Soil and Soilless Tomato Cultivation Promote Different Microbial Communities That Provide New Models for Future Crop Interventions
Abstract
:1. Introduction
2. Results
2.1. General Structure of the Tomato Root Associated Microbiome in the Seed, in Nursery and in Greenhouse in Soil and Soilless Cultivation Systems
2.1.1. Taxonomic Distributions of Bacterial Communities
2.1.2. Taxonomic Distribution of Fungal Communities

2.2. Analysis of Diversity Indices
2.2.1. Richness and Diversity of Microbial Communities
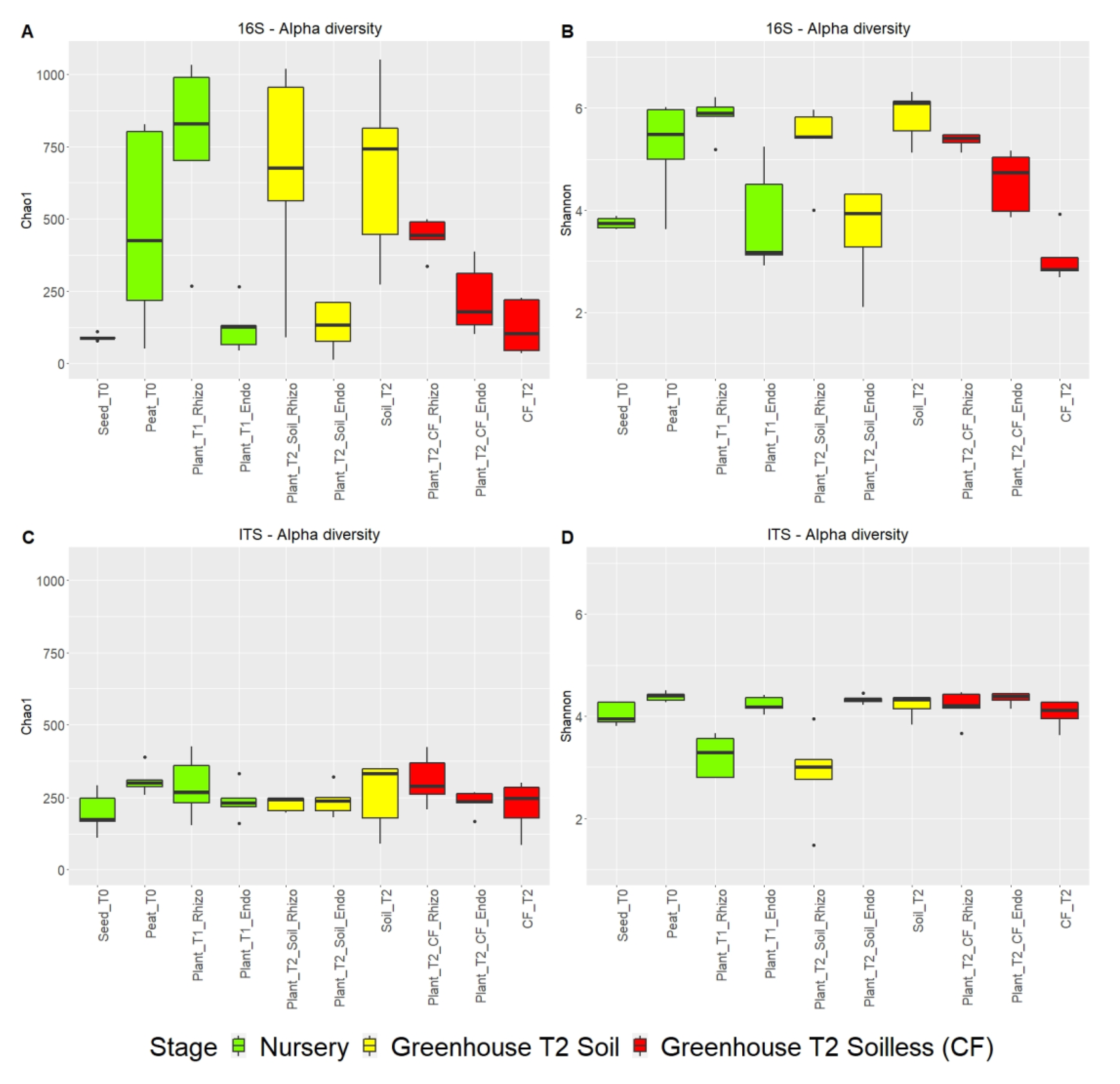
2.2.2. Comparison of Microbial Communities
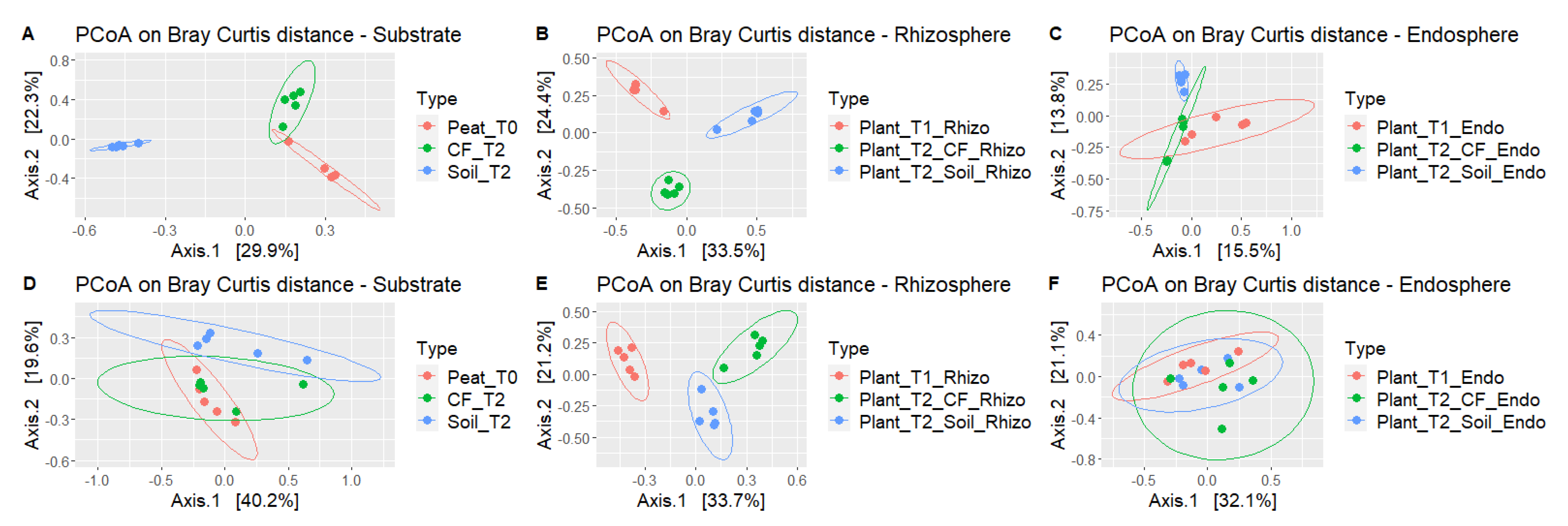
2.2.3. Tomatoes Grown in Soil and Soilless Media Attract Different Microbiome Communities
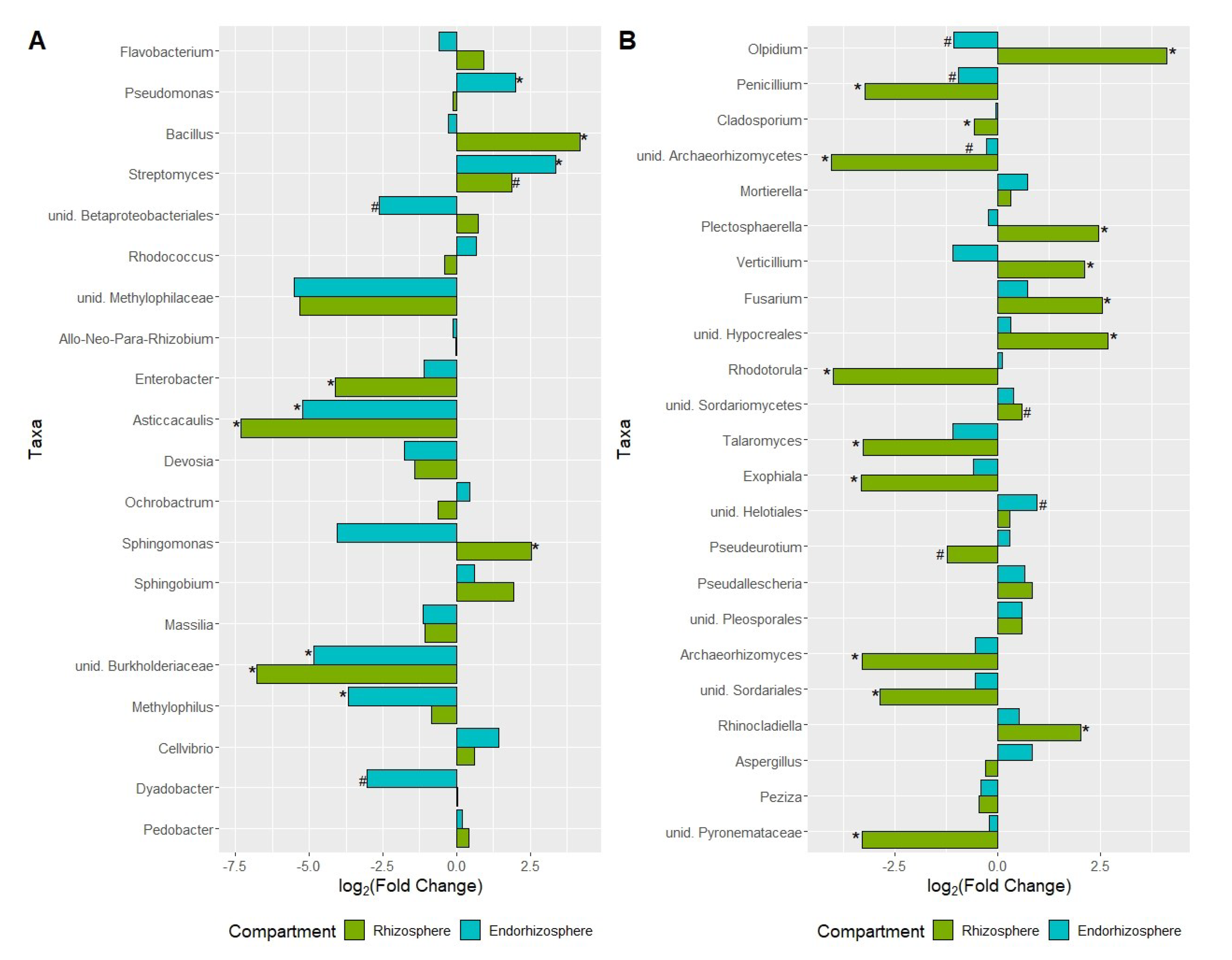
2.2.4. Network Analysis and Keystone Taxa Associated with Bacterial and Fungal Communities
