RNA-Seq Analysis of UPM-Exposed Epithelium Co-Cultivated with Macrophages and Dendritic Cells in Obstructive Lung Diseases
Abstract
:1. Introduction
2. Results
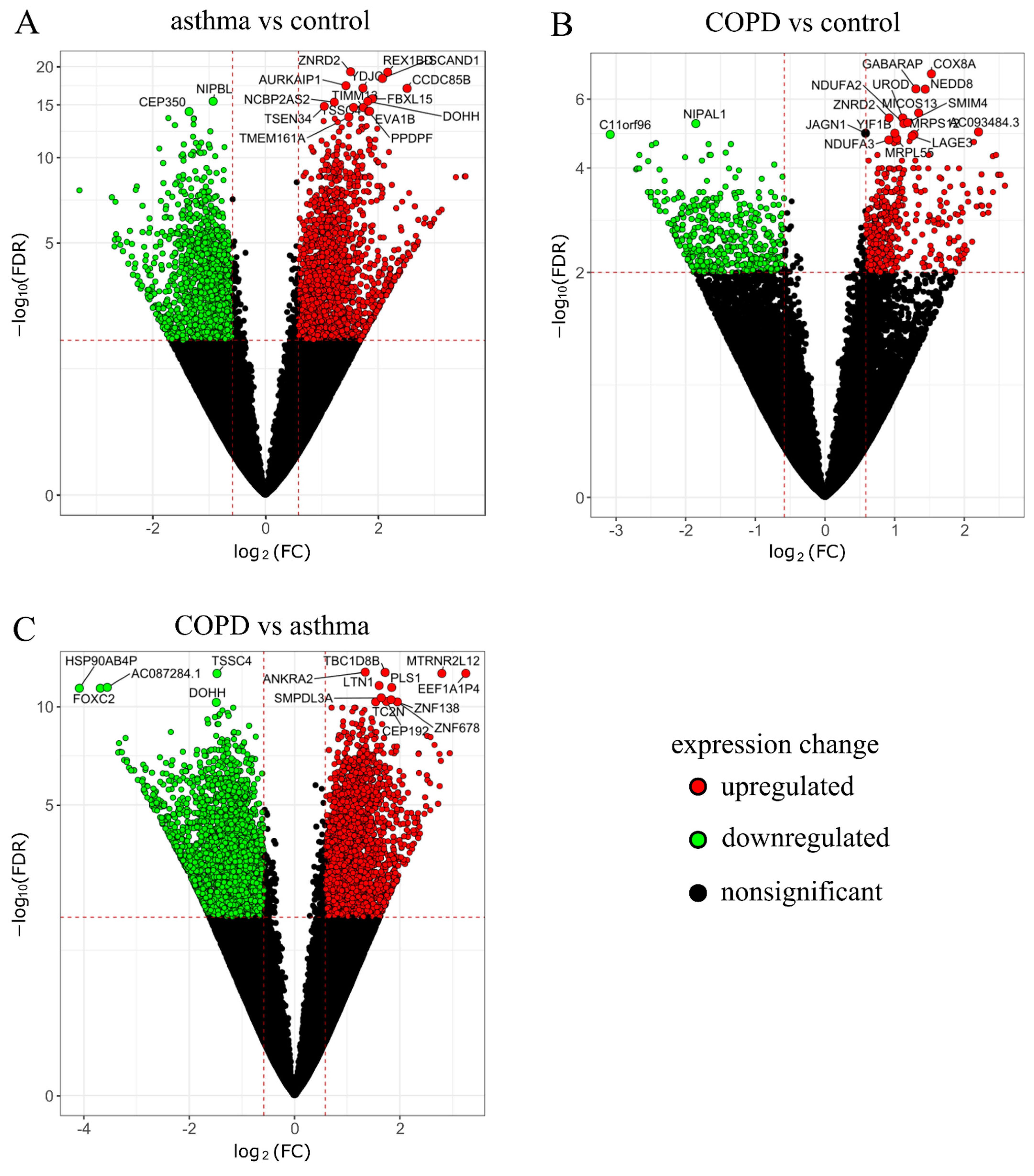
2.1. RNA-Seq Data Analysis
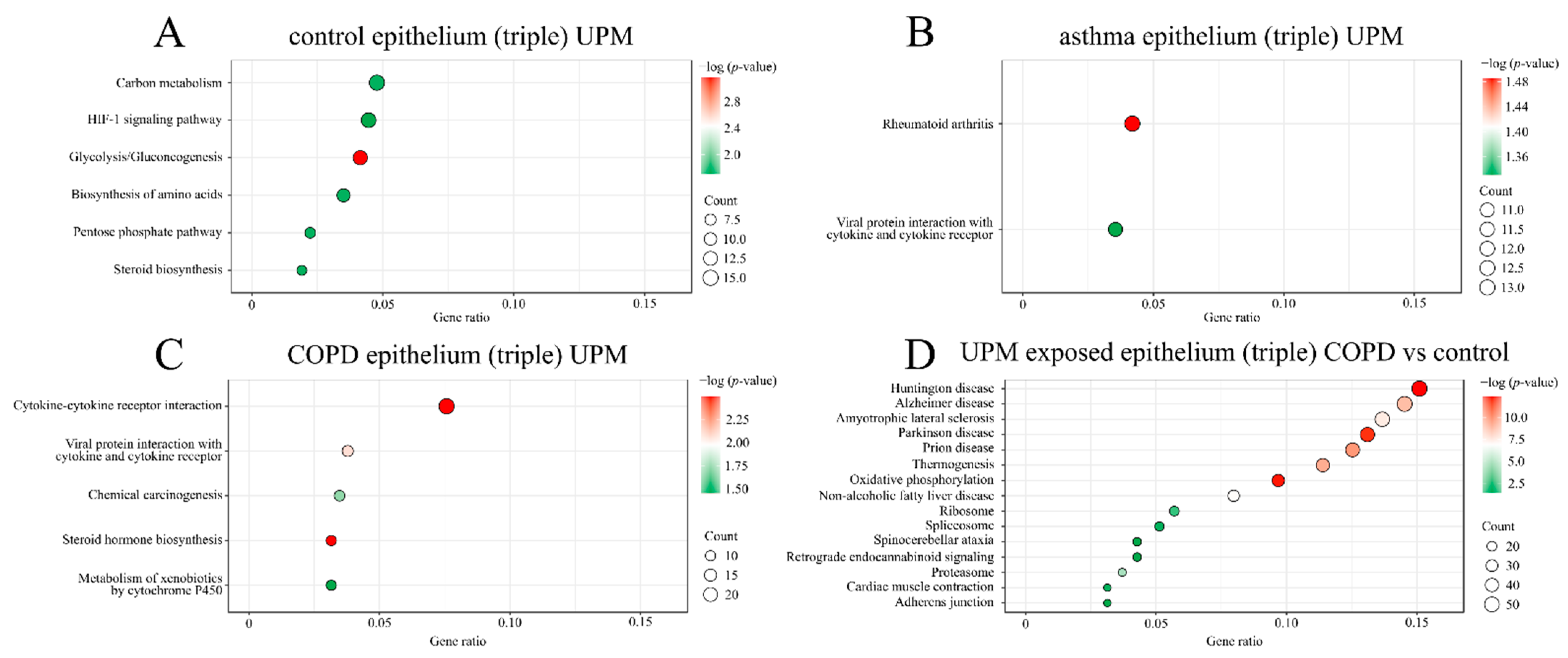
2.2. GO and KEGG Pathway Enrichment Analysis
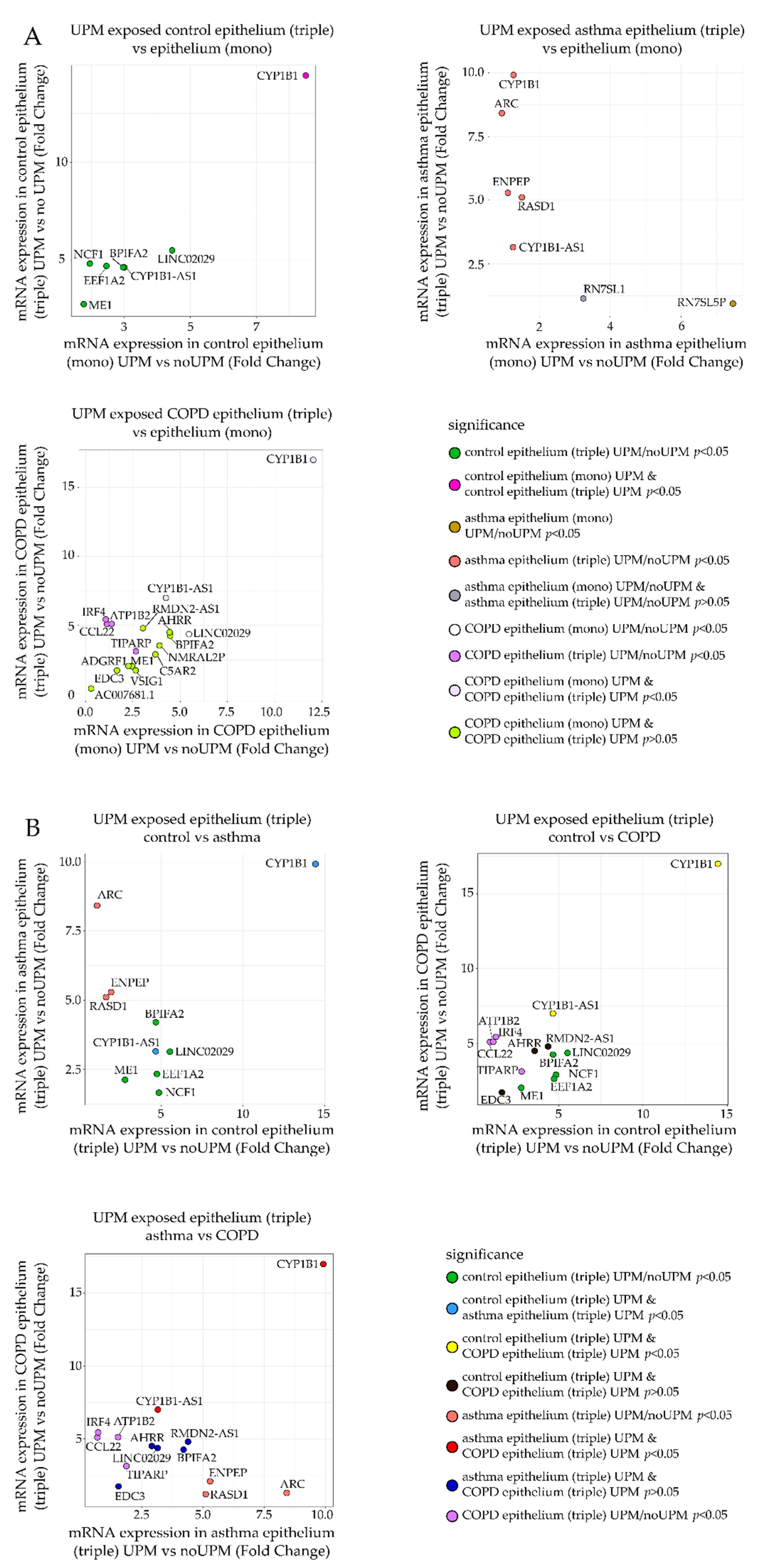
2.3. RT-qPCR Analysis
- (a)
- Fold change of mRNA expression between the UPM exposed and nonexposed epithelial cells from the triple co-culture (Figure 5A, Y-axis) was compared to the fold change of mRNA expression between the UPM and no UPM exposed epithelial cells from the monoculture (Figure 5A, X-axis). A separate plot (and gene selection) was prepared for each group (control/asthma/COPD).
- (b)
- Fold change of mRNA expression between the UPM exposed and nonexposed epithelial cells from the triple co-culture in one of the clinical group was plotted against the same value in other clinical groups (three panels: asthma–control, COPD–control, COPD–asthma).
