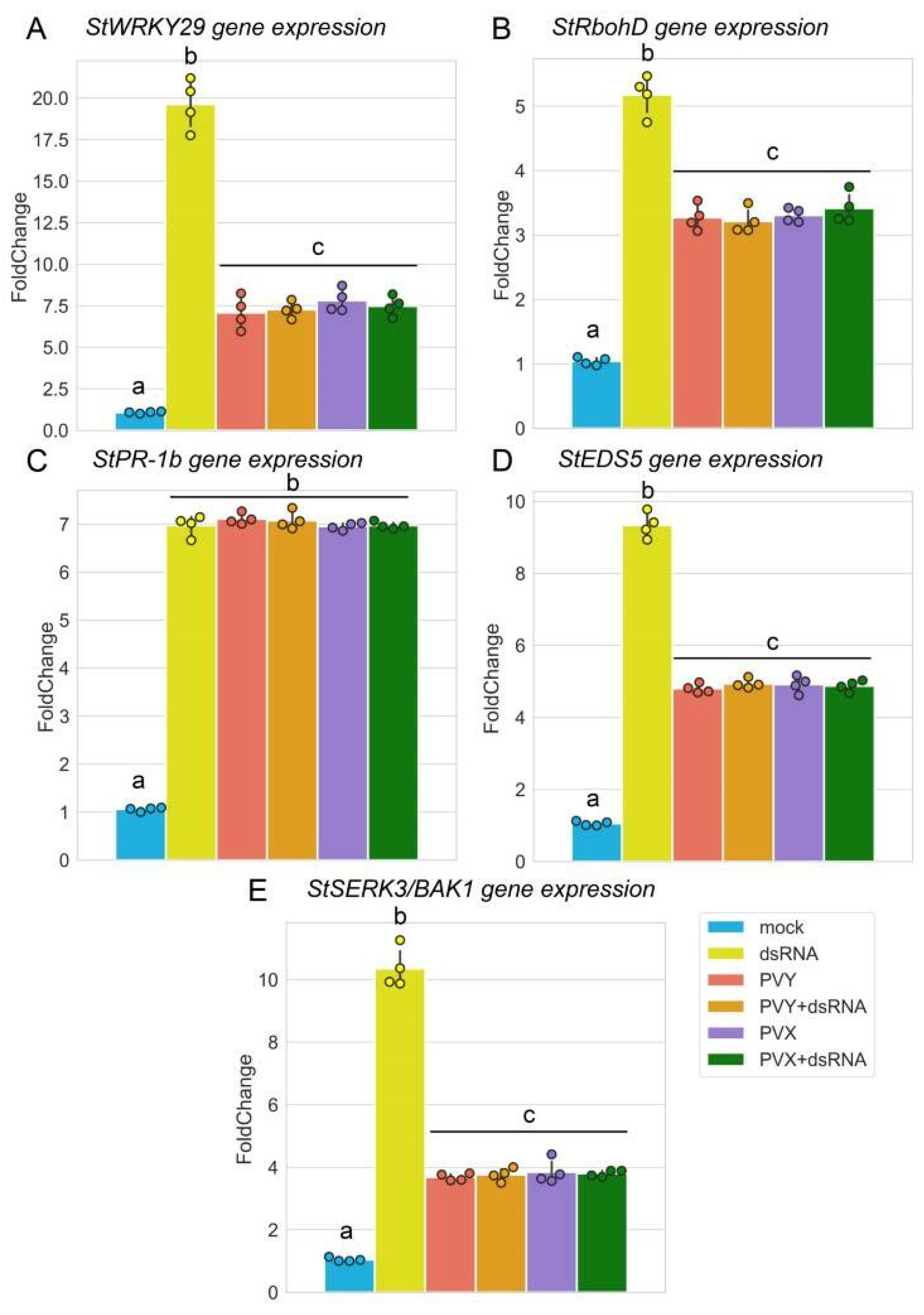
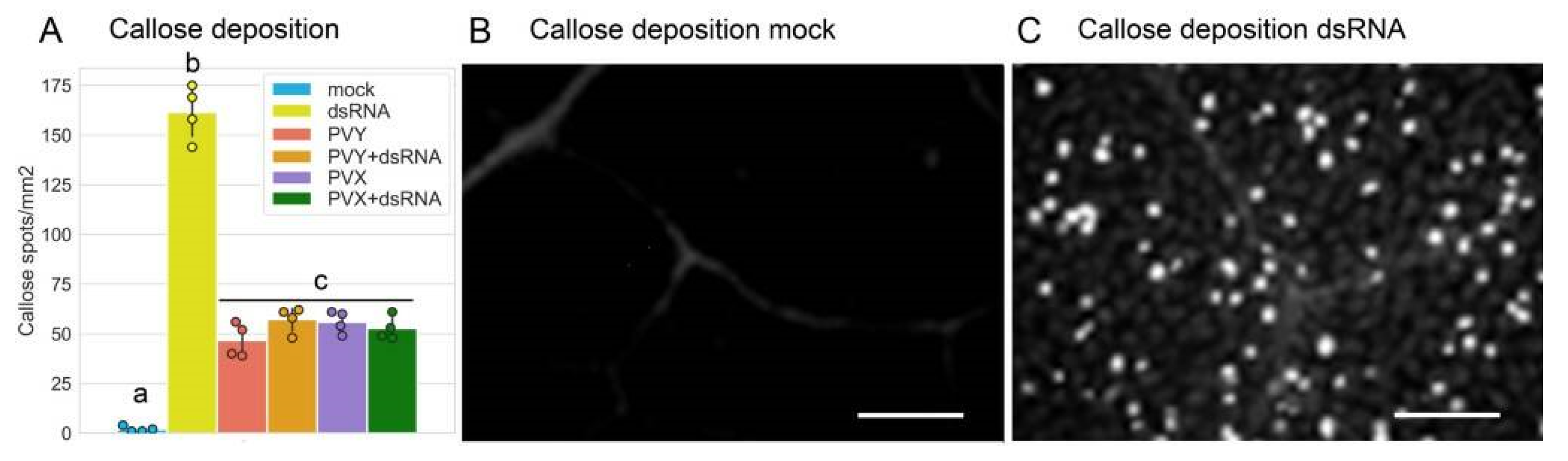
2.2. PVY-Specific dsRNA Induces Typical PTI Responses in Potato
2.3. Effect of PVY-Specific dsRNA on Poly(ADP-Ribose) Metabolism
3. Discussion
4. Materials and Methods
4.1. Virus, Plants, and Growth Conditions
4.2. Production and Purification of dsRNA
4.3. Exogenous dsRNA Application for Plant Protection against Virus Infection
4.4. Plant RNA Extraction and Northern Blot Analysis
4.5. Real Time Quantitative RT-PCR (RT-qPCR)
4.6. Callose Staining
4.7. Immunological Detection of Poly(ADP-Ribose) (PAR)
4.8. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ding, S.-W. RNA-Based Antiviral Immunity. Nat. Rev. Immunol. 2010, 10, 632–644. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Li, Y. Dissection of RNAi-Based Antiviral Immunity in Plants. Curr. Opin. Virol. 2018, 32, 88–99. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Li, Y.; Ding, S.-W. Small RNA-Based Antimicrobial Immunity. Nat. Rev. Immunol. 2019, 19, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Ghoshal, B.; Sanfaçon, H. Symptom Recovery in Virus-Infected Plants: Revisiting the Role of RNA Silencing Mechanisms. Virology 2015, 479–480, 167–179. [Google Scholar] [CrossRef]
- Baulcombe, D. RNA Silencing. Trends Biochem. Sci. 2005, 30, 290–293. [Google Scholar] [CrossRef]
- Mlotshwa, S.; Pruss, G.J.; Peragine, A.; Endres, M.W.; Li, J.; Chen, X.; Poethig, R.S.; Bowman, L.H.; Vance, V. DICER-LIKE2 Plays a Primary Role in Transitive Silencing of Transgenes in Arabidopsis. PLoS ONE 2008, 3, e1755. [Google Scholar] [CrossRef]
- Alvarado, V.; Scholthof, H.B. Plant Responses against Invasive Nucleic Acids: RNA Silencing and Its Suppression by Plant Viral Pathogens. Semin. Cell Dev. Biol. 2009, 20, 1032–1040. [Google Scholar] [CrossRef]
- Garcia-Ruiz, H.; Takeda, A.; Chapman, E.J.; Sullivan, C.M.; Fahlgren, N.; Brempelis, K.J.; Carrington, J.C. Arabidopsis RNA-Dependent RNA Polymerases and Dicer-like Proteins in Antiviral Defense and Small Interfering RNA Biogenesis during Turnip Mosaic Virus Infection. Plant Cell 2010, 22, 481–496. [Google Scholar] [CrossRef]
- Dubrovina, A.S.; Kiselev, K.V. Exogenous RNAs for Gene Regulation and Plant Resistance. Int. J. Mol. Sci. 2019, 20, 2282. [Google Scholar] [CrossRef]
- Das, P.R.; Sherif, S.M. Application of Exogenous DsRNAs-Induced RNAi in Agriculture: Challenges and Triumphs. Front. Plant Sci. 2020, 11, 946. [Google Scholar] [CrossRef]
