Altered Expression of TMEM43 Causes Abnormal Cardiac Structure and Function in Zebrafish
Abstract
1. Introduction
2. Results
2.1. Conservation and Expression of TMEM43 Ortholog in Zebrafish
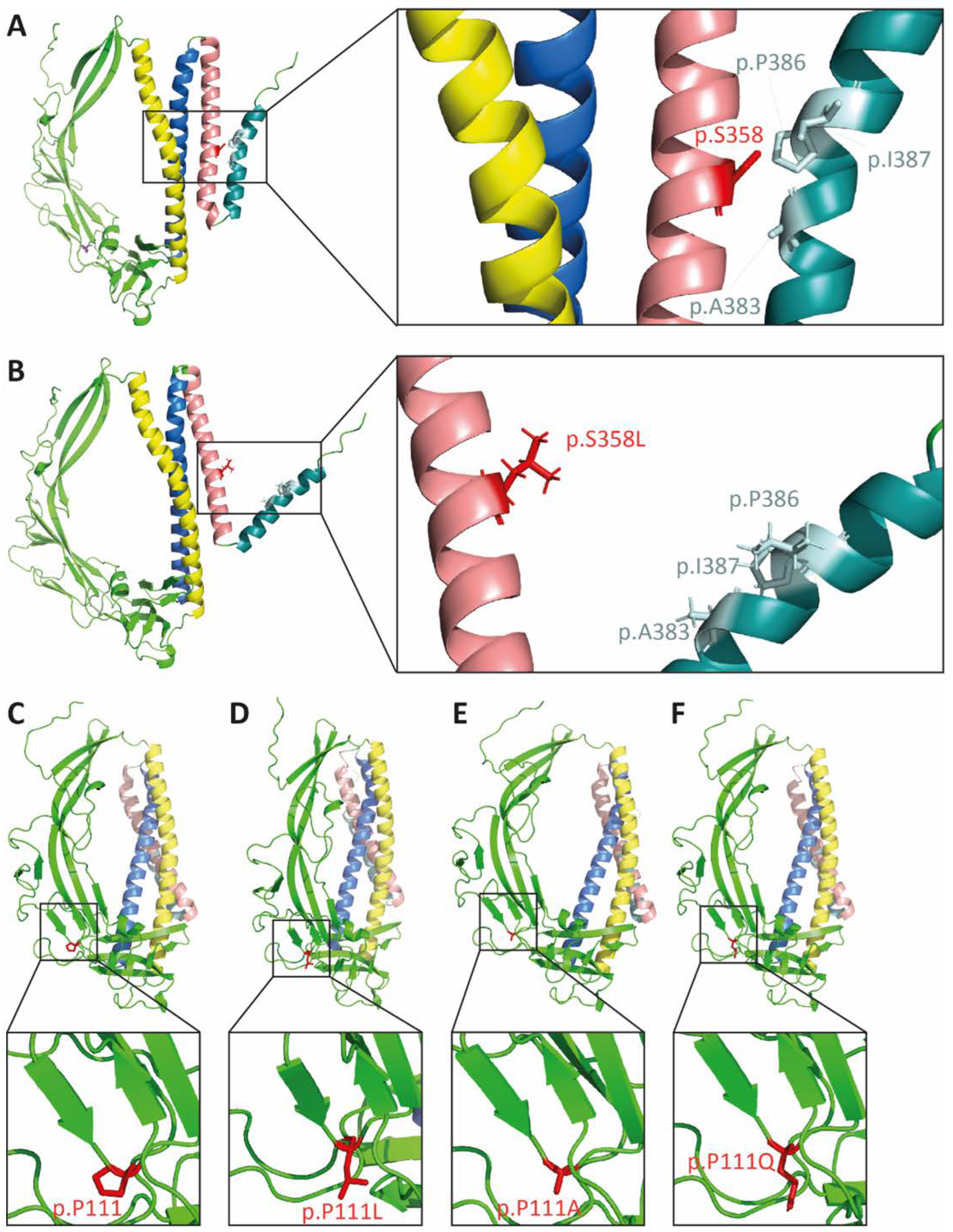
2.2. In Silico Modeling of the p.S358L Variant Predicts Conformational Change of TMEM43
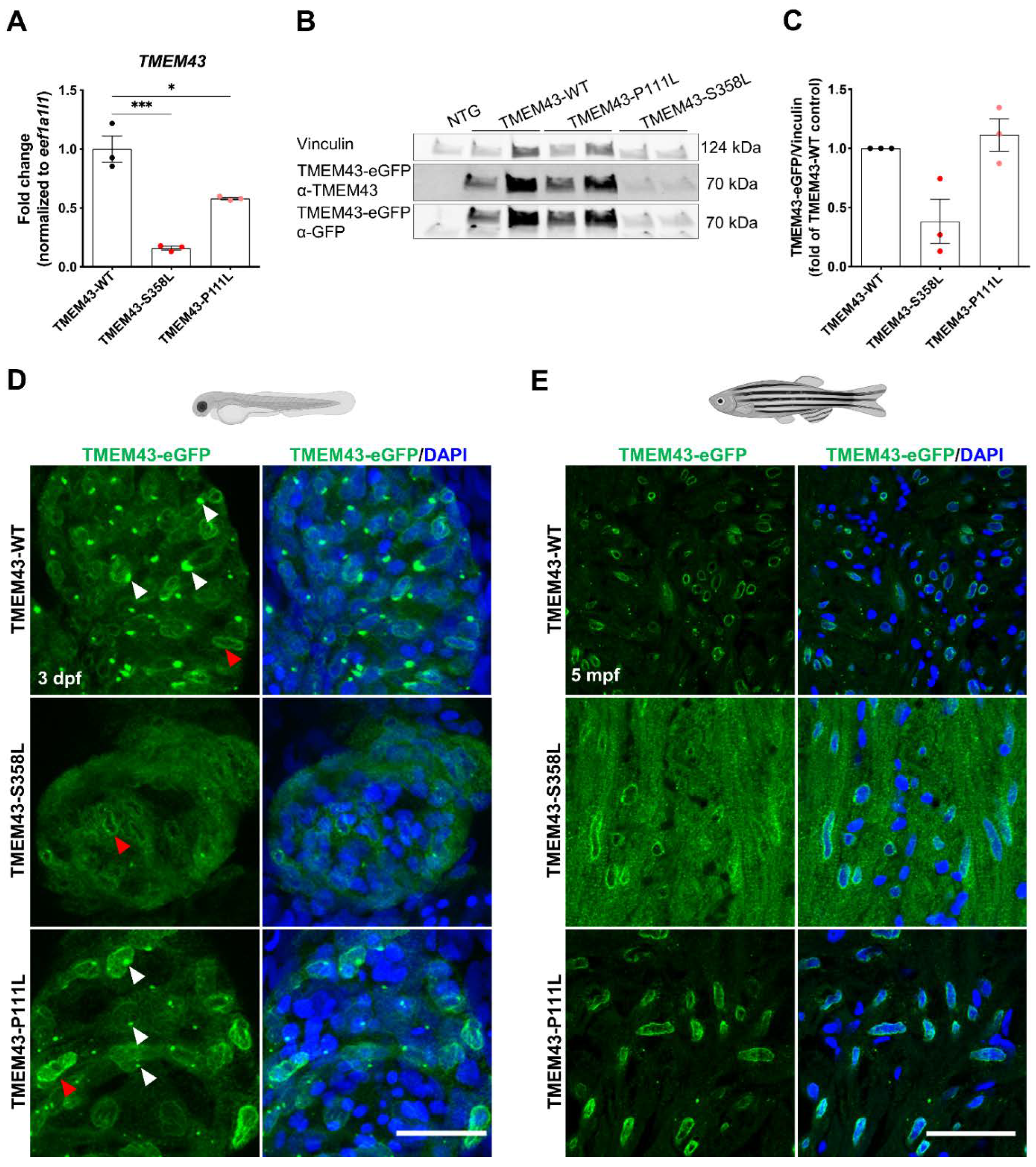
2.3. The p.S358L Variant Alters Cellular Localization of TMEM43 in Zebrafish Cardiomyocytes
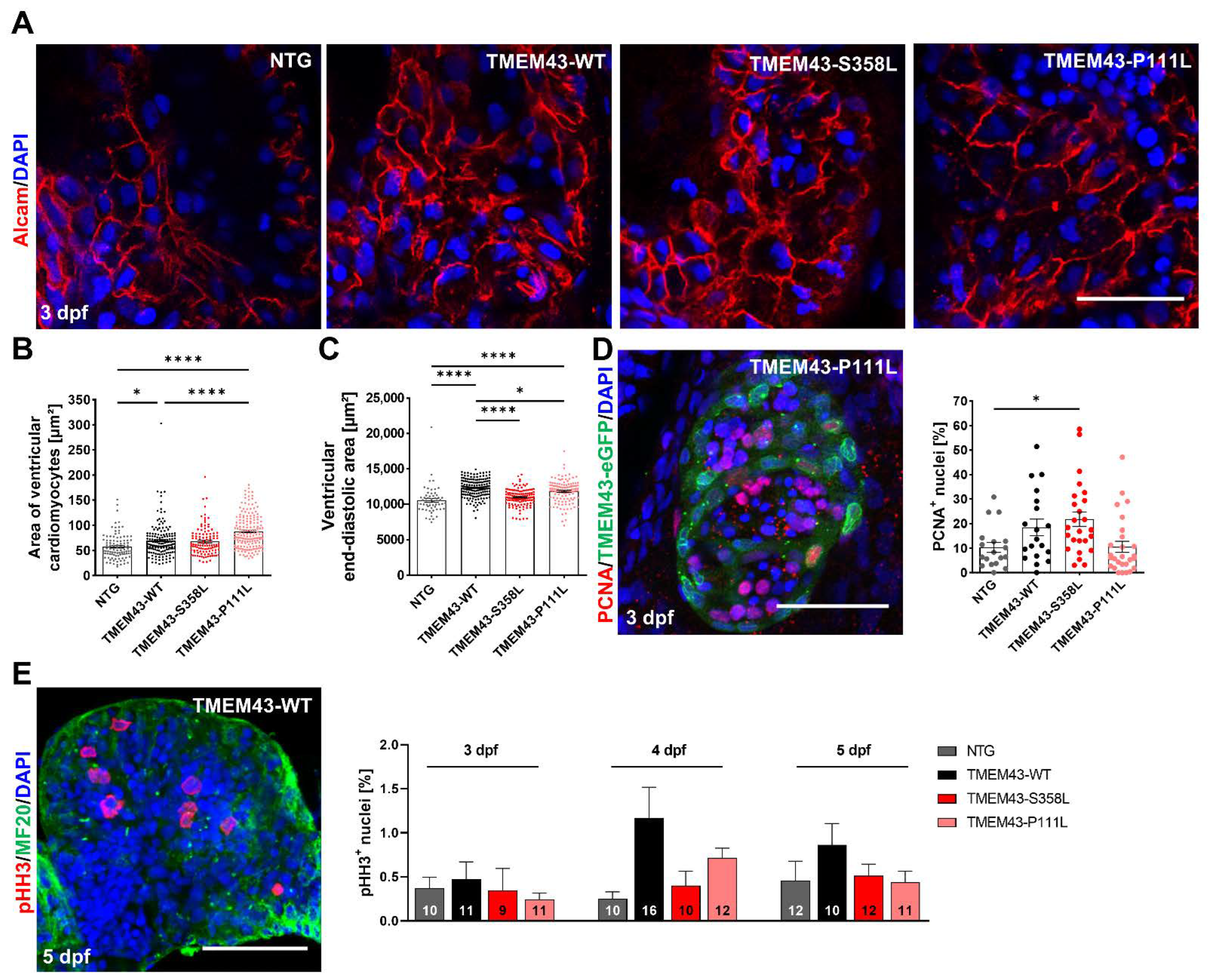
2.4. Mutant TMEM43 Impairs Cardiac Development and Alters Cardiac Function at 3 dpf
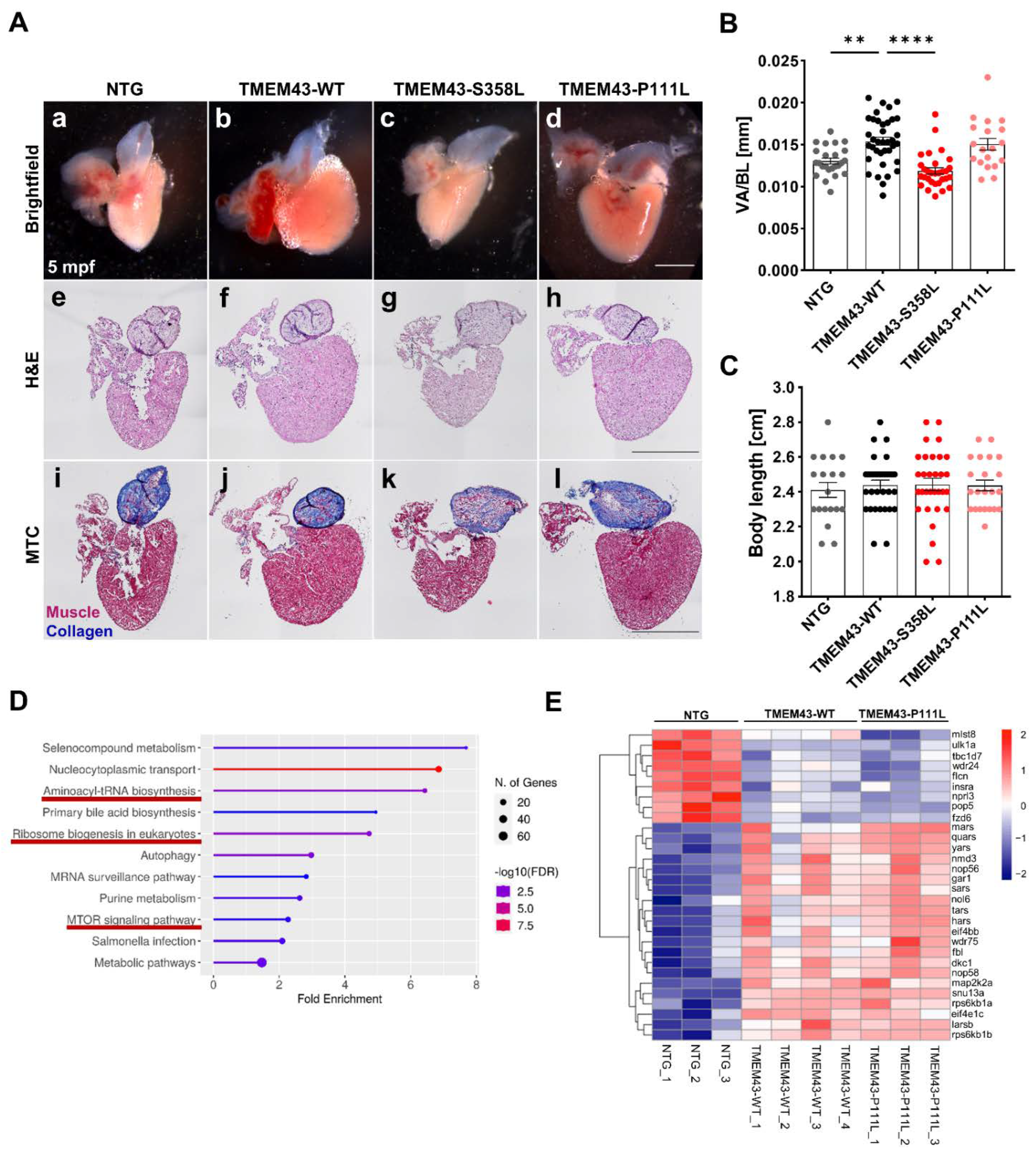
2.5. Cardiac-Restricted Overexpression of TMEM43-WT and TMEM43-P111L Results in Hypertrophied Embryonic and Adult Hearts
2.6. Transcriptomic Analysis Reveals Regulation of Various Pathways including Cell Growth and Hypertrophy
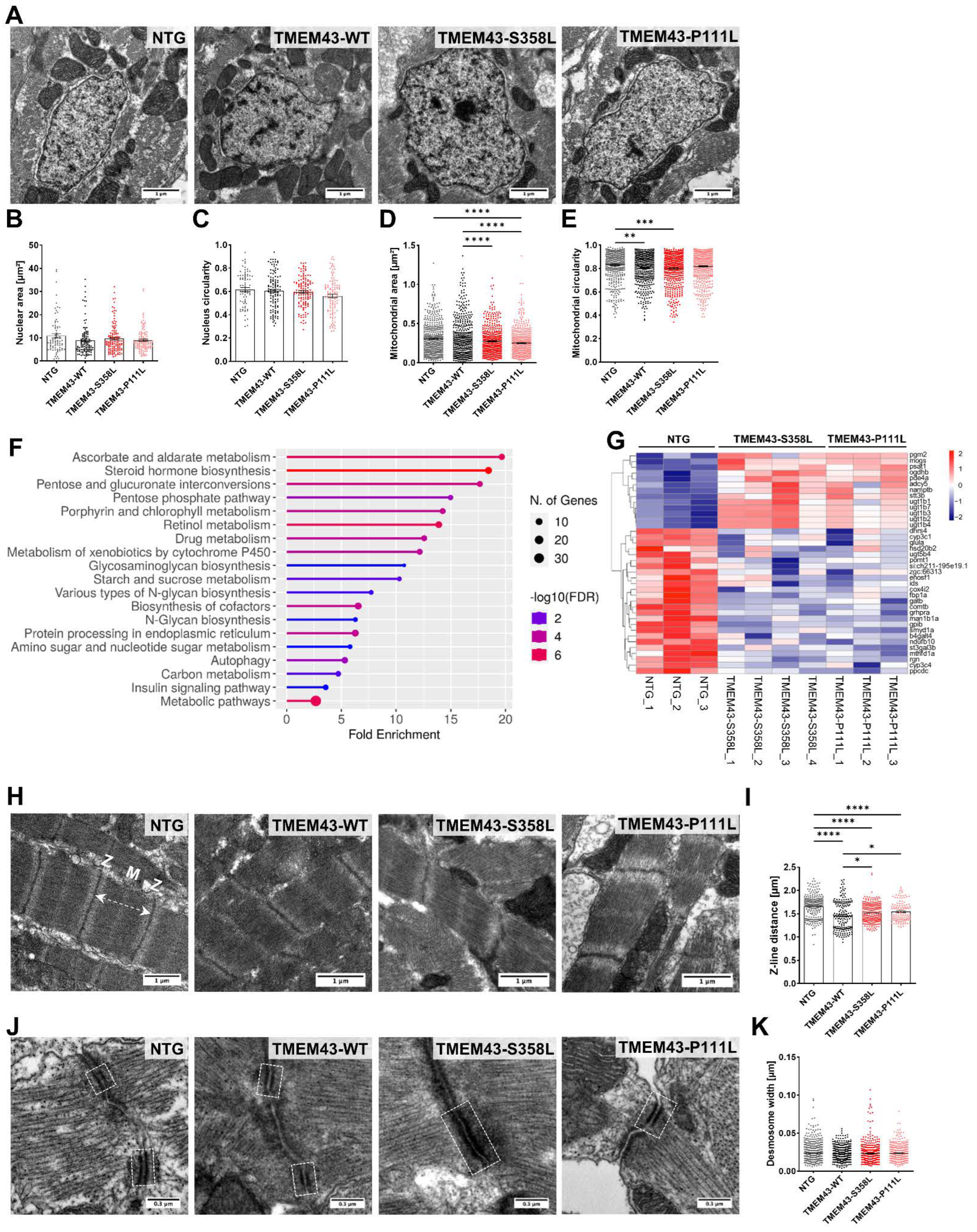
2.7. Altered Ultrastructure of Ventricular Tissue in Adult TMEM43 Expressing Transgenic Zebrafish
2.8. Generation of CRISPR/Cas9-Induced tmem43 Knock-Out Zebrafish Lines
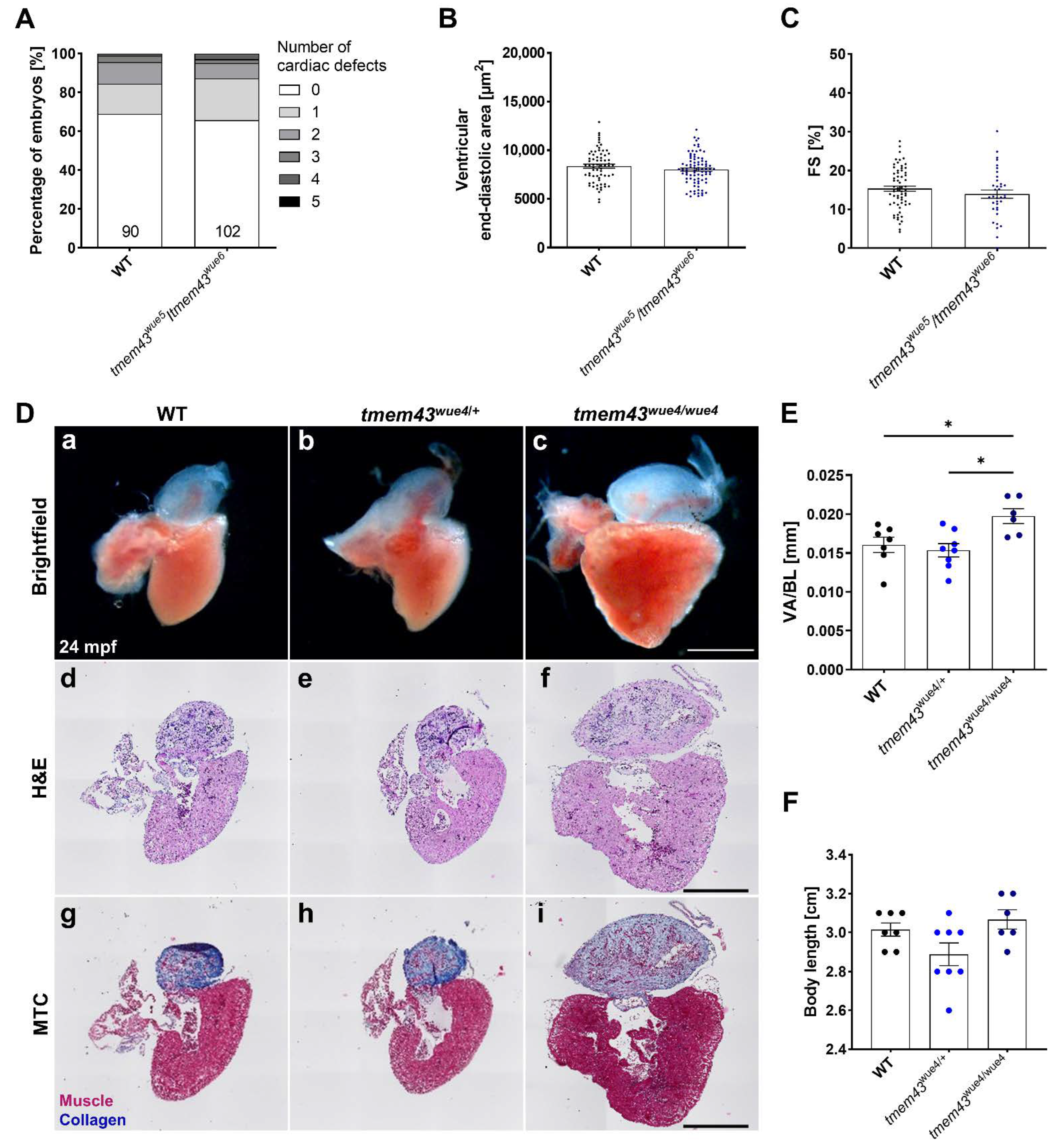
2.9. Homozygous tmem43 Mutants Develop a Late-Onset Cardiac Phenotype
3. Discussion
4. Materials and Methods
4.1. Zebrafish Husbandry
4.2. Generation of Transgenic Lines
4.3. Generation of tmem43 CRISPR Lines
4.4. Microinjection into Zebrafish Embryos
4.5. Whole-Mount In Situ Hybridization
4.6. In Vivo Imaging and Analysis
4.7. Surface ECG Recording and Analysis
4.8. Immunofluorescence Staining
4.9. Histologic Analyses
4.10. Transmission Electron Microscopy
4.11. Sanger Sequencing
4.12. RT-PCR
4.13. qPCR
4.14. RNA-Seq
4.15. Western Blot Analysis
4.16. Bioinformatical Analysis
4.17. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Corrado, D.; Basso, C.; Judge, D.P. Arrhythmogenic Cardiomyopathy. Circ. Res. 2017, 121, 784–802. [Google Scholar] [CrossRef] [PubMed]
- James, C.A.; Syrris, P.; Van Tintelen, J.P.; Calkins, H. The role of genetics in cardiovascular disease: Arrhythmogenic cardiomyopathy. Eur. Heart J. 2020, 41, 1393–1400. [Google Scholar] [CrossRef]
- Thiene, G.; Nava, A.; Corrado, D.; Rossi, L.; Pennelli, N. Right ventricular cardiomyopathy and sudden death in young people. N. Engl. J. Med. 1988, 318, 129–133. [Google Scholar] [CrossRef] [PubMed]