3. Discussion
3.1. Tomato Root Microbiome in Soil vs. Soilless Growing Conditions
3.2. Bacterial Communities Associated with the Root Environment of Tomato
3.3. Fungal Communities Associated with the Root Environment of Tomato
4. Materials and Methods
4.1. Experimental Design
4.2. Sample Preparation
- -
- Culture substrates and agricultural soil: 5 g aliquots were suspended in 20 mL of sterile saline buffer (0.85% NaCl) in sterile tubes and vortexed for 1 min.
- -
- Rhizosphere (R): tomato roots were vigorously shaken by hand to remove soil particles. Five grams of roots with firmly attached soil was taken and suspended in 20 mL of saline buffer in sterile tubes and vortexed for 5 min.
- -
- Samples of seeds (S) and root endosphere (E) were collected in sterile tubes with 20 mL of sterile saline buffer. The samples were then washed several times with sterile distilled water. Seeds and root samples were surface sterilized and treated according to the protocol described by Bragina et al. (2012) [70]. Sterility was assessed by placing the surface-sterilized seeds and roots on Potato Dextrose Agar plates (PDA, Oxoid, Milan, Italy) at 27 °C for 4 days. All samples (roots and seeds) were homogenized with a mortar and pestle and suspended in 20 mL of sterile saline buffer.
4.3. Metagenomics Analysis
4.3.1. DNA Isolation, PCR Amplification and Sequencing
4.3.2. Data Analysis of 16S rDNA and ITS Amplicon for Determination of Microbial Community Structure
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Food and Agriculture Organization of the United Nations FAOSTAT. Available online: http://www.fao.org/faostat/en/ (accessed on 13 December 2021).
