Microbial Community, Metabolic Potential and Seasonality of Endosphere Microbiota Associated with Leaves of the Bioenergy Tree Paulownia elongata × fortunei
Abstract
:1. Introduction
2. Results
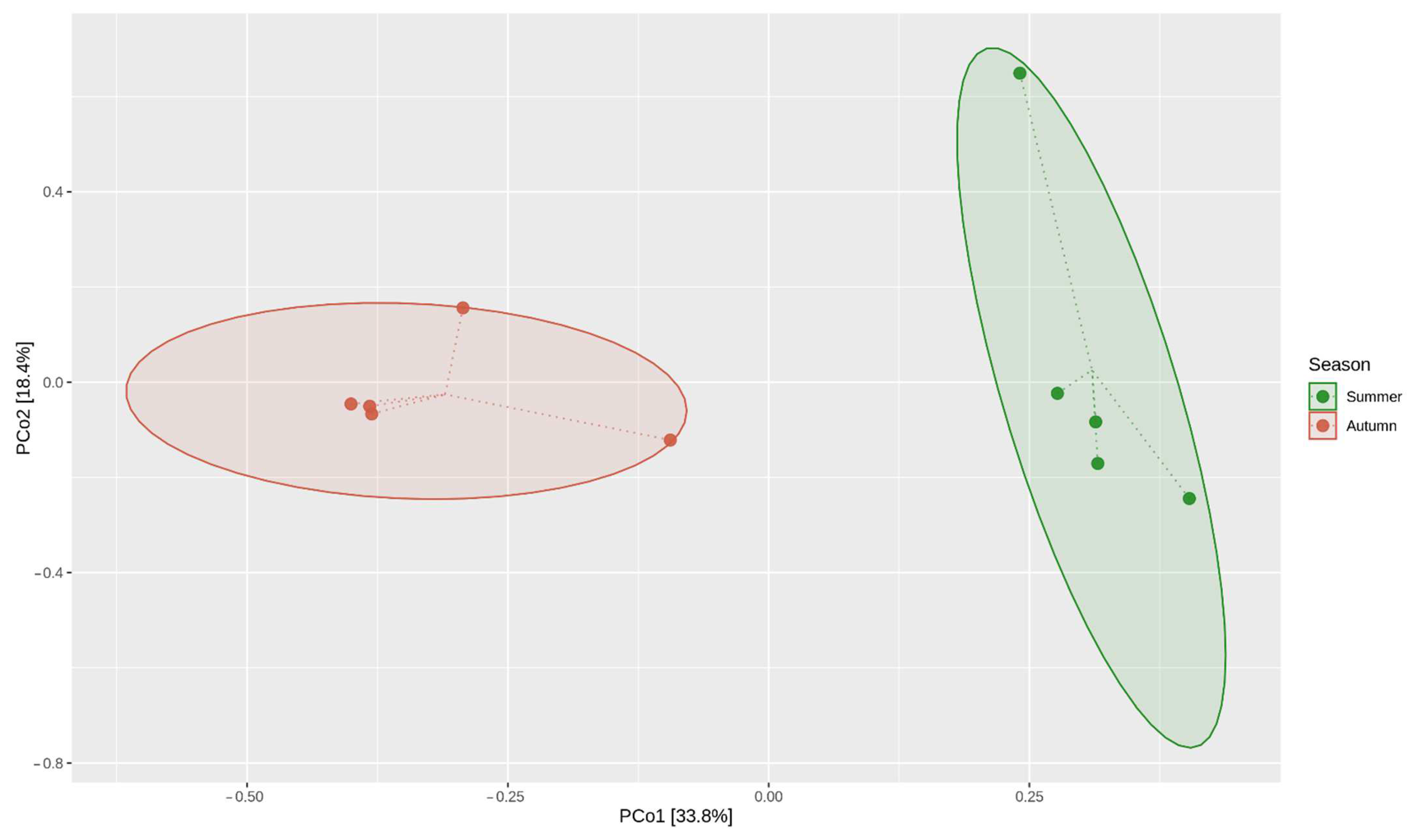
2.1. CLPP Analysis Using Biolog EcoPlates
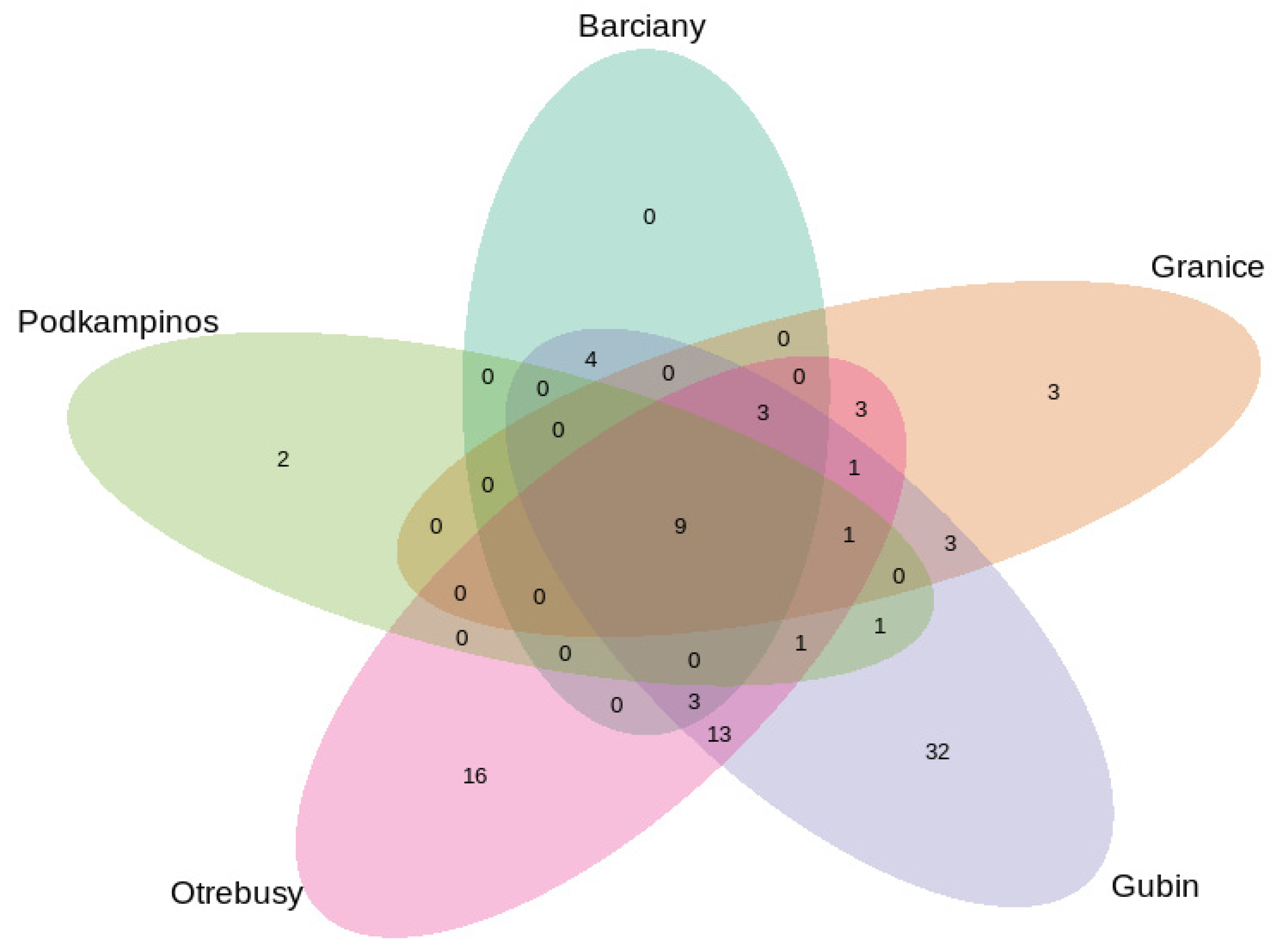
2.2. Structural Analysis of the Microbial Community
Bacterial Community Composition
3. Discussion
3.1. Community-Level Physiological Profiling
3.2. Microbial Structures
4. Materials and Methods
4.1. Microbial Community Structure Analysis
4.2. Microbial Community Analysis Using Biolog EcoPlatesTM
4.3. DNA Extraction, PCR and Illumina Amplicon Sequencing
4.4. Bioinformatic Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AWCD | The Average Colour development |
| AVS | Amplicon Sequence Variants |
| CLPP | Community Level Physiological Profiling |
| E | Shannon evenness index |
| H’ | Shannon diversity index |
| HSD test | Honestly Significant Difference test |
| IPM | Integrated Pest Management |
| PCoA | Principal coordinate analysis |
| R | Richness index |
| RDP | Ribosomal Database Project |
| TSA | Tryptic Soy Agar |
References
- Hassani, M.A.; Durán, P.; Hacquard, S. Microbial interactions within the plant holobiont. Microbiome 2018, 6, 58. [Google Scholar] [CrossRef] [PubMed]
- Vandenkoornhuyse, P.; Quaiser, A.; Duhamel, M.; Le Van, A.; Dufresne, A. The importance of the microbiome of the plant holobiont. New Phytol. 2015, 206, 1196–1206. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Yang, X.Y.; Yan, Z.Q.; Liu, Q.; Li, X.Z.; Chen, J.X.; Qin, B. Characterization of rhizosphere and endophytic bacterial communities from leaves, stems and roots of medicinal Stellera chamaejasme L. Syst. Appl. Microbiol. 2014, 37, 376–385. [Google Scholar] [CrossRef]
- Massoni, J.; Bortfeld-Miller, M.; Jardillier, L.; Salazar, G.; Sunagawa, S.; Vorholt, J.A. Consistent host and organ occupancy of phyllosphere bacteria in a community of wild herbaceous plant species. ISME J. 2020, 14, 245–258. [Google Scholar] [CrossRef]
- Agler, M.T.; Ruhe, J.; Kroll, S.; Morhenn, C.; Kim, S.T.; Weigel, D.; Kemen, E.M. Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biol. 2016, 14, e1002352. [Google Scholar] [CrossRef]
- Vorholt, J.A.; Vogel, C.; Carlström, C.I.; Müller, D.B. Establishing causality: Opportunities of synthetic communities for plant microbiome research. Cell Host Microbe 2017, 22, 142–155. [Google Scholar] [CrossRef] [PubMed]
- Berg, G.; Smalla, K. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 2009, 68, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Wieland, G.; Neumann, R.; Backhaus, H. Variation of microbial communities in soil, rhizosphere, and rhizoplane in response to crop species, soil type, and crop development. Appl. Environ. Microbiol. 2001, 67, 5849–5854. [Google Scholar] [CrossRef]
- Vorholt, J.A. Microbial life in the phyllosphere. Nat. Rev. Microbiol. 2012, 10, 828–840. [Google Scholar] [CrossRef]
- Bashir, I.; War, A.F.; Rafiq, I.; Reshi, Z.A.; Rashid, I.; Shouche, Y.S. Phyllosphere Microbiome: Diversity and Functions. Res. Microbiol. 2021, 254, 126888. [Google Scholar] [CrossRef]