3. Discussion
4. Materials and Methods
4.1. Patient Characteristics
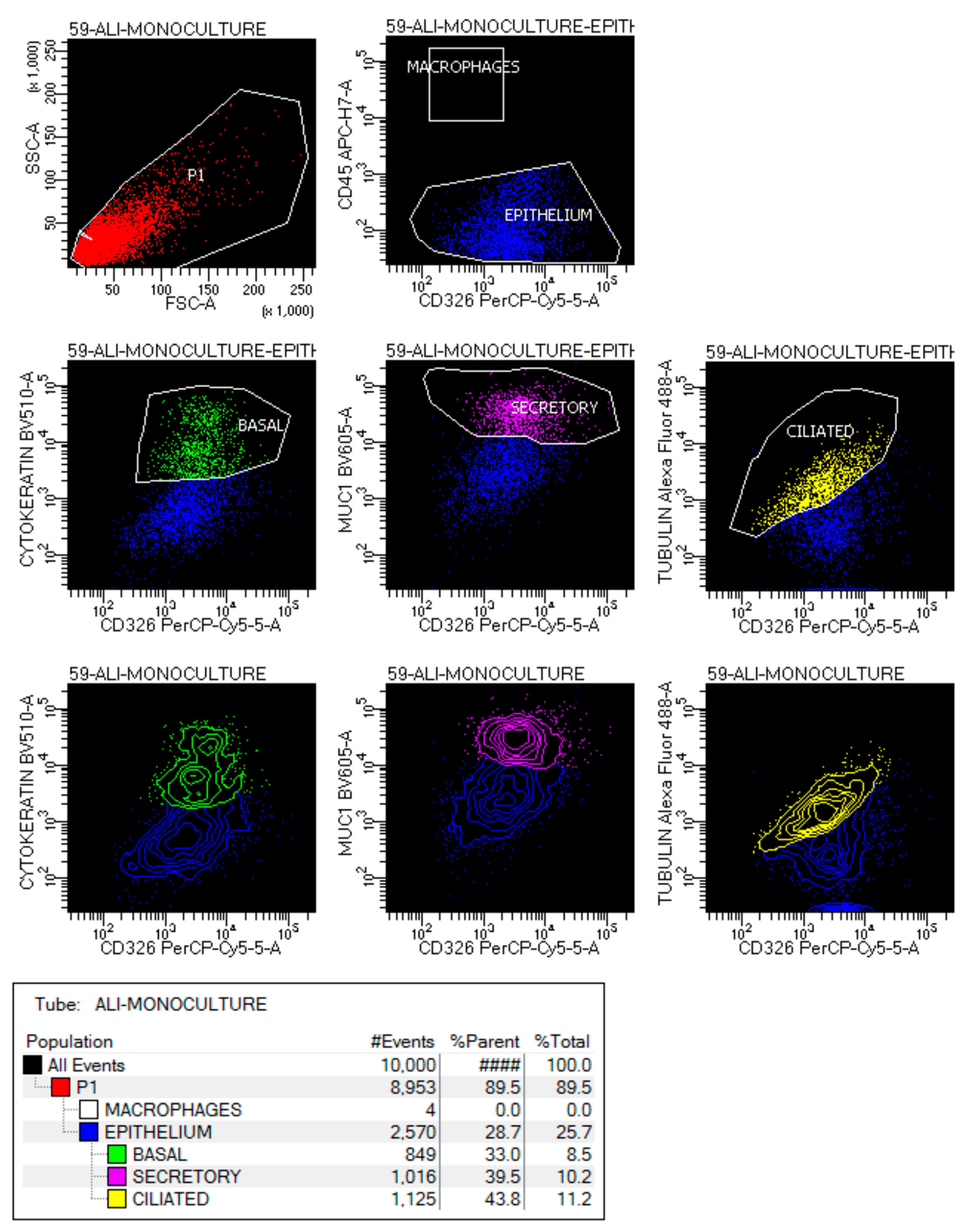
4.2. Flow Cytometry Analysis
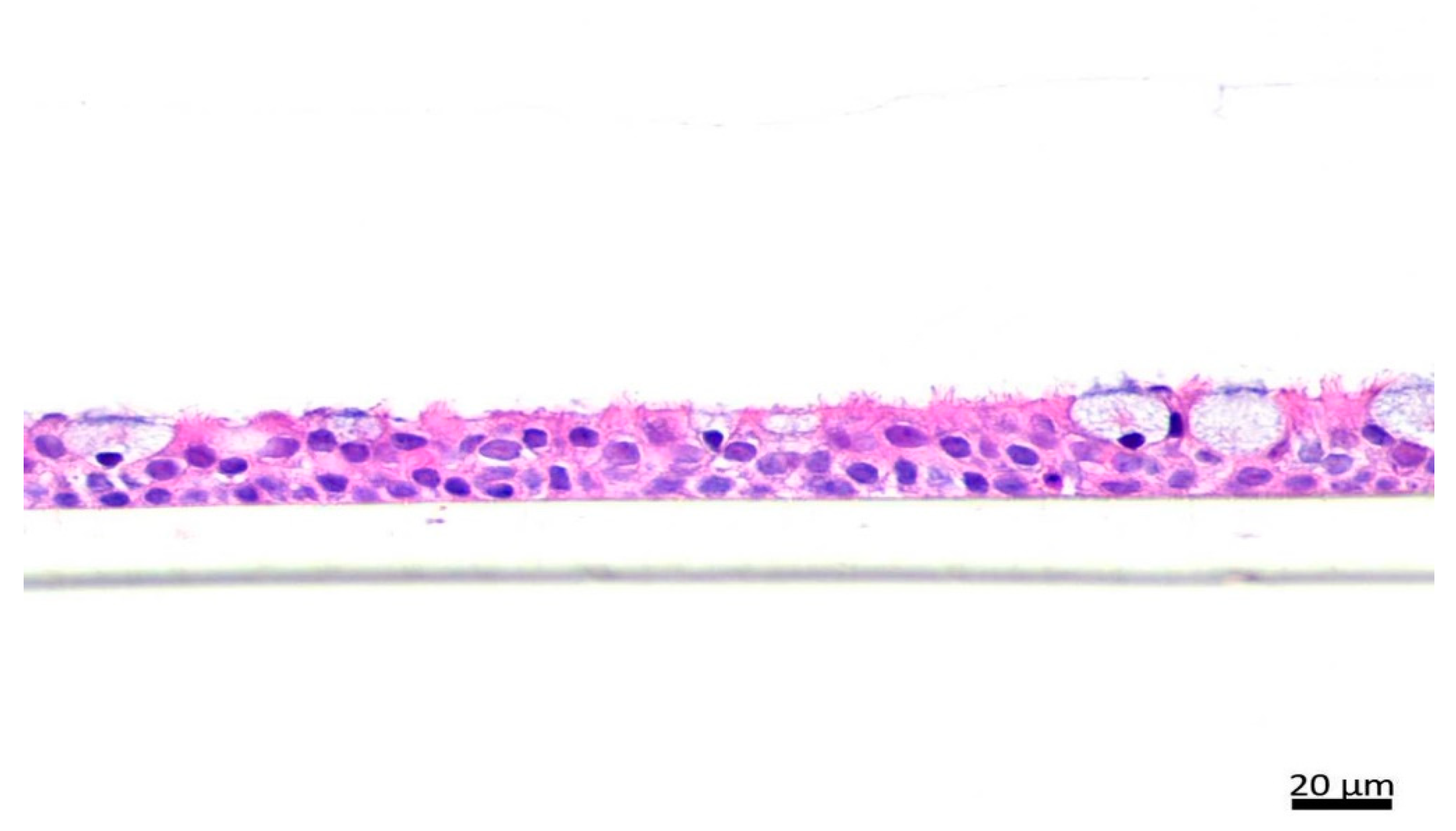
4.3. Hematoxylin and Eosin (H&E) Staining