- Dorais, M.; Papadopoulos, A.P.; Gosselin, A. Greenhouse tomato fruit quality. In Horticultural Reviews; Wiley: Hoboken, NJ, USA, 2010; pp. 239–319. [Google Scholar]
- Savvas, D.; Gianquinto, G.; Tuzel, Y.; Gruda, N. Chapter 12. Soilless culture. In Good Agricultural Practices for Greenhouse Vegetable Crops: Principles for Mediterranean Climate Areas; FAO: Rome, Italy, 2013. [Google Scholar]
- Gruda, N.; Qaryouti, M.M.; Leonardi, C. Growing media. In Good Agricultural Practices for Greenhouse Vegetable Crops: Principles for Mediterranean Climate Areas; FAO: Rome, Italy, 2013. [Google Scholar]
- Postma, J. The status of biological control of plant diseases in soilless cultivation. In Recent Developments in Management of Plant Diseases; Springer: Berlin, Germany, 2010. [Google Scholar]
- Vallance, J.; Déniel, F.; Le Floch, G.; Guérin-Dubrana, L.; Blancard, D.; Rey, P. Pathogenic and beneficial microorganisms in soilless cultures. Agron. Sustain. Dev. 2011, 31, 711–726. [Google Scholar] [CrossRef]
- Stanghellini, M.E.; Rasmussen, S.L. Identification and origin of plant pathogenic microorganisms in recirculating nutrient solutions. Adv. Sp. Res. 1994, 14, 349–355. [Google Scholar] [CrossRef]
- Dimartino, M.; Panebianco, S.; Vitale, A.; Castello, I.; Leonardi, C.; Cirvilleri, G.; Polizzi, G. Occurrence and pathogenicity of Pseudomonas fluorescens and P. putida on tomato plants in Italy. J. Plant Pathol. 2011, 93, 79–87. [Google Scholar] [CrossRef]
- Aiello, D.; Scuderi, G.; Vitale, A.; Firrao, G.; Polizzi, G.; Cirvilleri, G. A pith necrosis caused by Xanthomonas perforans on tomato plants. Eur. J. Plant Pathol. 2013, 137, 29–41. [Google Scholar] [CrossRef]
- Caruso, A.; Licciardello, G.; La Rosa, R.; Catara, V.; Bella, P. Mixed infection of Pectobacterium carotovorum subsp. carotovorum and P. carotovorum subsp. brasiliensis in tomato stem rot in Italy. J. Plant Pathol. 2016, 98, 661–665. [Google Scholar] [CrossRef]
- Mendes, R.; Kruijt, M.; De Bruijn, I.; Dekkers, E.; Van Der Voort, M.; Schneider, J.H.M.; Piceno, Y.M.; DeSantis, T.Z.; Andersen, G.L.; Bakker, P.A.H.M.; et al. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 2011, 332, 1092–1100. [Google Scholar] [CrossRef]
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; Van Der Putten, W.H. Going back to the roots: The microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789–799. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, Y.; Zhang, P.; Trivedi, P.; Riera, N.; Wang, Y.; Liu, X.; Fan, G.; Tang, J.; Coletta-Filho, H.D.; et al. The structure and function of the global citrus rhizosphere microbiome. Nat. Commun. 2018, 9, 4894. [Google Scholar] [CrossRef] [Green Version]
- Bulgarelli, D.; Schlaeppi, K.; Spaepen, S.; Van Themaat, E.V.L.; Schulze-Lefert, P. Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 2013, 64, 807–838. [Google Scholar] [CrossRef] [Green Version]
- Berg, G.; Rybakova, D.; Grube, M.; Köberl, M. The plant microbiome explored: Implications for experimental botany. J. Exp. Bot. 2016, 67, 955–1002. [Google Scholar] [CrossRef]
- Yan, Y.; Kuramae, E.E.; De Hollander, M.; Klinkhamer, P.G.L.; Van Veen, J.A. Functional traits dominate the diversity-related selection of bacterial communities in the rhizosphere. ISME J. 2017, 11, 56–66. [Google Scholar] [CrossRef] [Green Version]
- Mauch-Mani, B.; Lombardi, N.; Woo, S.L.; Vinale, F.; Turrà, D.; Marra, R. Editorial: The Plant Holobiont Volume II: Impacts of the Rhizosphere on Plant Health. Front. Plant Sci. 2021, 12, 809291. [Google Scholar] [CrossRef]
- Raaijmakers, J.M.; Paulitz, T.C.; Steinberg, C.; Alabouvette, C.; Moënne-Loccoz, Y. The rhizosphere: A playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil 2009, 321, 341–361. [Google Scholar] [CrossRef] [Green Version]
- Vandenkoornhuyse, P.; Quaiser, A.; Duhamel, M.; Le Van, A.; Dufresne, A. The importance of the microbiome of the plant holobiont. New Phytol. 2015, 206, 1196–1206. [Google Scholar] [CrossRef]
- Hardoim, P.R.; Overbeek, L.S.; Berg, G.; Pirttilä, A.M.; Compant, S.; Campisano, A.; Döring, M.; Sessitsch, A. The hidden world within plants: Ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef] [Green Version]