- Leveau, J.H. A brief from the leaf: Latest research to inform our understanding of the phyllosphere microbiome. Curr. Opin. Microbiol. 2019, 49, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Chaudhry, V.; Runge, P.; Sengupta, P.; Doehlemann, G.; Parker, J.E.; Kemen, E. Shaping the leaf microbiota: Plant–microbe–microbe interactions. J. Exp. Bot. 2021, 72, 36–56. [Google Scholar] [CrossRef] [PubMed]
- Lindow, S.E.; Brandl, M.T. Microbiology of the phyllosphere. Appl. Environ. Microbiol. 2003, 69, 1875–1883. [Google Scholar] [CrossRef] [PubMed]
- Hardoim, P.R.; Van Overbeek, L.S.; Berg, G.; Pirttilä, A.M.; Compant, S.; Campisano, A.; Sessitsch, A. The hidden world within plants: Ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef]
- Brader, G.; Compant, S.; Vescio, K.; Mitter, B.; Trognitz, F.; Ma, L.J.; Sessitsch, A. Ecology and genomic insights into plant-pathogenic and plant-nonpathogenic endophytes. Annu. Rev. Phytopathol. 2017, 55, 61–83. [Google Scholar] [CrossRef]
- Hassan, S.E.D. Plant growth-promoting activities for bacterial and fungal endophytes isolated from medicinal plant of Teucrium polium L. J. Adv. Res. 2017, 8, 687–695. [Google Scholar] [CrossRef]
- Woźniak, M.; Gałązka, A.; Tyśkiewicz, R.; Jaroszuk-Ściseł, J. Endophytic bacteria potentially promote plant growth by synthesizing different metabolites and their phenotypic/physiological profiles in the Biolog GEN III MicroPlateTM Test. Int. J. Mol. Sci. 2019, 20, 5283. [Google Scholar] [CrossRef]
- Cueva-Yesquén, L.G.; Goulart, M.C.; Attili de Angelis, D.; Nopper Alves, M.; Fantinatti-Garboggini, F. Multiple plant growth-promotion traits in endophytic bacteria retrieved in the vegetative stage from passionflower. Front. Plant Sci. 2020, 11, 2282. [Google Scholar] [CrossRef]
- Fouda, A.; Eid, A.M.; Elsaied, A.; El-Belely, E.F.; Barghoth, M.G.; Azab, E.; Hassan, S.E.D. Plant growth-promoting endophytic bacterial community inhabiting the leaves of Pulicaria incisa (Lam.) DC inherent to arid regions. Plants 2021, 10, 76. [Google Scholar] [CrossRef]
- Bringel, F.; Couée, I. Pivotal roles of phyllosphere microorganisms at the interface between plant functioning and atmospheric trace gas dynamics. Front. Microbiol. 2015, 6, 486. [Google Scholar] [CrossRef]
- Humphrey, P.T.; Nguyen, T.T.; Villalobos, M.M.; Whiteman, N.K. Diversity and abundance of phyllosphere bacteria are linked to insect herbivory. Mol. Ecol. 2014, 23, 1497–1515. [Google Scholar] [CrossRef] [PubMed]
- Rahman, L.; Shinwari, Z.K.; Iqrar, I.; Rahman, L.; Tanveer, F. An assessment on the role of endophytic microbes in the therapeutic potential of Fagonia indica. Ann. Clin. Microbiol. Antimicrob. 2017, 16, 53. [Google Scholar] [CrossRef] [PubMed]
- Caruso, G.; Abdelhamid, M.; Kalisz, A.; Sekara, A. Linking endophytic fungi to medicinal plants therapeutic activity. A case study on Asteraceae. Agriculture 2020, 10, 286. [Google Scholar] [CrossRef]
- Teimoori-Boghsani, Y.; Ganjeali, A.; Cernava, T.; Müller, H.; Asili, J.; Berg, G. Endophytic fungi of native Salvia abrotanoides plants reveal high taxonomic diversity and unique profiles of secondary metabolites. Front. Microbiol. 2020, 10, 3013. [Google Scholar] [CrossRef] [PubMed]
- Friesen, M.L.; Porter, S.S.; Stark, S.C.; von Wettberg, E.J.; Sachs, J.L.; Martinez-Romero, E. Microbially mediated plant functional traits. Annu. Rev. Ecol. Evol. Syst. 2011, 42, 23–46. [Google Scholar] [CrossRef]