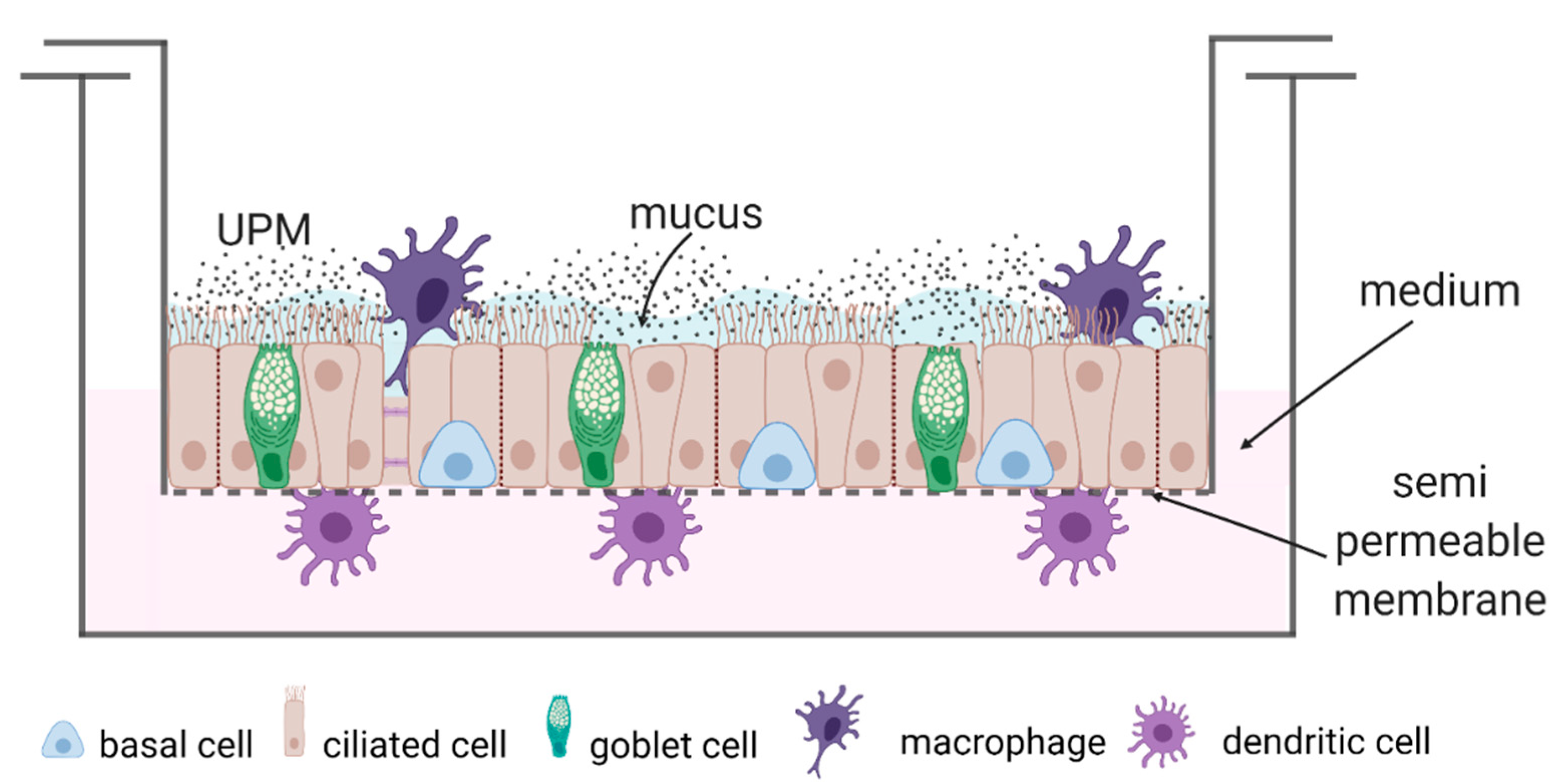
4.4. Cell Culture and Scheme of the Study
- (1)
- Epithelial cells (monoculture);
- (2)
- Epithelial cells + moMφs + moDCs (triple co-culture).