- Collinge, D.B.; Jørgensen, H.J.L.; Latz, M.A.C.; Manzotti, A.; Ntana, F.; Rojas, E.C.; Jensen, B. Searching for novel fungal biological control agents for plant disease control among endophytes. In Endophytes for a Growing World; Cambridge University Press: New York, NY, USA, 2019. [Google Scholar]
- Truyens, S.; Weyens, N.; Cuypers, A.; Vangronsveld, J. Bacterial seed endophytes: Genera, vertical transmission and interaction with plants. Environ. Microbiol. Rep. 2015, 7, 40–50. [Google Scholar] [CrossRef]
- Bergna, A.; Cernava, T.; Rändler, M.; Grosch, R.; Zachow, C.; Berg, G. Tomato seeds preferably transmit plant beneficial endophytes. Phytobiomes J. 2018, 2, 183–193. [Google Scholar] [CrossRef] [Green Version]
- Cavazos, B.R.; Bohner, T.F.; Donald, M.L.; Sneck, M.E.; Shadow, A.; Omacini, M.; Rudgers, J.A.; Miller, T.E.X. Testing the roles of vertical transmission and drought stress in the prevalence of heritable fungal endophytes in annual grass populations. New Phytol. 2018, 219, 1075–1084. [Google Scholar] [CrossRef] [Green Version]
- Rezki, S.; Campion, C.; Simoneau, P.; Jacques, M.A.; Shade, A.; Barret, M. Assembly of seed-associated microbial communities within and across successive plant generations. Plant Soil 2018, 422, 67–69. [Google Scholar] [CrossRef] [Green Version]
- Brader, G.; Compant, S.; Vescio, K.; Mitter, B.; Trognitz, F.; Ma, L.J.; Sessitsch, A. Ecology and genomic insights into plant-pathogenic and plant-nonpathogenic endophytes. Annu. Rev. Phytopathol. 2017, 55, 61–83. [Google Scholar] [CrossRef]
- Berg, G.; Krechel, A.; Ditz, M.; Sikora, R.A.; Ulrich, A.; Hallmann, J. Endophytic and ectophytic potato-associated bacterial communities differ in structure and antagonistic function against plant pathogenic fungi. FEMS Microbiol. Ecol. 2005, 51, 215–229. [Google Scholar] [CrossRef] [Green Version]
- Mendes, R.; Garbeva, P.; Raaijmakers, J.M. The rhizosphere microbiome: Significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 2013, 37, 634–663. [Google Scholar] [CrossRef]
- Poli, A.; Lazzari, A.; Prigione, V.; Voyron, S.; Spadaro, D.; Varese, G.C. Influence of plant genotype on the cultivable fungi associated to tomato rhizosphere and roots in different soils. Fungal Biol. 2016, 120, 862–872. [Google Scholar] [CrossRef] [Green Version]
- Dong, C.J.; Wang, L.L.; Li, Q.; Shang, Q.M. Bacterial communities in the rhizosphere, phyllosphere and endosphere of tomato plants. PLoS ONE 2019, 14, e0223847. [Google Scholar] [CrossRef] [PubMed]
- French, E.; Tran, T.; Iyer-Pascuzzi, A.S. Tomato genotype modulates selection and responses to root microbiota. Phytobiomes J. 2020, 4, 314–326. [Google Scholar] [CrossRef]
- Manzotti, A.; Bergna, A.; Burow, M.; Jørgensen, H.J.L.; Cernava, T.; Berg, G.; Collinge, D.B.; Jensen, B. Insights into the community structure and lifestyle of the fungal root endophytes of tomato by combining amplicon sequencing and isolation approaches with phytohormone profiling. FEMS Microbiol. Ecol. 2020, 96, fiaa052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taffner, J.; Bergna, A.; Cernava, T.; Berg, G. Tomato-associated Archaea ahow a cultivar-specific rhizosphere effect but an unspecific transmission by seeds. Phytobiomes J. 2020, 4, 133–141. [Google Scholar] [CrossRef] [Green Version]
- Poudel, R.; Jumpponen, A.; Kennelly, M.M.; Rivard, C.L.; Gomez-Montano, L.; Garrett, K.A. Rootstocks shape the rhizobiome: Rhizosphere and endosphere bacterial communities in the grafted tomato system. Appl. Environ. Microbiol. 2019, 85, e01765–e01818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allard, S.M.; Walsh, C.S.; Wallis, A.E.; Ottesen, A.R.; Brown, E.W.; Micallef, S.A. Solanum lycopersicum (tomato) hosts robust phyllosphere and rhizosphere bacterial communities when grown in soil amended with various organic and synthetic fertilizers. Sci. Total Environ. 2016, 573, 555–563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, B.; Zhang, C.; Ye, Y.; Wen, J.; Wu, Y.; Wang, H.; Li, H.; Cai, S.; Cai, W.; Cheng, Z.; et al. Beneficial traits of bacterial endophytes belonging to the core communities of the tomato root microbiome. Agric. Ecosyst. Environ. 2017, 247, 149–156. [Google Scholar] [CrossRef]