- Li, Y.; Wu, X.; Chen, T.; Wang, W.; Liu, G.; Zhang, W.; Zhang, G. Plant phenotypic traits eventually shape its microbiota: A common garden test. Front. Microbiol. 2018, 9, 2479. [Google Scholar] [CrossRef]
- Henry, L.P.; Bruijning, M.; Forsberg, S.K.; Ayroles, J.F. The microbiome extends host evolutionary potential. Nat. Commun. 2021, 12, 5141. [Google Scholar] [CrossRef]
- Porras-Alfaro, A.; Bayman, P. Hidden fungi, emergent properties: Endophytes and microbiomes. Annu. Rev. Phytopathol. 2011, 49, 291–315. [Google Scholar] [CrossRef]
- Robinson, R.J.; Fraaije, B.A.; Clark, I.M.; Jackson, R.W.; Hirsch, P.R.; Mauchline, T.H. Endophytic bacterial community composition in wheat (Triticum aestivum) is determined by plant tissue type, developmental stage and soil nutrient availability. Plant Soil 2016, 405, 381–396. [Google Scholar] [CrossRef]
- Noman, M.; Ahmed, T.; Ijaz, U.; Shahid, M.; Li, D.; Manzoor, I.; Song, F. Plant–Microbiome crosstalk: Dawning from composition and assembly of microbial community to improvement of disease resilience in plants. Int. J. Mol. Sci. 2021, 22, 6852. [Google Scholar] [CrossRef]
- Lucas-Borja, M.E.; Wic-Baena, C.; Moreno, J.L.; Dadi, T.; García, C.; Andrés-Abellán, M. Microbial activity in soils under fast-growing Paulownia (Paulownia elongata × fortunei) plantations in Mediterranean areas. Appl. Soil Ecol. 2011, 51, 42–51. [Google Scholar] [CrossRef]
- Woźniak, M.; Gałązka, A.; Grządziel, J.; Frąc, M. Microbial diversity of Paulownia spp. leaves—A new source of green manure. BioResources 2018, 13, 4807–4819. [Google Scholar] [CrossRef]
- Malhi, Y.; Franklin, J.; Seddon, N.; Solan, M.; Turner, M.G.; Field, C.B.; Knowlton, N. Climate change and ecosystems: Threats, opportunities and solutions. Philos. Trans. R. Soc. B 2020, 375, 20190104. [Google Scholar] [CrossRef]
- Muluneh, M.G. Impact of climate change on biodiversity and food security: A global perspective—A review article. Agric. Food Secur. 2021, 10, 36. [Google Scholar] [CrossRef]
- El-Showk, S.; El-Showk, N. The Paulownia Tree: An Alternative for Sustainable Forestry. The Farm. 2003. Available online: http://www.cropdevelopment.org/docs/PaulowniaBooklet_print.pdf (accessed on 29 June 2022).
- Icka, P.; Damo, R.; Icka, E. Paulownia tomentosa, a fast growing timber. Ann. Valahia Univ. Targoviste-Agric. 2016, 10, 14–19. [Google Scholar] [CrossRef]
- Woźniak, M.; Grządziel, J.; Gałązka, A.; Frąc, M. Metagenomic Analysis of Bacterial and Fungal Community Composition Associated with Paulownia elongate × Paulownia fortunei. BioResources 2019, 14, 8511–8529. [Google Scholar] [CrossRef]
- Popova, T.P.; Baykov, B.D. Antimicrobial activity of aqueous extracts of leaves and silage from Paulownia elongate. Am. J. Biol. Chem. Pharm. Sci. 2013, 1, 8–15. [Google Scholar]
- Eevers, N.; Beckers, B.; de Beeck, M.O.; White, J.C.; Vangronsveld, J.; Weyens, N. Comparison between cultivated and total bacterial communities associated with Cucurbita pepo using cultivation-dependent techniques and 454 pyrosequencing. Syst. Appl. Microbiol. 2016, 39, 58–66. [Google Scholar] [CrossRef]
- Szymańska, S.; Piernik, A.; Hrynkiewicz, K. Metabolic Potential of Microorganisms Associated with the Halophyte Aster tripolium L. in Saline Soils. Ecol. Quest. 2014. Available online: http://repozytorium.umk.pl/handle/item/1865 (accessed on 29 June 2022).