- Taliansky, M.; Samarskaya, V.; Zavriev, S.K.; Fesenko, I.; Kalinina, N.O.; Love, A.J. RNA-Based Technologies for Engineering Plant Virus Resistance. Plants Basel Switz. 2021, 10, 82. [Google Scholar] [CrossRef]
- Rego-Machado, C.M.; Nakasu, E.Y.T.; Silva, J.M.F.; Lucinda, N.; Nagata, T.; Inoue-Nagata, A.K. SiRNA Biogenesis and Advances in Topically Applied DsRNA for Controlling Virus Infections in Tomato Plants. Sci. Rep. 2020, 10, 22277. [Google Scholar] [CrossRef]
- Nilon, A.; Robinson, K.; Pappu, H.R.; Mitter, N. Current Status and Potential of RNA Interference for the Management of Tomato Spotted Wilt Virus and Thrips Vectors. Pathog. Basel Switz. 2021, 10, 320. [Google Scholar] [CrossRef] [PubMed]
- Necira, K.; Makki, M.; Sanz-García, E.; Canto, T.; Djilani-Khouadja, F.; Tenllado, F. Topical Application of Escherichia coli-Encapsulated dsRNA Induces Resistance in Nicotiana benthamiana to Potato Viruses and Involves RDR6 and Combined Activities of DCL2 and DCL4. Plants 2021, 10, 644. [Google Scholar] [CrossRef] [PubMed]
- Niehl, A.; Wyrsch, I.; Boller, T.; Heinlein, M. Double-Stranded RNAs Induce a Pattern-Triggered Immune Signaling Pathway in Plants. New Phytol. 2016, 211, 1008–1019. [Google Scholar] [CrossRef] [PubMed]
- Niehl, A.; Heinlein, M. Perception of Double-Stranded RNA in Plant Antiviral Immunity. Mol. Plant Pathol. 2019, 20, 1203–1210. [Google Scholar] [CrossRef]
- Huang, S.; Tang, Z.; Zhao, R.; Hong, Y.; Zhu, S.; Fan, R.; Ding, K.; Cao, M.; Luo, K.; Geng, M.; et al. Genome-Wide Identification of Cassava MeRboh Genes and Functional Analysis in Arabidopsis. Plant Physiol. Biochem. PPB 2021, 167, 296–308. [Google Scholar] [CrossRef]
- Chow, K.T.; Gale, M.; Loo, Y.-M. RIG-I and Other RNA Sensors in Antiviral Immunity. Annu. Rev. Immunol. 2018, 36, 667–694. [Google Scholar] [CrossRef]
- Hartmann, G. Nucleic Acid Immunity. Adv. Immunol. 2017, 133, 121–169. [Google Scholar] [CrossRef]
- Tan, S.-J.; Zhang, X.; Jin, X.-K.; Li, W.-W.; Li, J.-Y.; Wang, Q. Fatty Acid Binding Protein FABP3 from Chinese Mitten Crab Eriocheir Sinensis Participates in Antimicrobial Responses. Fish Shellfish Immunol. 2015, 43, 264–274. [Google Scholar] [CrossRef]
- Vabret, N.; Bhardwaj, N.; Greenbaum, B.D. Sequence-Specific Sensing of Nucleic Acids. Trends Immunol. 2017, 38, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.; Park, Y.-S.; Lee, S.; Song, G.C.; Ryu, C.-M. Bacterial RNAs Activate Innate Immunity in Arabidopsis. New Phytol. 2016, 209, 785–797. [Google Scholar] [CrossRef] [PubMed]