- Usero, F.M.; Armas, C.; Morillo, J.A.; Gallardo, M.; Thompson, R.B.; Pugnaire, F.I. Effects of soil microbial communities associated to different soil fertilization practices on tomato growth in intensive greenhouse agriculture. Appl. Soil Ecol. 2021, 162, 103896. [Google Scholar] [CrossRef]
- Li, J.G.; Ren, G.D.; Jia, Z.J.; Dong, Y.H. Composition and activity of rhizosphere microbial communities associated with healthy and diseased greenhouse tomatoes. Plant Soil 2014, 380, 337–347. [Google Scholar] [CrossRef]
- Tian, B.Y.; Cao, Y.; Zhang, K.Q. Metagenomic insights into communities, functions of endophytes, and their associates with infection by root-knot nematode, Meloidogyne incognita, in tomato roots. Sci. Rep. 2015, 5, 17087. [Google Scholar] [CrossRef] [Green Version]
- Larousse, M.; Rancurel, C.; Syska, C.; Palero, F.; Etienne, C.; Industri, B.; Nesme, X.; Bardin, M.; Galiana, E. Tomato root microbiota and Phytophthora parasitica-associated disease. Microbiome 2017, 5, 56. [Google Scholar] [CrossRef] [Green Version]
- Colagiero, M.; Rosso, L.C.; Catalano, D.; Schena, L.; Ciancio, A. Response of tomato rhizosphere bacteria to root-knot nematodes, fenamiphos and sampling time shows differential effects on low level taxa. Front. Microbiol. 2020, 11, 390. [Google Scholar] [CrossRef]
- Ottesen, A.R.; González Peña, A.; White, J.R.; Pettengill, J.B.; Li, C.; Allard, S.; Rideout, S.; Allard, M.; Hill, T.; Evans, P.; et al. Baseline survey of the anatomical microbial ecology of an important food plant: Solanum lycopersicum (tomato). BMC Microbiol. 2013, 13, 114. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.A.; Kim, Y.; Kim, J.M.; Chu, B.; Joa, J.H.; Sang, M.K.; Song, J.; Weon, H.Y. A preliminary examination of bacterial, archaeal, and fungal communities inhabiting different rhizocompartments of tomato plants under real-world environments. Sci. Rep. 2019, 9, 9300. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Z.; Lei, S.; Li, Y.; Huang, W.; Ma, R.; Xiong, J.; Zhang, T.; Jin, L.; Ul Haq, H.; Xu, X.; et al. Revealing the variation and stability of bacterial communities in tomato rhizosphere microbiota. Microorganisms 2020, 8, 170. [Google Scholar] [CrossRef] [Green Version]
- Chialva, M.; Salvioli di Fossalunga, A.; Daghino, S.; Ghignone, S.; Bagnaresi, P.; Chiapello, M.; Novero, M.; Spadaro, D.; Perotto, S.; Bonfante, P. Native soils with their microbiotas elicit a state of alert in tomato plants. New Phytol. 2018, 220, 170. [Google Scholar] [CrossRef] [Green Version]
- Gruda, N.; Schnitzler, W.H. Suitability of wood fiber substrates for production of vegetable transplants II. The effect of wood fiber substrates and their volume weights on the growth of tomato transplants. Sci. Hortic. 2004, 100, 333–340. [Google Scholar] [CrossRef]
- Spadaro, D.; Gullino, M.L. Innovations in sustainable agriculture. In Innovations in Sustainable Agriculture; Farooq, M., Pisante, M., Eds.; Springer: Berlin/Heidelberg, Germany, 2019; pp. 337–359. ISBN 9783030231699. [Google Scholar]
- Bulgarelli, D.; Garrido-Oter, R.; Münch, P.C.; Weiman, A.; Dröge, J.; Pan, Y.; McHardy, A.C.; Schulze-Lefert, P. Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe 2015, 17, 392–403. [Google Scholar] [CrossRef] [Green Version]
- Edwards, J.; Johnson, C.; Santos-Medellín, C.; Lurie, E.; Podishetty, N.K.; Bhatnagar, S.; Eisen, J.A.; Sundaresan, V.; Jeffery, L.D. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. USA 2015, 112, 911–920. [Google Scholar] [CrossRef] [Green Version]
- Fitzpatrick, C.R.; Copeland, J.; Wang, P.W.; Guttman, D.S.; Kotanen, P.M.; Johnson, M.T.J. Assembly and ecological function of the root microbiome across angiosperm plant species. Proc. Natl. Acad. Sci. USA 2018, 115, 1157–1165. [Google Scholar] [CrossRef] [Green Version]
- Reinhold-Hurek, B.; Bünger, W.; Burbano, C.S.; Sabale, M.; Hurek, T. Roots Shaping Their Microbiome: Global Hotspots for Microbial Activity. Annu. Rev. Phytopathol. 2015, 53, 403–424. [Google Scholar] [CrossRef]