- Frąc, M.; Oszust, K.; Lipiec, J. Community level physiological profiles (CLPP), characterization and microbial activity of soil amended with dairy sewage sludge. Sensors 2012, 12, 3253–3268. [Google Scholar] [CrossRef]
- Garland, J.L.; Mills, A.L. Classification and characterization of heterotrophic microbial communities on the basis of patterns of community-level sole-carbon-source utilization. Appl. Environ. Microbiol. 1991, 57, 2351–2359. [Google Scholar] [CrossRef]
- Insam, H. A new set of substrates proposed for community characterization in environmental samples. In Microbial Communities: Functional versus Structural Approaches; Insam, H., Rangger, A., Eds.; Springer: Berlin/Heidelberg, Germany, 1997; pp. 259–260. [Google Scholar]
- Ros, M.; Goberna, M.; Pascual, J.A.; Klammer, S.; Insam, H.J. 16S rDNA analysis reveals low microbial diversity in community level physiological profile assays. Microbiol. Methods 2008, 72, 221–226. [Google Scholar] [CrossRef] [PubMed]
- Akinsanya, M.A.; Goh, J.K.; Lim, S.P.; Ting, A.S.Y. Metagenomics study of endophytic bacteria in Aloe vera using next-generation technology. Genom. Data 2015, 6, 159–163. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Liu, P.; Ye, W. Illumina-based analysis of endophytic bacterial diversity of tree peony (Paeonia Sect. Moutan) roots and leaves. Braz. J. Microbiol. 2017, 48, 695–705. [Google Scholar] [CrossRef] [PubMed]
- Haonan, H.; Lei, F.; Fu, S.; Zou, K.; Zhang, S.; Liu, X.; Liang, Y. Endophytic bacterial communities of Ginkgo biloba leaves during leaf developmental period. Front. Microbiol. 2021, 12, 698703. [Google Scholar] [CrossRef]
- Cregger, M.A.; Veach, A.M.; Yang, Z.K.; Crouch, M.J.; Vilgalys, R.; Tuskan, G.A.; Schadt, C.W. The Populus holobiont: Dissecting the effects of plant niches and genotype on the microbiome. Microbiome 2018, 6, 31. [Google Scholar] [CrossRef]
- Zalewska, E.D.; Pogorzelec, M.; Król, E.D.; Serafin, A. Fungi inhabiting the aboveground organs of downy willow (Salix lapponum L.) and swamp willow (Salix myrtilloides L.). Acta Mycol. 2019, 54, 1134. [Google Scholar] [CrossRef]
- Dasgupta, M.G.; Burragoni, S.; Amrutha, S.; Muthupandi, M.; Parveen, A.B.M.; Sivakumar, V.; Ulaganathan, K. Diversity of bacterial endophyte in Eucalyptus clones and their implications in water stress tolerance. Microbiol. Res. 2020, 241, 126579. [Google Scholar] [CrossRef]
- Gryta, A.; Frąc, M.; Oszust, K. The Application of the Biolog EcoPlate Approach in Ecotoxicological Evaluation of Dairy Sewage Sludge. Appl. Biochem. Biotechnol. 2014, 174, 1434–1443. [Google Scholar] [CrossRef]