- Gerull, B.; Brodehl, A. Insights into Genetics and Pathophysiology of Arrhythmogenic Cardiomyopathy. Curr. Heart Fail. Rep. 2021, 18, 378–390. [Google Scholar] [CrossRef]
- Austin, K.M.; Trembley, M.A.; Chandler, S.F.; Sanders, S.P.; Saffitz, J.E.; Abrams, D.J.; Pu, W.T. Molecular mechanisms of arrhythmogenic cardiomyopathy. Nat. Rev. Cardiol. 2019, 16, 519–537. [Google Scholar] [CrossRef] [PubMed]
- Antonini-Canterin, F.; Di Nora, C. Arrhythmogenic right ventricular cardiomyopathy or athlete’s heart? Challenges in assessment of right heart morphology and function. Monaldi Arch. Chest Dis. 2019, 89, 1047. [Google Scholar] [CrossRef]
- Gerull, B.; Heuser, A.; Wichter, T.; Paul, M.; Basson, C.T.; McDermott, D.A.; Lerman, B.B.; Markowitz, S.M.; Ellinor, P.T.; MacRae, C.A.; et al. Mutations in the desmosomal protein plakophilin-2 are common in arrhythmogenic right ventricular cardiomyopathy. Nat. Genet. 2004, 36, 1162–1164. [Google Scholar] [CrossRef]
- Yang, Z.; Bowles, N.E.; Scherer, S.E.; Taylor, M.D.; Kearney, D.L.; Ge, S.; Nadvoretskiy, V.V.; DeFreitas, G.; Carabello, B.; Brandon, L.I.; et al. Desmosomal dysfunction due to mutations in desmoplakin causes arrhythmogenic right ventricular dysplasia/cardiomyopathy. Circ. Res. 2006, 99, 646–655. [Google Scholar] [CrossRef]
- Quarta, G.; Syrris, P.; Ashworth, M.; Jenkins, S.; Alapi, K.Z.; Morgan, J.; Muir, A.; Pantazis, A.; McKenna, W.J.; Elliott, P.M. Mutations in the Lamin A/C gene mimic arrhythmogenic right ventricular cardiomyopathy. Eur. Heart J. 2012, 33, 1128–1136. [Google Scholar] [CrossRef]
- Alastalo, T.P.; West, G.; Li, S.-P.; Keinänen, A.; Helenius, M.; Tyni, T.; Lapatto, R.; Turanlahti, M.; Heikkilä, P.; Kääriäinen, H.; et al. LMNA Mutation c.917T>G (p.L306R) Leads to Deleterious Hyper-Assembly of Lamin A/C and Associates with Severe Right Ventricular Cardiomyopathy and Premature Aging. Hum. Mutat. 2015, 36, 694–703. [Google Scholar] [CrossRef]
- Abdelfatah, N.; Chen, R.; Duff, H.J.; Seifer, C.M.; Buffo, I.; Huculak, C.; Clarke, S.; Clegg, R.; Jassal, D.; Gordon, P.M.; et al. Characterization of a Unique Form of Arrhythmic Cardiomyopathy Caused by Recessive Mutation in LEMD2. JACC Basic Transl. Sci. 2019, 4, 204–221. [Google Scholar] [CrossRef] [PubMed]