- Boller, T.; Felix, G. A Renaissance of Elicitors: Perception of Microbe-Associated Molecular Patterns and Danger Signals by Pattern-Recognition Receptors. Annu. Rev. Plant Biol. 2009, 60, 379–406. [Google Scholar] [CrossRef] [PubMed]
- Kørner, C.J.; Klauser, D.; Niehl, A.; Domínguez-Ferreras, A.; Chinchilla, D.; Boller, T.; Heinlein, M.; Hann, D.R. The Immunity Regulator BAK1 Contributes to Resistance against Diverse RNA Viruses. Mol. Plant-Microbe Interact. MPMI 2013, 26, 1271–1280. [Google Scholar] [CrossRef] [PubMed]
- Nicaise, V.; Candresse, T. Plum Pox Virus Capsid Protein Suppresses Plant Pathogen-Associated Molecular Pattern (PAMP)-Triggered Immunity. Mol. Plant Pathol. 2017, 18, 878–886. [Google Scholar] [CrossRef]
- Zvereva, A.S.; Golyaev, V.; Turco, S.; Gubaeva, E.G.; Rajeswaran, R.; Schepetilnikov, M.V.; Srour, O.; Ryabova, L.A.; Boller, T.; Pooggin, M.M. Viral Protein Suppresses Oxidative Burst and Salicylic Acid-Dependent Autophagy and Facilitates Bacterial Growth on Virus-Infected Plants. New Phytol. 2016, 211, 1020–1034. [Google Scholar] [CrossRef]
- Morozov, S.Y.; Solovyev, A.G.; Kalinina, N.O.; Taliansky, M.E. Double-Stranded RNAs in Plant Protection Against Pathogenic Organisms and Viruses in Agriculture. Acta Nat. 2019, 11, 13–21. [Google Scholar] [CrossRef]
- Hernández-Soto, A.; Chacón-Cerdas, R. RNAi Crop Protection Advances. Int. J. Mol. Sci. 2021, 22, 12148. [Google Scholar] [CrossRef]
- Torrance, L.; Talianksy, M.E. Potato Virus Y Emergence and Evolution from the Andes of South America to Become a Major Destructive Pathogen of Potato and Other Solanaceous Crops Worldwide. Viruses 2020, 12, 1430. [Google Scholar] [CrossRef]
- Kreuze, J.F.; Souza-Dias, J.A.C.; Jeevalatha, A.; Figueira, A.R.; Valkonen, J.P.T.; Jones, R.A.C. Viral Diseasesin Potato. In The Potato Crop: Its Agricultural, Nutritional and Social Contribution to Humankind; Campos, H., Ortiz, O., Eds.; Springer: Berlin/Heidelberg, Germany, 2020; pp. 389–430. [Google Scholar] [CrossRef]
- Sarowar, S.; Alam, S.T.; Makandar, R.; Lee, H.; Trick, H.N.; Dong, Y.; Shah, J. Targeting the Pattern-Triggered Immunity Pathway to Enhance Resistance to Fusarium Graminearum. Mol. Plant Pathol. 2019, 20, 626–640. [Google Scholar] [CrossRef]
- Makarova, S.; Makhotenko, A.; Spechenkova, N.; Love, A.J.; Kalinina, N.O.; Taliansky, M. Interactive Responses of Potato (Solanum tuberosum L.) Plants to Heat Stress and Infection with Potato Virus Y. Front. Microbiol. 2018, 9, 2582. [Google Scholar] [CrossRef] [PubMed]