- Woźniak, M.; Gałązka, A.; Siebielec, G.; Frąc, M. Can the Biological Activity of Abandoned Soils Be Changed by the Growth of Paulownia elongata× Paulownia fortunei?—Preliminary Study on a Young Tree Plantation. Agriculture 2022, 12, 128. [Google Scholar] [CrossRef]
- Németh, I.; Molnár, S.; Vaszita, E.; Molnár, M. The Biolog EcoPlate™ Technique for Assessing the Effect of Metal Oxide Nanoparticles on Freshwater Microbial Communities. Nanomaterials 2021, 11, 1777. [Google Scholar] [CrossRef]
- Oest, A.; Alsaffar, A.; Fenner, M.; Azzopardi, D.; Tiquia-Arashiro, S.M. Patterns of change in metabolic capabilities of sediment microbial communities in river and lake ecosystems. Int. J. Microbiol. 2018, 2018, 6234931. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.Y.; Wu, Y.H.; Zhuang, L.L.; Wang, X.X.; Hu, H.Y. Screening heterotrophic microalgal strains by using the Biolog method for biofuel production from organic wastewater. Algal Res. 2014, 6, 175–179. [Google Scholar] [CrossRef]
- Yadav, R.K.P.; Papatheodorou, E.M.; Karamanoli, K.; Constantinidou, H.I.A.; Vokou, D. Abundance and diversity of the phyllosphere bacterial communities of Mediterranean perennial plants that differ in leaf chemistry. Chemoecology 2008, 18, 217–226. [Google Scholar] [CrossRef]
- Banik, A.; Kumar, U.; Mukhopadhyay, S.K.; Dangar, T.K. Dynamics of endophytic and epiphytic bacterial communities of Indian cultivated and wild rice (Oryza spp.) genotypes. Ecol. Genet. Genom. 2017, 3, 7–17. [Google Scholar] [CrossRef]
- Candan, E.D.; İdil, N.; Candan, O. The microbial community in a green turtle nesting beach in the Mediterranean: Application of the Biolog EcoPlate approach for beach pollution. Environ. Sci. Pollut. Res. 2021, 28, 49685–49696. [Google Scholar] [CrossRef] [PubMed]
- Sofo, A.; Ricciuti, P. A standardized method for estimating the functional diversity of soil bacterial community by Biolog® EcoPlatesTM assay—The case study of a sustainable olive orchard. Appl. Sci. 2019, 9, 4035. [Google Scholar] [CrossRef]
- Dresler, S.; Hawrylak-Nowak, B.; Kováčik, J.; Woźniak, M.; Gałązka, A.; Staniak, M.; Sowa, I. Organic nitrogen modulates not only cadmium toxicity but also microbial activity in plants. J. Hazard. Mater. 2021, 402, 123887. [Google Scholar] [CrossRef]
- Wang, S.; Tang, J.Y.; Ma, J.; Li, X.D.; Li, Y.H. Moss habitats distinctly affect their associated bacterial community structures as revealed by the high-throughput sequencing method. World J. Microbiol. Biotechnol. 2018, 34, 58. [Google Scholar] [CrossRef] [PubMed]
- Dijkstra, P.; Thomas, S.C.; Heinrich, P.L.; Koch, G.W.; Schwartz, E.; Hungate, B.A. Effect of temperature on metabolic activity of intact microbial communities: Evidence for altered metabolic pathway activity but not for increased maintenance respiration and reduced carbon use efficiency. Soil Biol. Biochem. 2011, 43, 2023–2031. [Google Scholar] [CrossRef]