- Protonotarios, A.; Brodehl, A.; Asimaki, A.; Jager, J.; Quinn, E.; Stanasiuk, C.; Ratnavadivel, S.; Futema, M.; Akhtar, M.M.; Gossios, T.D.; et al. The Novel Desmin Variant p.Leu115Ile Is Associated With a Unique Form of Biventricular Arrhythmogenic Cardiomyopathy. Can. J. Cardiol. 2021, 37, 857–866. [Google Scholar] [CrossRef] [PubMed]
- Good, J.M.; Fellmann, F.; Bhuiyan, Z.A.; Rotman, S.; Pruvot, E.; Schläpfer, J. ACTN2 variant associated with a cardiac phenotype suggestive of left-dominant arrhythmogenic cardiomyopathy. Heart Rhythm Case Rep. 2020, 6, 15–19. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tiso, N.; Stephan, D.A.; Nava, A.; Bagattin, A.; Devaney, J.M.; Stanchi, F.; Larderet, G.; Brahmbhatt, B.; Brown, K.; Bauce, B.; et al. Identification of mutations in the cardiac ryanodine receptor gene in families affected with arrhythmogenic right ventricular cardiomyopathy type 2 (ARVD2). Hum. Mol. Genet. 2001, 10, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Van der Zwaag, P.A.; Van Rijsingen, I.A.; Asimaki, A.; Jongbloed, J.D.; Van Veldhuisen, D.J.; Wiesfeld, A.C.; Cox, M.G.P.J.; van Lochem, L.T.; de Boer, R.A.; Hofstra, R.M.W.; et al. Phospholamban R14del mutation in patients diagnosed with dilated cardiomyopathy or arrhythmogenic right ventricular cardiomyopathy: Evidence supporting the concept of arrhythmogenic cardiomyopathy. Eur. J. Heart Fail. 2012, 14, 1199–1207. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Q.; Cao, Q.; Zhou, Q.; Xie, J.; Shen, Y.; Wan, R.; Yu, J.; Yan, S.; Marian, A.J.; Hong, K. Arrhythmogenic cardiomyopathy in a patient with a rare loss-of-function KCNQ1 mutation. J. Am. Heart Assoc. 2015, 4, e001526. [Google Scholar] [CrossRef]
- Beffagna, G.; Occhi, G.; Nava, A.; Vitiello, L.; Ditadi, A.; Basso, C.; Bauce, B.; Carraro, G.; Thiene, G.; Towbin, J.A.; et al. Regulatory mutations in transforming growth factor-beta3 gene cause arrhythmogenic right ventricular cardiomyopathy type 1. Cardiovasc. Res. 2005, 65, 366–373. [Google Scholar] [CrossRef]
- Merner, N.D.; Hodgkinson, K.A.; Haywood, A.F.; Connors, S.; French, V.M.; Drenckhahn, J.-D.; Kupprion, C.; Ramadanova, K.; Thierfelder, L.; McKenna, W.; et al. Arrhythmogenic right ventricular cardiomyopathy type 5 is a fully penetrant, lethal arrhythmic disorder caused by a missense mutation in the TMEM43 gene. Am. J. Hum. Genet. 2008, 82, 809–821. [Google Scholar] [CrossRef]