4.5. Particle Preparation
4.6. RNA Isolation
4.7. RNA-Seq Analysis
4.8. Bioinformatic Analysis
4.9. qRT-PCR Measurements
4.10. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Acronym | Full Gene Name |
|---|---|
| 18s rRNA | 18s ribosomal RNA |
| AHRR | aryl-hydrocarbon receptor repressor |
| ARC | activity regulated cytoskeleton associated protein |
| ATP1B2 | ATPase Na+/K+ transporting subunit beta 2 |
| BPIFA2 | BPI fold containing family A member 2 |
| CCL22 | C-C motif chemokine ligand 22 |
| CYP1B1 | cytochrome P450 family 1 subfamily B member 1 |
| CYP1B1-AS1 | CYP1B1 antisense RNA 1 |
| EDC3 | enhancer of mRNA decapping 3 |
| EEF1A2 | eukaryotic translation elongation factor 1 alpha 2 |
| ENPEP | glutamyl aminopeptidase |
| IRF4 | interferon regulatory factor 4 |
| LINC02029 | Long non-coding RNA |
| ME1 | malic enzyme 1 |
| NCF1 | neutrophil cytosolic factor 1 |
| RASD1 | ras related dexamethasone induced 1 |
| RMDN2-AS1 | RMDN2 antisense RNA 1 |
| TIPARP | TCDD inducible poly(ADP-ribose) polymerase |
| Epithelium (mono) + UPM | Epithelium (trio) + UPM | |
|---|---|---|
| AHRR | 0.640 | 0.144 |
| ARC | 0.957 | 0.047 # |
| ATP1B2 | 0.154 | 0.597 |
| BPIFA2 | 0.025 # | 0.006 # |
| CCL22 | 0.214 | 0.109 |
| CYP1B1 | 0.224 | 0.167 |
| CYP1B1-AS1 | 0.023 * | 0.435 |
| EDC3 | 0.065 | 0.655 |
| EEF1A2 | 0.745 | 0.318 |
| ENPEP | 0.028 * | 0.150 |
| IRF4 | 0.407 | 0.913 |
| LINC02029 | 0.777 | 0.530 |
| ME1 | 0.093 | 0.868 |
| NCF1 | 0.806 | 0.353 |
| RASD1 | 0.394 | 0.663 |
| RMDN2-AS1 | 0.091 | 0.040 |
| TIPARP | 0.049 | 0.029 # |
| Control n = 8 | Asthma n = 10 | COPD n = 8 | Overall p-Value & | Pairwise p-Value * | |||
|---|---|---|---|---|---|---|---|
| Asthma vs. Control | COPD vs. Control | Asthma vs. COPD | |||||
| Age (years) | 38.5 (32.5–48) | 55 (38–62) | 62 (59.5–72.5) | 0.005 | 0.138 | 0.0002 | 0.138 |
| Gender (F/M) | 6/2 | 3/10 | 5/3 | 0.046 | |||
| BMI (kg/m2) | 22.1 (20.7–24.1) | 26.9 (26–27.7) | 28 (25.4–30.3) | 0.002 | 0.002 | 0.0003 | 0.696 |
| Atopy (n) | 3 | 8 | 2 | 0.03 | |||
| Smoking exposure (pack-years) | 0 (0–0) | 0 (0–4) | 32.5 (22.5–50) | 0.0002 | 0.277 | 0.0002 | 0.0003 |
| FEV1 (% predicted) | 103 (81–111) | 81 (75–94) | 61.5 (51.5–76.5) | 0.006 | 0.043 | 0.004 | 0.034 |
| FEV1/VC (%) | 106 (81.8–112) | 82 (75–86) | 54 (50–68) | 0.0003 | 0.02 | 0.0006 | 0.0004 |
| FeNO (ppb) | 11.0 (9.3–12.6) | 52.3 (31.3–77.6) | 17.4 (12.6–26.1) | 0.0047 | 0.03 | 0.333 | 0.003 |
| ACT (points) | N.A. | 20.5 (17–25) | N.A. | N.A | N.A. | N.A. | N.A. |
| ICS treatment (n) | N.A. | 6 | 1 | N.A | N.A. | N.A. | N.A. |
| CAT (points) | N.A. | N.A. | 10.5 (8–15) | N.A | N.A. | N.A. | N.A. |
| mMRC (points) | N.A. | N.A. | 1.5 (1–3) | N.A | N.A. | N.A. | N.A. |
| Gene Symbol | Forward Primer | Reverse Primer | Probe | Product Size |
|---|---|---|---|---|
| LINC02029 | TGCCCCCACG AGGTACAC | CAGGACCCAAA GAAGGAATGAT | 6-FAM-TCCCGGGA AACAAA-MGB | 58 |
| Gene symbol | Entrez Gene ID | |||
| 18s rRNA | Hs99999901_s1 | 187 | ||
| AHRR | Hs01005075_m1 | 98 | ||
| ARC | Hs01045540_g1 | 92 | ||
| ATP1B2 | Hs01020302_g1 | 81 | ||
| BPIFA2 | Hs00395980_m1 | 68 | ||
| CCL22 | Hs01574247_m1 | 88 | ||
| CYP1B1 | Hs00164383_m1 | 118 | ||
| CYP1B1-AS1 | Hs00381672_m1 | 80 | ||
| EDC3 | Hs00257810_m1 | 122 | ||
| EEF1A2 | Hs00951278_m1 | 80 | ||
| ENPEP | Hs00989749_m1 | 70 | ||
| IRF4 | Hs00180031_m1 | 88 | ||
| ME1 | Hs00159110_m1 | 73 | ||
| NCF1 | Hs00165362_m1 | 113 | ||
| RASD1 | Hs02568415_s1 | 159 | ||
| RMDN2-AS1 | Hs04409587_s1 | 86 | ||
| TIPARP | Hs00296054_m1 | 80 | ||

References
- WHO. Ambient Air Pollution: A Global Assessment of Exposure and Burden of Disease; WHO: Geneva, Switzerland, 2016. [Google Scholar]
- WHO. Air Quality Guidelines—Global Update 2005; WHO: Geneva, Switzerland, 2005. [Google Scholar]
- Takizawa, H.; Ohtoshi, T.; Kawasaki, S.; Abe, S.; Sugawara, I.; Nakahara, K.; Matsushima, K.; Kudoh, S. Diesel Exhaust Particles Activate Human Bronchial Epithelial Cells to Express Inflammatory Mediators in the Airways: A Review. Respirology 2000, 5, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Karagulian, F.; Belis, C.A.; Dora, C.F.C.; Prüss-Ustün, A.M.; Bonjour, S.; Adair-Rohani, H.; Amann, M. Contributions to Cities’ Ambient Particulate Matter (PM): A Systematic Review of Local Source Contributions at Global Level. Atmos. Environ. 2015, 120, 475–483. [Google Scholar] [CrossRef]
- Yue, H.; Yun, Y.; Gao, R.; Li, G.; Sang, N. Winter Polycyclic Aromatic Hydrocarbon-Bound Particulate Matter from Peri-Urban North China Promotes Lung Cancer Cell Metastasis. Environ. Sci. Technol. 2015, 49, 14484–14493. [Google Scholar] [CrossRef]
- Paplinska-Goryca, M.; Misiukiewicz-Stepien, P.; Proboszcz, M.; Nejman-Gryz, P.; Gorska, K.; Zajusz-Zubek, E.; Krenke, R. Interactions of Nasal Epithelium with Macrophages and Dendritic Cells Variously Alter Urban PM-Induced Inflammation in Healthy, Asthma and COPD. Sci. Rep. 2021, 11, 13259. [Google Scholar] [CrossRef] [PubMed]