- Lin, Y.T.; Jia, Z.; Wang, D.; Chiu, C.Y. Effects of temperature on the composition and diversity of bacterial communities in bamboo soils at different elevations. Biogeosciences 2017, 14, 4879–4889. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, B.; Miao, R.; Zhou, J.; Ye, L.; Jia, D.; Peng, W.; Yan, L.; Zhang, X.; Li, X.; et al. Influence of temperature on the bacterial community in substrate and extracellular enzyme activity of Auricularia cornea. Mycobiology 2018, 46, 224–235. [Google Scholar] [CrossRef]
- Redford, A.J.; Bowers, R.M.; Knight, R.; Linhart, Y.; Fierer, N. The ecology of the phyllosphere: Geographic and phylogenetic variability in the distribution of bacteria on tree leaves. Environ. Microbiol. 2010, 12, 2885–2893. [Google Scholar] [CrossRef]
- Liu, H.; Carvalhais, L.C.; Crawford, M.; Singh, E.; Dennis, P.G.; Pieterse, C.M.; Schenk, P.M. Inner plant values: Diversity, colonization and benefits from endophytic bacteria. Front. Microbiol. 2017, 8, 2552. [Google Scholar] [CrossRef]
- Liu, J.; Abdelfattah, A.; Norelli, J.; Burchard, E.; Schena, L.; Droby, S.; Wisniewski, M. Apple endophytic microbiota of different rootstock/scion combinations suggests a genotype-specific influence. Microbiome 2018, 6, 18. [Google Scholar] [CrossRef]
- Ulrich, K.; Becker, R.; Behrendt, U.; Kube, M.; Ulrich, A. A comparative analysis of ash leaf-colonizing bacterial communities identifies putative antagonists of Hymenoscyphus fraxineus. Front. Microbiol. 2020, 11, 966. [Google Scholar] [CrossRef]
- Mocali, S.; Bertelli, E.; Di Cello, F.; Mengoni, A.; Sfalanga, A.; Viliani, F.; Fani, R. Fluctuation of bacteria isolated from elm tissues during different seasons and from different plant organs. Res. Microbiol. 2003, 154, 105–114. [Google Scholar] [CrossRef]
- Shen, S.Y.; Fulthorpe, R. Seasonal variation of bacterial endophytes in urban trees. Front. Microbiol. 2015, 6, 427. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Thorngate, J.H.; Richardson, P.M.; Mills, D.A. Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc. Natl. Acad. Sci. USA 2014, 111, E139–E148. [Google Scholar] [CrossRef]
- Adams, P.D.; Kloepper, J.W. Effect of host genotype on indigenous bacterial endophytes of cotton (Gossypium hirsutum L.). Plant Soil 2002, 240, 181–189. [Google Scholar] [CrossRef]
- Kumar, M.; Kumar, A.; Sahu, K.P.; Patel, A.; Reddy, B.; Sheoran, N.; Rathour, R. Deciphering core-microbiome of rice leaf endosphere: Revelation by metagenomic and microbiological analysis of aromatic and non-aromatic genotypes grown in three geographical zones. Microbiol. Res. 2021, 246, 126704. [Google Scholar] [CrossRef]