- Milting, H.; Klauke, B.; Christensen, A.H.; Müsebeck, J.; Walhorn, V.; Grannemann, S.; Münnich, T.; Šarić, T.; Rasmussen, T.B.; Jensen, H.K.; et al. The TMEM43 Newfoundland mutation p.S358L causing ARVC-5 was imported from Europe and increases the stiffness of the cell nucleus. Eur. Heart J. 2015, 36, 872–881. [Google Scholar] [CrossRef]
- Christensen, A.H.; Andersen, C.B.; Tybjaerg-Hansen, A.; Haunso, S.; Svendsen, J.H. Mutation analysis and evaluation of the cardiac localization of TMEM43 in arrhythmogenic right ventricular cardiomyopathy. Clin. Genet. 2011, 80, 256–264. [Google Scholar] [CrossRef]
- Baskin, B.; Skinner, J.R.; Sanatani, S.; Terespolsky, D.; Krahn, A.D.; Ray, P.N.; Scherer, S.W.; Hamilton, R.M. TMEM43 mutations associated with arrhythmogenic right ventricular cardiomyopathy in non-Newfoundland populations. Hum. Genet. 2013, 132, 1245–1252. [Google Scholar] [CrossRef] [PubMed]
- Shinomiya, H.; Kato, H.; Kuramoto, Y.; Watanabe, N.; Tsuruda, T.; Arimura, T.; Miyashita, Y.; Miyasaka, Y.; Mashimo, T.; Takuwa, A.; et al. Aberrant accumulation of TMEM43 accompanied by perturbed transmural gene expression in arrhythmogenic cardiomyopathy. FASEB J. 2021, 35, e21994. [Google Scholar] [CrossRef] [PubMed]
- Honda, T.; Kanai, Y.; Ohno, S.; Ando, H.; Honda, M.; Niwano, S.; Ishii, M. Fetal arrhythmogenic right ventricular cardiomyopathy with double mutations in TMEM43. Pediatr. Int. 2016, 58, 409–411. [Google Scholar] [CrossRef] [PubMed]
- Bao, J.; Wang, J.; Yao, Y.; Wang, Y.; Fan, X.; Sun, K.; He, D.S.; Marcus, F.I.; Zhang, S.; Hui, R.; et al. Correlation of ventricular arrhythmias with genotype in arrhythmogenic right ventricular cardiomyopathy. Circ. Cardiovasc. Genet. 2013, 6, 552–556. [Google Scholar] [CrossRef] [PubMed]
- Mukai, T.; Mori-Yoshimura, M.; Nishikawa, A.; Hokkoku, K.; Sonoo, M.; Nishino, I.; Takahashi, Y. Emery-Dreifuss muscular dystrophy-related myopathy with TMEM43 mutations. Muscle Nerve 2019, 59, E5–E7. [Google Scholar] [CrossRef]
- Liang, W.C.; Mitsuhashi, H.; Keduka, E.; Nonaka, I.; Noguchi, S.; Nishino, I.; Hayashi, Y.K. TMEM43 mutations in Emery-Dreifuss muscular dystrophy-related myopathy. Ann. Neurol. 2011, 69, 1005–1013. [Google Scholar] [CrossRef]