- Hirota, J.A.; Marchant, D.J.; Singhera, G.K.; Moheimani, F.; Dorscheid, D.R.; Carlsten, C.; Sin, D.; Knight, D. Urban Particulate Matter Increases Human Airway Epithelial Cell IL-1β Secretion Following Scratch Wounding and H1N1 Influenza A Exposure in Vitro. Exp. Lung Res. 2015, 41, 353–362. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, U.S.; Rastogi, N.; McWhinney, R.D.; Urch, B.; Chow, C.-W.; Evans, G.J.; Scott, J.A. The Combined Effects of Physicochemical Properties of Size-Fractionated Ambient Particulate Matter on in Vitro Toxicity in Human A549 Lung Epithelial Cells. Toxicol. Rep. 2014, 1, 145–156. [Google Scholar] [CrossRef]
- Chirino, Y.I.; Sánchez-Pérez, Y.; Osornio-Vargas, Á.R.; Morales-Bárcenas, R.; Gutiérrez-Ruíz, M.C.; Segura-García, Y.; Rosas, I.; Pedraza-Chaverri, J.; García-Cuellar, C.M. PM10 Impairs the Antioxidant Defense System and Exacerbates Oxidative Stress Driven Cell Death. Toxicol. Lett. 2010, 193, 209–216. [Google Scholar] [CrossRef]
- Michael, S.; Montag, M.; Dott, W. Pro-Inflammatory Effects and Oxidative Stress in Lung Macrophages and Epithelial Cells Induced by Ambient Particulate Matter. Environ. Pollut. 2013, 183, 19–29. [Google Scholar] [CrossRef]
- Kim, H.J.; Choi, M.G.; Park, M.K.; Seo, Y.R. Predictive and Prognostic Biomarkers of Respiratory Diseases Due to Particulate Matter Exposure. J. Cancer Prev. 2017, 22, 6–15. [Google Scholar] [CrossRef]
- Moore, E.; Chatzidiakou, L.; Kuku, M.-O.; Jones, R.L.; Smeeth, L.; Beevers, S.; Kelly, F.J.; Barratt, B.; Quint, J.K. Global Associations between Air Pollutants and Chronic Obstructive Pulmonary Disease Hospitalizations. A Systematic Review. Ann. Am. Thorac. Soc. 2016, 13, 1814–1827. [Google Scholar] [CrossRef]
- Orellano, P.; Quaranta, N.; Reynoso, J.; Balbi, B.; Vasquez, J. Effect of Outdoor Air Pollution on Asthma Exacerbations in Children and Adults: Systematic Review and Multilevel Meta-Analysis. PLoS ONE 2017, 12, e0174050. [Google Scholar] [CrossRef] [PubMed]
- Loxham, M.; Morgan-Walsh, R.J.; Cooper, M.J.; Blume, C.; Swindle, E.J.; Dennison, P.W.; Howarth, P.H.; Cassee, F.R.; Teagle, D.A.H.; Palmer, M.R.; et al. The Effects on Bronchial Epithelial Mucociliary Cultures of Coarse, Fine, and Ultrafine Particulate Matter from an Underground Railway Station. Toxicol. Sci. 2015, 145, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Howe, K.L.; Achuthan, P.; Allen, J.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R.; Bhai, J.; et al. Ensembl 2021. Nucleic Acids Res. 2021, 49, D884–D891. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Chen, H.; Wan, P.; Song, S.; Chen, N. Downregulation of Enhancer RNA EMX2OS is Associated with Poor Prognosis in Kidney Renal Clear Cell Carcinoma. Aging 2020, 12, 25865–25877. [Google Scholar] [CrossRef]
- Richter, G.M.; Kruppa, J.; Munz, M.; Wiehe, R.; Häsler, R.; Franke, A.; Martins, O.; Jockel-Schneider, Y.; Bruckmann, C.; Dommisch, H.; et al. A Combined Epigenome- and Transcriptome-Wide Association Study of the Oral Masticatory Mucosa Assigns CYP1B1 a Central Role for Epithelial Health in Smokers. Clin. Epigenet. 2019, 11, 105. [Google Scholar] [CrossRef]
- Stueve, T.R.; Li, W.-Q.; Shi, J.; Marconett, C.N.; Zhang, T.; Yang, C.; Mullen, D.; Yan, C.; Wheeler, W.; Hua, X.; et al. Epigenome-Wide Analysis of DNA Methylation in Lung Tissue Shows Concordance with Blood Studies and Identifies Tobacco Smoke-Inducible Enhancers. Hum. Mol. Genet. 2017, 26, 3014–3027. [Google Scholar] [CrossRef]
- Nebert, D.W.; Dalton, T.P.; Okey, A.B.; Gonzalez, F.J. Role of Aryl Hydrocarbon Receptor-Mediated Induction of the CYP1 Enzymes in Environmental Toxicity and Cancer. J. Biol. Chem. 2004, 279, 23847–23850. [Google Scholar] [CrossRef]
- Rajput, N.; Lakhani, A. Measurements of Polycyclic Aromatic Hydrocarbons at an Industrial Site in India. Environ. Monit. Assess. 2009, 150, 273–284. [Google Scholar] [CrossRef]
- Borgie, M.; Ledoux, F.; Verdin, A.; Cazier, F.; Greige, H.; Shirali, P.; Courcot, D.; Dagher, Z. Genotoxic and Epigenotoxic Effects of Fine Particulate Matter from Rural and Urban Sites in Lebanon on Human Bronchial Epithelial Cells. Environ. Res. 2015, 136, 352–362. [Google Scholar] [CrossRef]
- Grimaldi, G.; Rajendra, S.; Matthews, J. The Aryl Hydrocarbon Receptor Regulates the Expression of TIPARP and Its Cis Long Non-Coding RNA, TIPARP-AS. Biochem. Biophys. Res. Commun. 2018, 495, 2356–2362. [Google Scholar] [CrossRef]
- Matthews, J. AHR Toxicity and Signaling: Role of TIPARP and ADP-Ribosylation. Curr. Opin. Toxicol. 2017, 2, 50–57. [Google Scholar] [CrossRef]
- MacPherson, L.; Tamblyn, L.; Rajendra, S.; Bralha, F.; McPherson, J.P.; Matthews, J. 2,3,7,8-Tetrachlorodibenzo-p-Dioxin Poly(ADP-Ribose) Polymerase (TiPARP, ARTD14) Is a Mono-ADP-Ribosyltransferase and Repressor of Aryl Hydrocarbon Receptor Transactivation. Nucleic Acids Res. 2013, 41, 1604–1621. [Google Scholar] [CrossRef]