- Qiao, J.; Yu, X.; Liang, X.; Liu, Y.; Borriss, R.; Liu, Y. Addition of plant-growth-promoting Bacillus subtilis PTS-394 on tomato rhizosphere has no durable impact on composition of root microbiome. BMC Microbiol. 2017, 17, 131. [Google Scholar] [CrossRef]
- Lee, S.A.; Park, J.; Chu, B.; Kim, J.M.; Joa, J.H.; Sang, M.K.; Song, J.; Weon, H.Y. Comparative analysis of bacterial diversity in the rhizosphere of tomato by culture-dependent and -independent approaches. J. Microbiol. 2016, 54, 823–831. [Google Scholar] [CrossRef]
- Sessitsch, A.; Hardoim, P.; Döring, J.; Weilharter, A.; Krause, A.; Woyke, T.; Mitter, B.; Hauberg-Lotte, L.; Friedrich, F.; Rahalkar, M.; et al. Functional characteristics of an endophyte community colonizing rice roots as revealed by metagenomic analysis. Mol. Plant-Microbe Interact. 2012, 25, 28–36. [Google Scholar] [CrossRef] [Green Version]
- Bünger, W.; Jiang, X.; Müller, J.; Hurek, T.; Reinhold-Hurek, B. Novel cultivated endophytic Verrucomicrobia reveal deep-rooting traits of bacteria to associate with plants. Sci. Rep. 2020, 10, 8692. [Google Scholar] [CrossRef] [PubMed]
- Kwak, M.J.; Kong, H.G.; Choi, K.; Kwon, S.K.; Song, J.Y.; Lee, J.; Lee, P.A.; Choi, S.Y.; Seo, M.; Lee, H.J.; et al. Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nat. Biotechnol. 2018, 36, 1100–1109. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Li, H.; Wang, B.; Ding, J.; Liu, Y.; Wei, Y.; Li, J.; Ding, G.C. Taxonomic and functional diversity of rhizosphere microbiome recruited from compost synergistically determined by plant species and compost. Front. Microbiol. 2022, 12, 798746. [Google Scholar] [CrossRef] [PubMed]
- Davis, K.E.R.; Sangwan, P.; Janssen, P.H. Acidobacteria, Rubrobacteridae and Chloroflexi are abundant among very slow-growing and mini-colony-forming soil bacteria. Environ. Microbiol. 2011, 13, 798–805. [Google Scholar] [CrossRef]
- Jacobsen, B.J.; Zidack, N.K.; Larson, B.J. The role of Bacillus-based biological control agents in integrated pest management systems: Plant diseases. Phytopathology 2004, 94, 1272–1275. [Google Scholar] [CrossRef] [Green Version]
- Weller, D.M. Pseudomonas biocontrol agents of soilborne pathogens: Looking back over 30 years. Phytopathology 2007, 97, 250–256. [Google Scholar] [CrossRef] [Green Version]
- Ciancio, A.; Pieterse, C.M.J.; Mercado-Blanco, J. Editorial: Harnessing useful rhizosphere microorganisms for pathogen and pest biocontrol-second edition. Front. Microbiol. 2019, 10, 1935. [Google Scholar] [CrossRef] [Green Version]
- Woo, S.L.; Pepe, O. Microbial consortia: Promising probiotics as plant biostimulants for sustainable agriculture. Front. Plant Sci. 2018, 9, 1801. [Google Scholar] [CrossRef]
- Ghadamgahi, F.; Tarighi, S.; Taheri, P.; Varma Saripella, G.; Anzalone, A.; Kalyandurg, P.B.; Catara, V.; Ortiz, R.; Vetukuri, R.R. Plant growth-promoting activity of Pseudomonas aeruginosa FG106 and its ability to act as a biocontrol agent against potato, tomato and taro pathogens. Biology 2022, 11, 140. [Google Scholar] [CrossRef]
- Köhl, J.; Kolnaar, R.; Ravensberg, W.J. Mode of action of microbial biological control agents against plant diseases: Relevance beyond efficacy. Front. Plant Sci. 2019, 10, 845. [Google Scholar] [CrossRef] [Green Version]
- Sang, M.K.; Kim, K.D. The volatile-producing Flavobacterium johnsoniae strain GSE09 shows biocontrol activity against Phytophthora capsici in pepper. J. Appl. Microbiol. 2012, 113, 383–398. [Google Scholar] [CrossRef]
- Youseif, S.H. Genetic diversity of plant growth promoting rhizobacteria and their effects on the growth of maize plants under greenhouse conditions. Ann. Agric. Sci. 2018, 63, 25–35. [Google Scholar] [CrossRef]
- Zhou, X.; Wang, J.T.; Wang, W.H.; Tsui, C.K.; Cai, L. Changes in bacterial and fungal microbiomes associated with tomatoes of healthy and infected by Fusarium oxysporum f. sp. lycopersici. Microb. Ecol. 2021, 81, 1004–1017. [Google Scholar] [CrossRef]
- Lay, C.Y.; Hamel, C.; St-Arnaud, M. Taxonomy and pathogenicity of Olpidium brassicae and its allied species. Fungal Biol. 2018, 122, 837–846. [Google Scholar] [CrossRef]
- Dong, C.; Wang, L.; Li, Q.; Shang, Q. Epiphytic and endophytic fungal communities of tomato plants. Hortic. Plant J. 2021, 7, 38–48. [Google Scholar] [CrossRef]