- Jang, M.W.; Oh, D.-Y.; Yi, E.; Liu, X.; Ling, J.; Kim, N.; Sharma, K.; Kim, T.Y.; Lee, S.; Kim, A.-R.; et al. A nonsense TMEM43 variant leads to disruption of connexin-linked function and autosomal dominant auditory neuropathy spectrum disorder. Proc. Natl. Acad. Sci. USA 2021, 118, e2019681118. [Google Scholar] [CrossRef]
- Bengtsson, L.; Otto, H. LUMA interacts with emerin and influences its distribution at the inner nuclear membrane. J. Cell Sci. 2008, 121 Pt 4, 536–548. [Google Scholar] [CrossRef]
- Franke, W.W.; Dörflinger, Y.; Kuhn, C.; Zimbelmann, R.; Winter-Simanowski, S.; Frey, N.; Heid, H. Protein LUMA is a cytoplasmic plaque constituent of various epithelial adherens junctions and composite junctions of myocardial intercalated disks: A unifying finding for cell biology and cardiology. Cell Tissue Res. 2014, 357, 159–172. [Google Scholar] [CrossRef]
- Padron-Barthe, L.; Villalba-Orero, M.; Gómez-Salinero, J.M.; Domínguez, F.; Román, M.; Larrasa-Alonso, J.; Ortiz-Sánchez, P.; Martínez, F.; López-Olañeta, M.; Bonzón-Kulichenko, E.; et al. Severe Cardiac Dysfunction and Death Caused by Arrhythmogenic Right Ventricular Cardiomyopathy Type 5 Are Improved by Inhibition of Glycogen Synthase Kinase-3beta. Circulation 2019, 140, 1188–1204. [Google Scholar] [CrossRef]
- Stroud, M.J.; Fang, X.; Zhang, J.; Guimaraes-Camboa, N.; Veevers, J.; Dalton, N.D.; Gu, Y.; Bradford, W.H.; Peterson, K.L.; Evans, S.M.; et al. Luma is not essential for murine cardiac development and function. Cardiovasc. Res. 2018, 114, 378–388. [Google Scholar] [CrossRef] [PubMed]
- Rouhi, L.; Cheedipudi, S.M.; Chen, S.N.; Fan, S.; Lombardi, R.; Chen, X.; Coarfa, C.; Robertson, M.J.; Gurha, P.; Marian, A.J. Haploinsufficiency of Tmem43 in cardiac myocytes activates the DNA damage response pathway leading to a late-onset senescence-associated pro-fibrotic cardiomyopathy. Cardiovasc. Res. 2021, 117, 2377–2394. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Jiang, C.; Li, Y.; Yang, D.; Ma, Y.; Zhang, B.; Li, X.; Zhang, P.; Hu, X.; Zhao, X.; et al. TMEM43-S358L mutation enhances NF-kappaB-TGFbeta signal cascade in arrhythmogenic right ventricular dysplasia/cardiomyopathy. Protein Cell 2019, 10, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Gu, Q.; Xu, F.; Orgil, B.O.; Khuchua, Z.; Munkhsaikhan, U.; Johnson, J.N.; Alberson, N.R.; Pierre, J.F.; Black, D.D.; Dong, D.; et al. Systems genetics analysis defines importance of TMEM43/LUMA for cardiac- and metabolic-related pathways. Physiol. Genom. 2022, 54, 22–35. [Google Scholar] [CrossRef]