- Rissel, D.; Peiter, E. Poly(ADP-Ribose) Polymerases in Plants and Their Human Counterparts: Parallels and Peculiarities. Int. J. Mol. Sci. 2019, 20, 1638. [Google Scholar] [CrossRef] [PubMed]
- Gibson, B.A.; Kraus, W.L. New Insights into the Molecular and Cellular Functions of Poly(ADP-Ribose) and PARPs. Nat. Rev. Mol. Cell Biol. 2012, 13, 411–424. [Google Scholar] [CrossRef] [PubMed]
- Briggs, A.G.; Bent, A.F. Poly(ADP-Ribosyl)Ation in Plants. Trends Plant Sci. 2011, 16, 372–380. [Google Scholar] [CrossRef]
- Yan, S.; Wang, W.; Marqués, J.; Mohan, R.; Saleh, A.; Durrant, W.E.; Song, J.; Dong, X. Salicylic Acid Activates DNA Damage Responses to Potentiate Plant Immunity. Mol. Cell 2013, 52, 602–610. [Google Scholar] [CrossRef]
- Song, J.; Bent, A.F. Microbial Pathogens Trigger Host DNA Double-Strand Breaks Whose Abundance Is Reduced by Plant Defense Responses. PLoS Pathog. 2014, 10, e1004030. [Google Scholar] [CrossRef]
- Feng, B.; Liu, C.; de Oliveira, M.V.V.; Intorne, A.C.; Li, B.; Babilonia, K.; de Souza Filho, G.A.; Shan, L.; He, P. Protein Poly(ADP-Ribosyl)Ation Regulates Arabidopsis Immune Gene Expression and Defense Responses. PLoS Genet. 2015, 11, e1004936. [Google Scholar] [CrossRef]
- Love, A.J.; Yu, C.; Petukhova, N.V.; Kalinina, N.O.; Chen, J.; Taliansky, M.E. Cajal Bodies and Their Role in Plant Stress and Disease Responses. RNA Biol. 2017, 14, 779–790. [Google Scholar] [CrossRef]
- Briggs, A.G.; Adams-Phillips, L.C.; Keppler, B.D.; Zebell, S.G.; Arend, K.C.; Apfelbaum, A.A.; Smith, J.A.; Bent, A.F. A Transcriptomics Approach Uncovers Novel Roles for Poly(ADP-Ribosyl)Ation in the Basal Defense Response in Arabidopsis Thaliana. PLoS ONE 2017, 12, e0190268. [Google Scholar] [CrossRef]
- Naveed, K.; Mitter, N.; Harper, A.; Dhingra, A.; Pappu, H.R. Comparative Analysis of Virus-Specific Small RNA Profiles of Three Biologically Distinct Strains of Potato Virus Y in Infected Potato (Solanum tuberosum) Cv. Russet Burbank. Virus Res. 2014, 191, 153–160. [Google Scholar] [CrossRef]
- Lacaze, A.; Joly, D.L. Structural Specificity in Plant–Filamentous Pathogen Interactions. Mol. Plant Pathol. 2020, 21, 1513–1525. [Google Scholar] [CrossRef] [PubMed]
- Otulak-Kozieł, K.; Kozieł, E.; Valverde, R.A. The Respiratory Burst Oxidase Homolog D (RbohD) Cell and Tissue Distribution in Potato-Potato Virus Y (PVYNTN) Hypersensitive and Susceptible Reactions. Int. J. Mol. Sci. 2019, 20, 2741. [Google Scholar] [CrossRef] [PubMed]