- Bragina, A.; Berg, C.; Cardinale, M.; Shcherbakov, A.; Chebotar, V.; Berg, G. Sphagnum mosses harbour highly specific bacterial diversity during their whole lifecycle. ISME J. 2012, 6, 802–813. [Google Scholar] [CrossRef]
- Wassermann, B.; Müller, H.; Berg, G. An apple a day: Which bacteria do we eat with organic and conventional apples? Front. Microbiol. 2019, 10, 1629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pẽa, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols; Wiley: Hoboken, NJ, USA, 1990. [Google Scholar]
- Lundberg, D.S.; Yourstone, S.; Mieczkowski, P.; Jones, C.D.; Dangl, J.L. Practical innovations for high-throughput amplicon sequencing. Nat. Methods 2013, 10, 999–1002. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, 2584. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, 590–596. [Google Scholar] [CrossRef]
- Kõljalg, U.; Nilsson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.F.S.; Bahram, M.; Bates, S.T.; Bruns, T.D.; Bengtsson-Palme, J.; Callaghan, T.M.; et al. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 2013, 22, 5217–5277. [Google Scholar] [CrossRef] [Green Version]
- Team, R.C. R development core team. RA Lang Env. Stat Comput 2013, 55, 275–286. [Google Scholar]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2. Wiley Interdiscip. Rev. Comput. Stat. 2011, 3, 180–185. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [Green Version]
- Harrell, F.E., Jr.; Harrell, M.F.E., Jr. Package ‘hmisc’. CRAN2018 2019, 2019, 235–236. [Google Scholar]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Berry, D.; Widder, S. Deciphering microbial interactions and detecting keystone species with co-occurrence networks. Front. Microbiol. 2014, 5, 219. [Google Scholar] [CrossRef] [Green Version]
- Gómez-Lama Cabanás, C.; Mercado-Blanco, J. What determines successful colonization and expression of biocontrol traits at the belowground level. In How Research Can Stimulate the Development of Commercial Biological Control Against Plant Diseases; De Cal, A., Melgarejo, P., Magan, N., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 31–46. ISBN 978-3-030-53238-3. [Google Scholar]




| Community | Samples | Nodes | Edges | Positive Edges | Negative Edges | Average Clustering Coefficient |
|---|---|---|---|---|---|---|
| Bacteria | Plant_T2_Soil_Rhizo_Endo | 224 | 14,081 | 13,828 | 253 | 0.29 |
| Plant_T2_CF_Rhizo_Endo | 217 | 4472 | 3904 | 568 | 0.41 | |
| Fungi | Plant_T2_Soil_Rhizo_Endo | 307 | 5988 | 4929 | 1059 | 0.31 |
| Plant_T2_CF_Rhizo_Endo | 484 | 9955 | 9771 | 184 | 0.34 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anzalone, A.; Mosca, A.; Dimaria, G.; Nicotra, D.; Tessitori, M.; Privitera, G.F.; Pulvirenti, A.; Leonardi, C.; Catara, V. Soil and Soilless Tomato Cultivation Promote Different Microbial Communities That Provide New Models for Future Crop Interventions. Int. J. Mol. Sci. 2022, 23, 8820. https://doi.org/10.3390/ijms23158820
Anzalone A, Mosca A, Dimaria G, Nicotra D, Tessitori M, Privitera GF, Pulvirenti A, Leonardi C, Catara V. Soil and Soilless Tomato Cultivation Promote Different Microbial Communities That Provide New Models for Future Crop Interventions. International Journal of Molecular Sciences. 2022; 23(15):8820. https://doi.org/10.3390/ijms23158820
Chicago/Turabian StyleAnzalone, Alice, Alexandros Mosca, Giulio Dimaria, Daniele Nicotra, Matilde Tessitori, Grete Francesca Privitera, Alfredo Pulvirenti, Cherubino Leonardi, and Vittoria Catara. 2022. "Soil and Soilless Tomato Cultivation Promote Different Microbial Communities That Provide New Models for Future Crop Interventions" International Journal of Molecular Sciences 23, no. 15: 8820. https://doi.org/10.3390/ijms23158820
APA StyleAnzalone, A., Mosca, A., Dimaria, G., Nicotra, D., Tessitori, M., Privitera, G. F., Pulvirenti, A., Leonardi, C., & Catara, V. (2022). Soil and Soilless Tomato Cultivation Promote Different Microbial Communities That Provide New Models for Future Crop Interventions. International Journal of Molecular Sciences, 23(15), 8820. https://doi.org/10.3390/ijms23158820