- Ratnavadivel, S.; de Toledo, M.S.; Rasmussen, T.B.; Šarić, T.; Gummert, J.; Zenke, M.; Milting, H. Human pluripotent stem cell line (HDZi001-A) derived from a patient carrying the ARVC-5 associated mutation TMEM43-p.S358L. Stem. Cell Res. 2020, 48, 101957. [Google Scholar] [CrossRef]
- Alirezaie, N.; Kernohan, K.D.; Hartley, T.; Majewski, J.; Hocking, T.D. ClinPred: Prediction Tool to Identify Disease-Relevant Nonsynonymous Single-Nucleotide Variants. Am. J. Hum. Genet. 2018, 103, 474–483. [Google Scholar] [CrossRef]
- Urasaki, A.; Morvan, G.; Kawakami, K. Functional dissection of the Tol2 transposable element identified the minimal cis-sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics 2006, 174, 639–649. [Google Scholar] [CrossRef]
- Kawakami, K.; Takeda, H.; Kawakami, N.; Kobayashi, M.; Matsuda, N.; Mishina, M. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev. Cell 2004, 7, 133–144. [Google Scholar] [CrossRef]
- Mayer, C.; Grummt, I. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene 2006, 25, 6384–6391. [Google Scholar] [CrossRef]
- Hwang, W.Y.; Fu, Y.; Reyon, D.; Maeder, M.L.; Tsai, S.Q.; Sander, J.D.; Peterson, R.T.; Yeh, J.-R.J.; Joung, J.K. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 227–229. [Google Scholar] [CrossRef]
- Sander, J.D.; Maeder, M.L.; Reyon, D.; Voytas, D.F.; Joung, J.K.; Dobbs, D. ZiFiT (Zinc Finger Targeter): An updated zinc finger engineering tool. Nucleic Acids Res. 2010, 38, W462–W468. [Google Scholar] [CrossRef] [PubMed]
- Sander, J.D.; Zaback, P.; Joung, J.K.; Voytas, D.F.; Dobbs, D. Zinc Finger Targeter (ZiFiT): An engineered zinc finger/target site design tool. Nucleic Acids Res. 2007, 35, W599–W605. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.Y.; Partridge, A.W.; Daniels, C.; Du, K.; Lukacs, G.; Deber, C.M. Destabilization of the transmembrane domain induces misfolding in a phenotypic mutant of cystic fibrosis transmembrane conductance regulator. J. Biol. Chem. 2005, 280, 4968–4974. [Google Scholar] [CrossRef] [PubMed]
- Schlebach, J.P.; Narayan, M.; Alford, C.; Mittendorf, K.F.; Carter, B.D.; Li, J.; Sanders, C.R. Conformational Stability and Pathogenic Misfolding of the Integral Membrane Protein PMP22. J. Am. Chem. Soc. 2015, 137, 8758–8768. [Google Scholar] [CrossRef]