- Pajerowska, K.M.; Parker, J.E.; Gebhardt, C. Potato Homologs of Arabidopsis Thaliana Genes Functional in Defense Signaling--Identification, Genetic Mapping, and Molecular Cloning. Mol. Plant-Microbe Interact. MPMI 2005, 18, 1107–1119. [Google Scholar] [CrossRef] [PubMed]
- Nietzschmann, L.; Gorzolka, K.; Smolka, U.; Matern, A.; Eschen-Lippold, L.; Scheel, D.; Rosahl, S. Early Pep-13-Induced Immune Responses Are SERK3A/B-Dependent in Potato. Sci. Rep. 2019, 9, 18380. [Google Scholar] [CrossRef]
- Cheng, X.; Wang, A. Multifaceted defense and counter-defense in co-evolutionary arms race between plants and viruses. Commun. Integr. Biol. 2017, 10, 508–525. [Google Scholar] [CrossRef]
- Love, A.J.; Geri, C.; Laird, J.; Carr, C.; Yun, B.-W.; Loake, G.J.; Tada, Y.; Sadanandom, A.; Milner, J.J. Cauliflower Mosaic Virus Protein P6 Inhibits Signaling Responses to Salicylic Acid and Regulates Innate Immunity. PLoS ONE 2012, 7, e47535. [Google Scholar] [CrossRef]
- Adams-Phillips, L.; Wan, J.; Tan, X.; Dunning, F.M.; Meyers, B.C.; Michelmore, R.W.; Bent, A.F. Discovery of ADP-Ribosylation and Other Plant Defense Pathway Elements through Expression Profiling of Four Different Arabidopsis-Pseudomonas R-Avr Interactions. Mol. Plant-Microbe Interact. MPMI 2008, 21, 646–657. [Google Scholar] [CrossRef]
- Adams-Phillips, L.; Briggs, A.G.; Bent, A.F. Disruption of Poly(ADP-Ribosyl)Ation Mechanisms Alters Responses of Arabidopsis to Biotic Stress. Plant Physiol. 2010, 152, 267–280. [Google Scholar] [CrossRef]
- Sekine, K.-T.; Kawakami, S.; Hase, S.; Kubota, M.; Ichinose, Y.; Shah, J.; Kang, H.-G.; Klessig, D.F.; Takahashi, H. High Level Expression of a Virus Resistance Gene, RCY1, Confers Extreme Resistance to Cucumber Mosaic Virus in Arabidopsis Thaliana. Mol. Plant-Microbe Interact. MPMI 2008, 21, 1398–1407. [Google Scholar] [CrossRef]
- Gu, Z.; Pan, W.; Chen, W.; Lian, Q.; Wu, Q.; Lv, Z.; Cheng, X.; Ge, X. New Perspectives on the Plant PARP Family: Arabidopsis PARP3 Is Inactive, and PARP1 Exhibits Predominant Poly (ADP-Ribose) Polymerase Activity in Response to DNA Damage. BMC Plant Biol. 2019, 19, 364. [Google Scholar] [CrossRef]
- Qin, L.; Mo, N.; Muhammad, T.; Liang, Y. Genome-Wide Analysis of DCL, AGO, and RDR Gene Families in Pepper (Capsicum annuum L.). Int. J. Mol. Sci. 2018, 19, 1038. [Google Scholar] [CrossRef] [PubMed]