- Ou, T.; Xu, W.F.; Wang, F.; Strobel, G.; Zhou, Z.Y.; Xiang, Z.H.; Xie, J. A microbiome study reveals seasonal variation in endophytic bacteria among different mulberry cultivars. Comput. Struct. Biotechnol. J. 2019, 17, 1091–1100. [Google Scholar] [CrossRef]
- Tong, W.; Yu, J.; Wu, Q.; Hu, L.; Tabys, D.; Wang, Y.; Bennetzen, J.L. Black tea quality is highly affected during processing by its leaf surface microbiome. J. Agric. Food Chem. 2021, 69, 7115–7126. [Google Scholar] [CrossRef] [PubMed]
- Eid, A.M.; Fouda, A.; Abdel-Rahman, M.A.; Salem, S.S.; Elsaied, A.; Oelmüller, R.; Hijri, M.; Bhowmik, A.; Eikelish, A.; Hassan, S.E.D. Harnessing bacterial endophytes for promotion of plant growth and biotechnological applications: An overview. Plants 2021, 10, 935. [Google Scholar] [CrossRef]
- Yang, C.H.; Crowley, D.E.; Borneman, J.; Keen, N.T. Microbial phyllosphere populations are more complex than previously realized. Proc. Natl. Acad. Sci. USA 2001, 98, 3889–3894. [Google Scholar] [CrossRef]
- Zinniel, D.K.; Lambrecht, P.; Harris, N.B.; Feng, Z.; Kuczmarski, D.; Higley, P.; Ishimaru, C.A.; Arunakumari, A.; Barletta, R.G.; Vidaver, A.K. Isolation and characterization of endophytic colonizing bacteria from agronomic crops and prairie plants. Appl. Environ. Microbiol. 2002, 68, 2198–2208. [Google Scholar] [CrossRef]
- Oszust, K.; Frąc, M. First report on the microbial communities of the wild and planted raspberry rhizosphere–A statement on the taxa, processes and a new indicator of functional diversity. Ecol. Indic. 2021, 121, 107117. [Google Scholar] [CrossRef]
- Chelius, M.K.; Triplett, E.W. The Diversity of Archaea and Bacteria in Association with the Roots of Zea mays L. Microb. Ecol. 2001, 41, 252–263. [Google Scholar] [CrossRef] [PubMed]
- Beckers, B.; Op De Beeck, M.; Thijs, S.; Truyens, S.; Weyens, N.; Boerjan, W.; Vangronsveld, J. Performance of 16S rDNA primer pairs in the study of rhizosphere and endosphere bacterial microbiomes in metabarcoding studies. Front. Microbiol. 2016, 7, 650. [Google Scholar] [CrossRef]
- Bodenhausen, N.; Horton, M.W.; Bergelson, J. Bacterial communities associated with the leaves and the roots of Arabidopsis thaliana. PLoS ONE 2013, 8, e56329. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2016. [Google Scholar]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Cui, Y.; Li, X.; Yao, M. Microeco: An R package for data mining in microbial community ecology. FEMS Microbiol. Ecol. 2021, 97, fiaa255. [Google Scholar] [CrossRef]
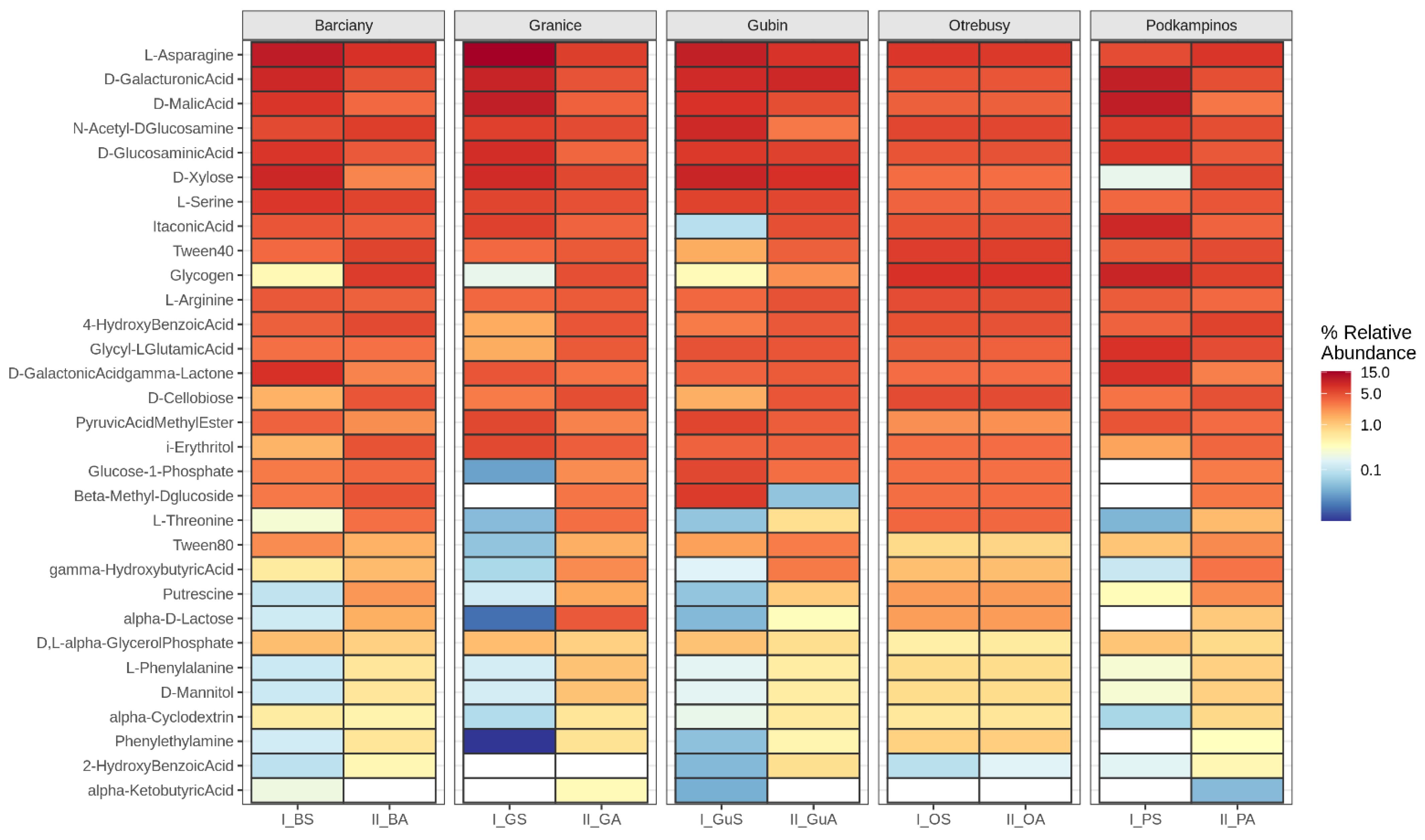
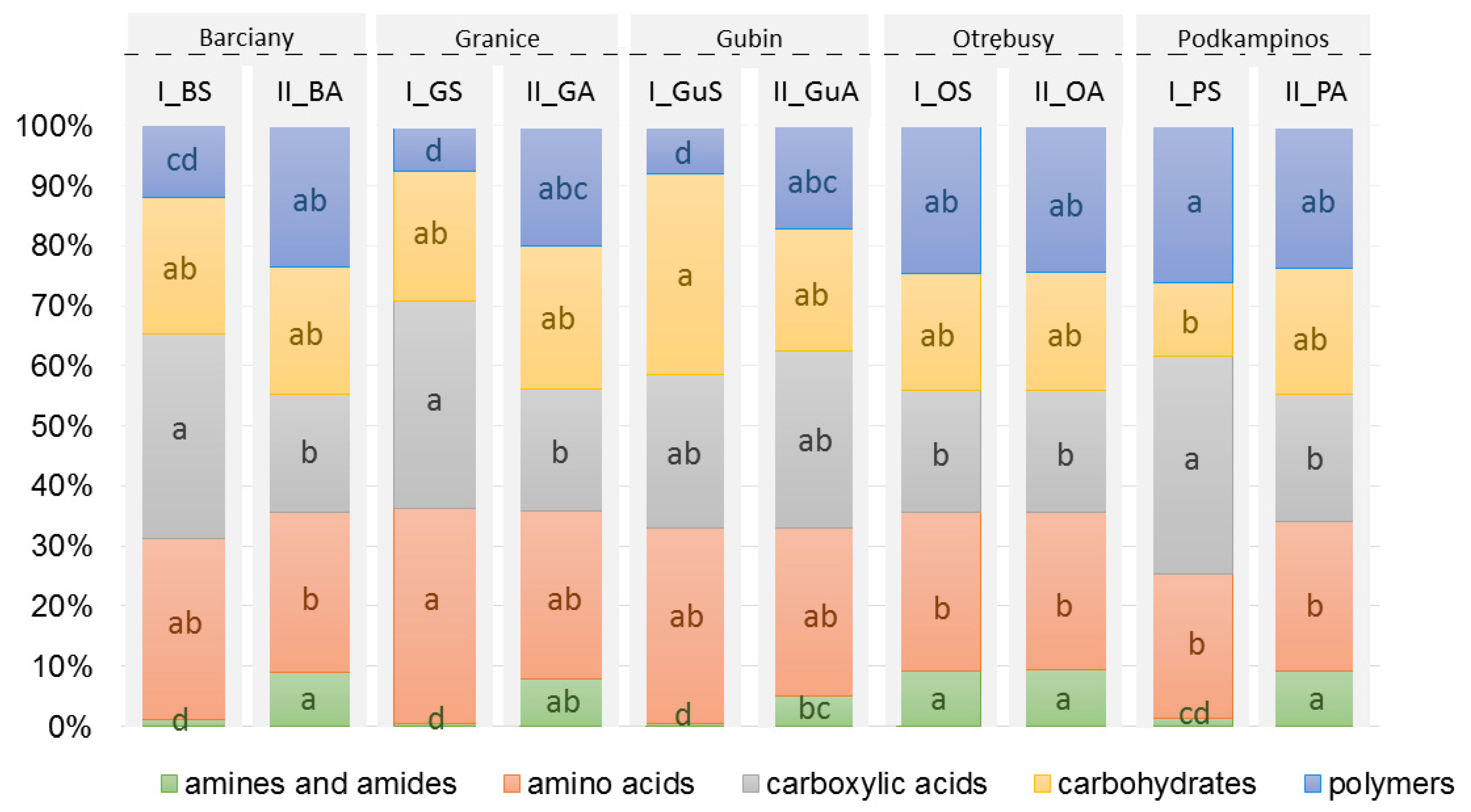
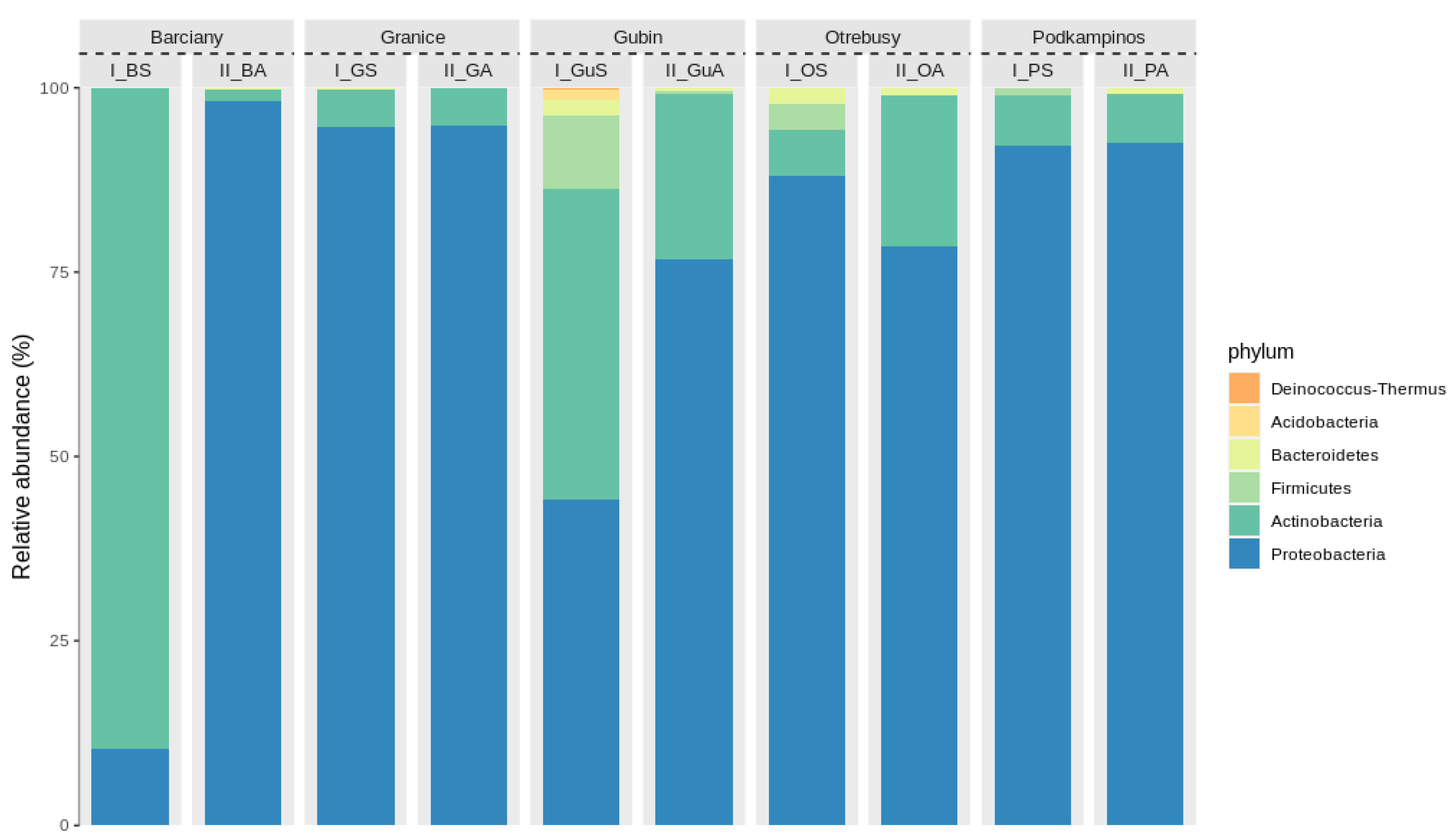
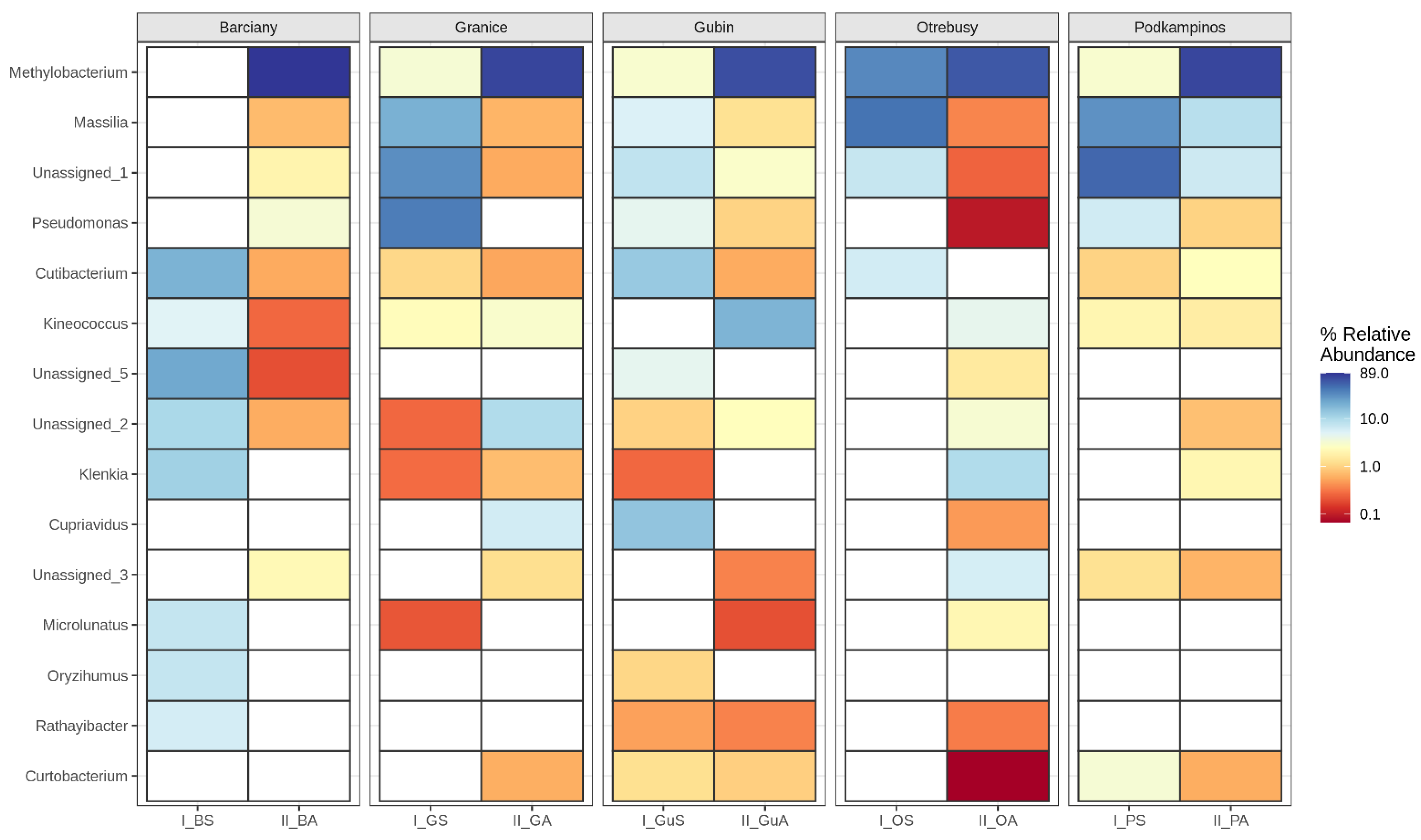
| No. | Location | Sample | AWCD | Richness (R) | Shannon Diversity (H’) | Shannon Evenness (E) |