- Tapia, O.; Fong, L.G.; Huber, M.D.; Young, S.G.; Gerace, L. Nuclear envelope protein Lem2 is required for mouse development and regulates MAP and AKT kinases. PLoS ONE 2015, 10, e0116196. [Google Scholar]
- Ross, J.A.; Arcos-Villacis, N.; Battey, E.; Boogerd, C.; Marhuenda, E.; Hodzic, D.; Prin, F.; Mohun, T.; Catibog, N.; Tapia, O.; et al. Lem2 is essential for cardiac development by maintaining nuclear integrity. bioRxiv 2022. [Google Scholar]
- Spudich, J.A. Three perspectives on the molecular basis of hypercontractility caused by hypertrophic cardiomyopathy mutations. Pflug. Arch. 2019, 471, 701–717. [Google Scholar] [CrossRef]
- Vander Roest, A.S.; Liu, C.; Morck, M.M.; Kooiker, K.B.; Jung, G.; Song, D.; Dawood, A.; Jhingran, A.; Pardon, G.; Ranjbarvarziri, S.; et al. Hypertrophic cardiomyopathy beta-cardiac myosin mutation (P710R) leads to hypercontractility by disrupting super relaxed state. Proc. Natl. Acad. Sci. USA 2021, 118, e2025030118. [Google Scholar] [CrossRef]
- Clay, S.A.; Domeier, T.L.; Hanft, L.M.; McDonald, K.S.; Krenz, M. Elevated Ca2+ transients and increased myofibrillar power generation cause cardiac hypercontractility in a model of Noonan syndrome with multiple lentigines. Am. J. Physiol. Heart Circ. Physiol. 2015, 308, H1086–H1095. [Google Scholar] [CrossRef]
- Prelich, G. Gene overexpression: Uses, mechanisms, and interpretation. Genetics 2012, 190, 841–854. [Google Scholar]
- Sarkar, S.; Leaman, D.W.; Gupta, S.; Sil, P.; Young, D.; Morehead, A.; Mukherjee, D.; Ratliff, N.; Sun, Y.; Rayborn, M.; et al. Cardiac overexpression of myotrophin triggers myocardial hypertrophy and heart failure in transgenic mice. J. Biol. Chem. 2004, 279, 20422–20434. [Google Scholar] [CrossRef] [PubMed]
- Collins, M.M.; Ahlberg, G.; Hansen, C.V.; Guenther, S.; Marín-Juez, R.; Sokol, A.M.; El-Sammak, H.; Piesker, J.; Hellsten, Y.; Olesen, M.S.; et al. Early sarcomere and metabolic defects in a zebrafish pitx2c cardiac arrhythmia model. Proc. Natl. Acad. Sci. USA 2019, 116, 24115–24121. [Google Scholar] [CrossRef] [PubMed]
- Yelon, D.; Horne, S.A.; Stainier, D.Y.R. Restricted expression of cardiac myosin genes reveals regulated aspects of heart tube assembly in zebrafish. Dev. Biol. 1999, 214, 23–37. [Google Scholar] [CrossRef] [PubMed]