- Boccara, M.; Sarazin, A.; Thiébeauld, O.; Jay, F.; Voinnet, O.; Navarro, L.; Colot, V. The Arabidopsis MiR472-RDR6 Silencing Pathway Modulates PAMP- and Effector-Triggered Immunity through the Post-Transcriptional Control of Disease Resistance Genes. PLoS Pathog. 2014, 10, e1003883. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Pignatta, D.; Bendix, C.; Brunkard, J.O.; Cohn, M.M.; Tung, J.; Sun, H.; Kumar, P.; Baker, B. MicroRNA Regulation of Plant Innate Immune Receptors. Proc. Natl. Acad. Sci. USA 2012, 109, 1790–1795. [Google Scholar] [CrossRef] [PubMed]
- Shivaprasad, P.V.; Chen, H.-M.; Patel, K.; Bond, D.M.; Santos, B.A.C.M.; Baulcombe, D.C. A MicroRNA Superfamily Regulates Nucleotide Binding Site-Leucine-Rich Repeats and Other MRNAs. Plant Cell 2012, 24, 859–874. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.-J.; Donahue, K.; Koh, Y.; Martin, R.R.; Choi, M.-Y. Microbial-Based Double-Stranded RNA Production to Develop Cost-Effective RNA Interference Application for Insect Pest Management. Int. J. Insect Sci. 2019, 11, 1179543319840323. [Google Scholar] [CrossRef]
- Taliansky, M.; Kim, S.H.; Mayo, M.A.; Kalinina, N.O.; Fraser, G.; McGeachy, K.D.; Barker, H. Escape of a plant virus from amplicon-mediated RNA silencing is associated with biotic or abiotic stress. Plant J. 2004, 39, 194–205. [Google Scholar] [CrossRef]
- Nicot, N.; Hausman, J.-F.; Hoffmann, L.; Evers, D. Housekeeping Gene Selection for Real-Time RT-PCR Normalization in Potato during Biotic and Abiotic Stress. J. Exp. Bot. 2005, 56, 2907–2914. [Google Scholar] [CrossRef]
- Baebler, Š.; Stare, K.; Kovač, M.; Blejec, A.; Prezelj, N.; Stare, T.; Kogovšek, P.; Pompe-Novak, M.; Rosahl, S.; Ravnikar, M.; et al. Dynamics of Responses in Compatible Potato—Potato Virus Y Interaction Are Modulated by Salicylic Acid. PLoS ONE 2011, 6, e29009. [Google Scholar] [CrossRef]
- Hauck, P.; Thilmony, R.; He, S.Y. A Pseudomonas Syringae Type III Effector Suppresses Cell Wall-Based Extracellular Defense in Susceptible Arabidopsis Plants. Proc. Natl. Acad. Sci. USA 2003, 100, 8577–8582. [Google Scholar] [CrossRef]
- De Block, M.; Verduyn, C.; De Brouwer, D.; Cornelissen, M. Poly(ADP-Ribose) Polymerase in Plants Affects Energy Homeostasis, Cell Death and Stress Tolerance. Plant J. Cell Mol. Biol. 2005, 41, 95–106. [Google Scholar] [CrossRef]
- Affar, E.B.; Duriez, P.J.; Shah, R.G.; Winstall, E.; Germain, M.; Boucher, C.; Bourassa, S.; Kirkland, J.B.; Poirier, G.G. Immunological Determination and Size Characterization of Poly(ADP-Ribose) Synthesized in Vitro and in Vivo. Biochim. Biophys. Acta 1999, 1428, 137–146. [Google Scholar] [CrossRef]