|---|---|---|---|---|---|---|
| 1. | Barciany | I_BS | 0.592 ± 0.066 d | 13.000 ± 4.000 d | 2.634 ± 0.225 cd | 0.788 ± 0.071 bc |
| 2. | II_BA | 1.047 ± 0.046 ab | 24.667 ± 1.155 ab | 3.175 ± 0.025 a | 0.937 ± 0.012 a | |
| 3. | Granice | I_GS | 0.448 ± 0.097 d | 12.333 ± 1.528 d | 2.529 ± 0.113 d | 0.790 ± 0.075 bc |
| 4. | II_GA | 1.179 ± 0.042 a | 27.333 ± 2.082 a | 3.218 ± 0.051 a | 0.962 ± 0.007 a | |
| 5. | Gubin | I_GuS | 1.007 ± 0.064 ab | 17.667 ± 0.577 cd | 2.823 ± 0.047 bc | 0.867 ± 0.064 abc |
| 6. | II_GuA | 0.835 ± 0.027 c | 20.333 ± 1.155 bc | 3.038 ± 0.02 ab | 0.913 ± 0.025 ab | |
| 7. | Otrebusy | I_OS | 0.909 ± 0.108 c | 24.667 ± 1.528 ab | 3.134 ± 0.041 a | 0.928 ± 0.02 a |
| 8. | II_OA | 0.927 ± 0.109 c | 25.000 ± 1.732 ab | 3.148 ± 0.033 a | 0.929 ± 0.014 a | |
| 9. | Podkampinos | I_PS | 0.414 ± 0.057 d | 14.333 ± 3.215 cd | 2.663 ± 0.032 cd | 0.860 ± 0.043 abc |
| 10. | II_PA | 1.215 ± 0.074 a | 24.667 ± 2.517 ab | 3.160 ± 0.079 a | 0.939 ± 0.012 a |
| No. | Location | Sampling Season | Abbreviations |
|---|---|---|---|
| 1. | Barciany | Summer | I_BS |
| 2. | Autumn | II_BA | |
| 3. | Granice | Summer | I_GS |
| 4. | Autumn | II_GA | |
| 5. | Gubin | Summer | I_GuS |
| 6. | Autumn | II_GuA | |
| 7. | Otrebusy | Summer | I_OS |
| 8. | Autumn | II_OA | |
| 9. | Podkampinos | Summer | I_PS |
| 10. | Autumn | II_PA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Woźniak, M.; Gałązka, A.; Marzec-Grządziel, A.; Frąc, M. Microbial Community, Metabolic Potential and Seasonality of Endosphere Microbiota Associated with Leaves of the Bioenergy Tree Paulownia elongata × fortunei. Int. J. Mol. Sci. 2022, 23, 8978. https://doi.org/10.3390/ijms23168978
Woźniak M, Gałązka A, Marzec-Grządziel A, Frąc M. Microbial Community, Metabolic Potential and Seasonality of Endosphere Microbiota Associated with Leaves of the Bioenergy Tree Paulownia elongata × fortunei. International Journal of Molecular Sciences. 2022; 23(16):8978. https://doi.org/10.3390/ijms23168978
Chicago/Turabian StyleWoźniak, Małgorzata, Anna Gałązka, Anna Marzec-Grządziel, and Magdalena Frąc. 2022. "Microbial Community, Metabolic Potential and Seasonality of Endosphere Microbiota Associated with Leaves of the Bioenergy Tree Paulownia elongata × fortunei" International Journal of Molecular Sciences 23, no. 16: 8978. https://doi.org/10.3390/ijms23168978
APA StyleWoźniak, M., Gałązka, A., Marzec-Grządziel, A., & Frąc, M. (2022). Microbial Community, Metabolic Potential and Seasonality of Endosphere Microbiota Associated with Leaves of the Bioenergy Tree Paulownia elongata × fortunei. International Journal of Molecular Sciences, 23(16), 8978. https://doi.org/10.3390/ijms23168978