- Ahmed, S.; Bott, D.; Gomez, A.; Tamblyn, L.; Rasheed, A.; Cho, T.; MacPherson, L.; Sugamori, K.S.; Yang, Y.; Grant, D.M.; et al. Loss of the Mono-ADP-Ribosyltransferase, Tiparp, Increases Sensitivity to Dioxin-Induced Steatohepatitis and Lethality. J. Biol. Chem. 2015, 290, 16824–16840. [Google Scholar] [CrossRef] [PubMed]
- Holmes, R.S.; Spradling-Reeves, K.D.; Cox, L.A. Mammalian Glutamyl Aminopeptidase Genes (ENPEP) and Proteins: Comparative Studies of a Major Contributor to Arterial Hypertension. J. Data Min. Genom. Proteom. 2017, 8, 2. [Google Scholar] [CrossRef]
- Qi, F.; Qian, S.; Zhang, S.; Zhang, Z. Single Cell RNA Sequencing of 13 Human Tissues Identify Cell Types and Receptors of Human Coronaviruses. Biochem. Biophys. Res. Commun. 2020, 526, 135–140. [Google Scholar] [CrossRef] [PubMed]
- The UniProt Consortium. UniProt: The Universal Protein Knowledgebase. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Prokopovic, V.; Popovic, M.; Andjelkovic, U.; Marsavelski, A.; Raskovic, B.; Gavrovic-Jankulovic, M.; Polovic, N. Isolation, Biochemical Characterization and Anti-Bacterial Activity of BPIFA2 Protein. Arch. Oral Biol. 2014, 59, 302–309. [Google Scholar] [CrossRef]
- Kang, D.; Jung, I.B.; Lee, S.Y.; Park, S.J.; Kwon, S.J.; Park, D.H.; Son, J.W. Particulate Matter Less than 10 Μm (PM10) Activates Cancer Related Genes in Lung Epithelial Cells. Inhal. Toxicol. 2020, 32, 487–493. [Google Scholar] [CrossRef]
- Chen, Z.; Newgard, C.B.; Kim, J.S.; IIkayeva, O.; Alderete, T.L.; Thomas, D.C.; Berhane, K.; Breton, C.; Chatzi, L.; Bastain, T.M.; et al. Near-Roadway Air Pollution Exposure and Altered Fatty Acid Oxidation among Adolescents and Young Adults—The Interplay with Obesity. Environ. Int. 2019, 130, 104935. [Google Scholar] [CrossRef]
- Reyes-Caballero, H.; Rao, X.; Sun, Q.; Warmoes, M.O.; Lin, P.; Sussan, T.E.; Park, B.; Fan, T.W.-M.; Maiseyeu, A.; Rajagopalan, S.; et al. Air Pollution-Derived Particulate Matter Dysregulates Hepatic Krebs Cycle, Glucose and Lipid Metabolism in Mice. Sci. Rep. 2019, 9, 17423. [Google Scholar] [CrossRef]
- Chen, H.; Li, Z.; Dong, L.; Wu, Y.; Shen, H.; Chen, Z. Lipid Metabolism in Chronic Obstructive Pulmonary Disease. Int. J. Chronic Obstr. Pulm. Dis. 2019, 14, 1009–1018. [Google Scholar] [CrossRef] [PubMed]
- Moré, J.M.; Voelker, D.R.; Silveira, L.J.; Edwards, M.G.; Chan, E.D.; Bowler, R.P. Smoking Reduces Surfactant Protein D and Phospholipids in Patients with and without Chronic Obstructive Pulmonary Disease. BMC Pulm. Med. 2010, 10, 53. [Google Scholar] [CrossRef] [PubMed]
- Leclercq, B.; Happillon, M.; Antherieu, S.; Hardy, E.M.; Alleman, L.Y.; Grova, N.; Perdrix, E.; Appenzeller, B.M.; Lo Guidice, J.-M.; Coddeville, P.; et al. Differential Responses of Healthy and Chronic Obstructive Pulmonary Diseased Human Bronchial Epithelial Cells Repeatedly Exposed to Air Pollution-Derived PM. Environ. Pollut. 2016, 218, 1074–1088. [Google Scholar] [CrossRef] [PubMed]
- Gambhir, L.; Sharma, V.; Kandwal, P.; Saxena, S. Perturbation in Cellular Redox Homeostasis: Decisive Regulator of T Cell Mediated Immune Responses. Int. Immunopharmacol. 2019, 67, 449–457. [Google Scholar] [CrossRef]
- Wunnapuk, K.; Pothirat, C.; Manokeaw, S.; Phetsuk, N.; Chaiwong, W.; Phuackchantuck, R.; Prapamontol, T. PM10-Related DNA Damage, Cytokinetic Defects, and Cell Death in COPD Patients from Chiang Dao District, Chiang Mai, Thailand. Environ. Sci. Pollut. Res. Int. 2019, 26, 25326–25340. [Google Scholar] [CrossRef]
- Lai, C.-H.; Lee, C.-N.; Bai, K.-J.; Yang, Y.-L.; Chuang, K.-J.; Wu, S.-M.; Chuang, H.-C. Protein Oxidation and Degradation Caused by Particulate Matter. Sci. Rep. 2016, 6, 33727. [Google Scholar] [CrossRef]
- Wei, J.; Rahman, S.; Ayaub, E.A.; Dickhout, J.G.; Ask, K. Protein Misfolding and Endoplasmic Reticulum Stress in Chronic Lung Disease. Chest 2013, 143, 1098–1105. [Google Scholar] [CrossRef]
- Huang, K.-L.; Liu, S.-Y.; Chou, C.C.K.; Lee, Y.-H.; Cheng, T.-J. The Effect of Size-Segregated Ambient Particulate Matter on Th1/Th2-like Immune Responses in Mice. PLoS ONE 2017, 12, e0173158. [Google Scholar] [CrossRef]
- Boxall, C.; Holgate, S.T.; Davies, D.E. The Contribution of Transforming Growth Factor-β and Epidermal Growth Factor Signalling to Airway Remodelling in Chronic Asthma. Eur. Respir. J. 2006, 27, 208–229. [Google Scholar] [CrossRef]
- Druilhe, A.; Zahm, J.-M.; Benayoun, L.; El Mehdi, D.; Grandsaigne, M.; Dombret, M.-C.; Mosnier, I.; Feger, B.; Depondt, J.; Aubier, M.; et al. Epithelium Expression and Function of Retinoid Receptors in Asthma. Am. J. Respir. Cell Mol. Biol. 2008, 38, 276–282. [Google Scholar] [CrossRef]
- GINA. Global Strategy for Asthma Management and Prevention 2018; GINA: Milwaukee, WI, USA, 2018. [Google Scholar]
- Global Initiative for Chronic Obstructive Lung Disease (GOLD). Global Strategy for the Diagnosis, Management and Prevention of COPD 2018; GOLD: Geneva, Switzerland, 2018; Available online: www.goldcopd.org (accessed on 12 July 2022).