- Van Rossum, D.; Schuurmans, F.P.; Gillis, M.; Muyotcha, A.; Van Verseveld, H.W.; Stouthamer, A.H.; Boogerd, F.C. Genetic and Phenetic Analyses of Bradyrhizobium Strains Nodulating Peanut (Arachis hypogaea L.) Roots. Appl. Environ. Microbiol. 1995, 61, 1599–1609. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Chen, J.; Hiruki, C. Optimization and application of a multiplex RT-PCR system for simultaneous detection of five potato viruses using 18S rRNA as an internal control. Plant Dis. 2006, 90, 185–189. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Glushkevich, A.; Spechenkova, N.; Fesenko, I.; Knyazev, A.; Samarskaya, V.; Kalinina, N.O.; Taliansky, M.; Love, A.J. Transcriptomic Reprogramming, Alternative Splicing and RNA Methylation in Potato (Solanum tuberosum L.) Plants in Response to Potato Virus Y Infection. Plants 2022, 11, 635. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Samarskaya, V.O.; Spechenkova, N.; Markin, N.; Suprunova, T.P.; Zavriev, S.K.; Love, A.J.; Kalinina, N.O.; Taliansky, M. Impact of Exogenous Application of Potato Virus Y-Specific dsRNA on RNA Interference, Pattern-Triggered Immunity and Poly(ADP-ribose) Metabolism. Int. J. Mol. Sci. 2022, 23, 7915. https://doi.org/10.3390/ijms23147915
Samarskaya VO, Spechenkova N, Markin N, Suprunova TP, Zavriev SK, Love AJ, Kalinina NO, Taliansky M. Impact of Exogenous Application of Potato Virus Y-Specific dsRNA on RNA Interference, Pattern-Triggered Immunity and Poly(ADP-ribose) Metabolism. International Journal of Molecular Sciences. 2022; 23(14):7915. https://doi.org/10.3390/ijms23147915
Chicago/Turabian StyleSamarskaya, Viktoriya O., Nadezhda Spechenkova, Nikolay Markin, Tatyana P. Suprunova, Sergey K. Zavriev, Andrew J. Love, Natalia O. Kalinina, and Michael Taliansky. 2022. "Impact of Exogenous Application of Potato Virus Y-Specific dsRNA on RNA Interference, Pattern-Triggered Immunity and Poly(ADP-ribose) Metabolism" International Journal of Molecular Sciences 23, no. 14: 7915. https://doi.org/10.3390/ijms23147915
APA StyleSamarskaya, V. O., Spechenkova, N., Markin, N., Suprunova, T. P., Zavriev, S. K., Love, A. J., Kalinina, N. O., & Taliansky, M. (2022). Impact of Exogenous Application of Potato Virus Y-Specific dsRNA on RNA Interference, Pattern-Triggered Immunity and Poly(ADP-ribose) Metabolism. International Journal of Molecular Sciences, 23(14), 7915. https://doi.org/10.3390/ijms23147915