- Paplinska-Goryca, M.; Misiukiewicz-Stepien, P.; Nejman-Gryz, P.; Proboszcz, M.; Mlacki, M.; Gorska, K.; Krenke, R. Epithelial-Macrophage-Dendritic Cell Interactions Impact Alarmins Expression in Asthma and COPD. Clin. Immunol. 2020, 215, 108421. [Google Scholar] [CrossRef] [PubMed]
- Blom, R.A.M.; Erni, S.T.; Krempaská, K.; Schaerer, O.; van Dijk, R.M.; Amacker, M.; Moser, C.; Hall, S.R.R.; von Garnier, C.; Blank, F. A Triple Co-Culture Model of the Human Respiratory Tract to Study Immune-Modulatory Effects of Liposomes and Virosomes. PLoS ONE 2016, 11, e0163539. [Google Scholar] [CrossRef] [PubMed]
- EN 12341:2014; London Ambient Air. Standard Gravimetric Measurement Method for the Determination of the PM10 or PM2,5 Mass Concentration of Suspended Particulate Matter. BSI British Standards Institution: London, UK, 2014.
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce Framework for Analyzing Next-Generation DNA Sequencing Data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup the Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Kopylova, E.; Noé, L.; Touzet, H. SortMeRNA: Fast and Accurate Filtering of Ribosomal RNAs in Metatranscriptomic Data. Bioinformatics 2012, 28, 3211–3217. [Google Scholar] [CrossRef]
- Zerbino, D.R.; Achuthan, P.; Akanni, W.; Amode, M.R.; Barrell, D.; Bhai, J.; Billis, K.; Cummins, C.; Gall, A.; Girón, C.G.; et al. Ensembl 2018. Nucleic Acids Res. 2018, 46, D754–D761. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python Framework to Work with High-Throughput Sequencing Data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Zhu, A.; Ibrahim, J.G.; Love, M.I. Heavy-Tailed Prior Distributions for Sequence Count Data: Removing the Noise and Preserving Large Differences. Bioinformatics 2019, 35, 2084–2092. [Google Scholar] [CrossRef] [PubMed]
- The Gene Ontology Consortium. The Gene Ontology Resource: 20 Years and Still GOing Strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New Perspectives on Genomes, Pathways, Diseases and Drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes among Gene Clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]



| Control n = 4 | Asthma n = 4 | COPD n = 4 | Overall p-Value | |
|---|---|---|---|---|
| Age (years) | 36 (27–44.5) | 61 (38–71) | 67 (62–72.5) | 0.06 |
| Gender (F/M) | 4/0 | 1/3 | 2/2 | 0.09 |
| BMI (kg/m2) | 22.4 (20.3–23.1) | 27.2 (26–30.1) | 28 (25.8–30.9) | 0.025 * |
| Atopy (n) | 2 | 3 | 0 | 0.03 |
| Smoking exposure (pack-years) | 0 (0–3.5) | 0 (0–0.75) | 25 (20–52) | 0.015 * |
| FEV1 (% predicted) | 105.5 (101–109.5) | 84 (81–100) | 53 (47–61) | 0.018 * |
| FEV1/VC (%) | 100.5 (98.5–106.5) | 76.3 (70–80.8) | 53 (47–61) | 0.013 * |
| FeNO (ppb) | 9.3 (9.3–9.3) | 47.5 (29.6–67.7) | 22.4 (13.9–37.3) | 0.124 |
| ACT (points) | N.A. | 19 (10–22) | N.A. | N.A. |
| ICS treatment (n) | N.A. | 2 | 0 | N.A. |
| CAT (points) | N.A. | N.A. | 11 (8–17) | N.A. |
| mMRC (points) | N.A. | N.A. | 3 (1–3) | N.A. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Misiukiewicz-Stępien, P.; Mierzejewski, M.; Zajusz-Zubek, E.; Goryca, K.; Adamska, D.; Szeląg, M.; Krenke, R.; Paplińska-Goryca, M. RNA-Seq Analysis of UPM-Exposed Epithelium Co-Cultivated with Macrophages and Dendritic Cells in Obstructive Lung Diseases. Int. J. Mol. Sci. 2022, 23, 9125. https://doi.org/10.3390/ijms23169125
Misiukiewicz-Stępien P, Mierzejewski M, Zajusz-Zubek E, Goryca K, Adamska D, Szeląg M, Krenke R, Paplińska-Goryca M. RNA-Seq Analysis of UPM-Exposed Epithelium Co-Cultivated with Macrophages and Dendritic Cells in Obstructive Lung Diseases. International Journal of Molecular Sciences. 2022; 23(16):9125. https://doi.org/10.3390/ijms23169125
Chicago/Turabian StyleMisiukiewicz-Stępien, Paulina, Michał Mierzejewski, Elwira Zajusz-Zubek, Krzysztof Goryca, Dorota Adamska, Michał Szeląg, Rafał Krenke, and Magdalena Paplińska-Goryca. 2022. "RNA-Seq Analysis of UPM-Exposed Epithelium Co-Cultivated with Macrophages and Dendritic Cells in Obstructive Lung Diseases" International Journal of Molecular Sciences 23, no. 16: 9125. https://doi.org/10.3390/ijms23169125