- Ng, W.A.; Grupp, I.L.; Subramaniam, A.; Robbins, J. Cardiac myosin heavy chain mRNA expression and myocardial function in the mouse heart. Circ. Res. 1991, 68, 1742–1750. [Google Scholar] [CrossRef] [PubMed]
- Zorzi, A.; Rigato, I.; Pilichou, K.; Perazzolo Marra, M.; Migliore, F.; Mazzotti, E.; Gregori, D.; Thiene, G.; Daliento, L.; Iliceto, S.; et al. Phenotypic expression is a prerequisite for malignant arrhythmic events and sudden cardiac death in arrhythmogenic right ventricular cardiomyopathy. Europace 2016, 18, 1086–1094. [Google Scholar] [CrossRef]
- Basso, C.; Thiene, G.; Corrado, D.; Angelini, A.; Nava, A.; Valente, M. Arrhythmogenic right ventricular cardiomyopathy. Dysplasia, dystrophy, or myocarditis? Circulation 1996, 94, 983–991. [Google Scholar] [CrossRef]
- Rizzo, S.; Lodder, E.M.; Verkerk, A.O.; Wolswinkel, R.; Beekman, L.; Pilichou, K.; Basso, C.; Remme, C.A.; Thiene, G.; Bezzina, C.R. Intercalated disc abnormalities, reduced Na(+) current density, and conduction slowing in desmoglein-2 mutant mice prior to cardiomyopathic changes. Cardiovasc. Res. 2012, 95, 409–418. [Google Scholar] [CrossRef]
- Noorman, M.; Hakim, S.; Kessler, E.; A Groeneweg, J.; Cox, M.G.; Asimaki, A.; van Rijen, H.V.; van Stuijvenberg, L.; Chkourko, H.; van der Heyden, M.A.; et al. Remodeling of the cardiac sodium channel, connexin43, and plakoglobin at the intercalated disk in patients with arrhythmogenic cardiomyopathy. Heart Rhythm 2013, 10, 412–419. [Google Scholar] [CrossRef]
- Cerrone, M.; Montnach, J.; Lin, X.; Zhao, Y.-T.; Zhang, M.; Agullo-Pascual, E.; Leo-Macias, A.; Alvarado, F.J.; Dolgalev, I.; Karathanos, T.V.; et al. Plakophilin-2 is required for transcription of genes that control calcium cycling and cardiac rhythm. Nat. Commun. 2017, 8, 106. [Google Scholar] [CrossRef]
- van Opbergen, C.J.M.; Noorman, M.; Pfenniger, A.; Copier, J.S.; Vermij, S.H.; Li, Z.; van der Nagel, R.; Zhang, M.; de Bakker, J.M.T.; Glass, A.M.; et al. Plakophilin-2 Haploinsufficiency Causes Calcium Handling Deficits and Modulates the Cardiac Response Towards Stress. Int. J. Mol. Sci. 2019, 20, 4076. [Google Scholar] [CrossRef]
- Brodehl, A.; Rezazadeh, S.; Williams, T.; Munsie, N.M.; Liedtke, D.; Oh, T.; Ferrier, R.; Shen, Y.; Jones, S.J.; Stiegler, A.L.; et al. Mutations in ILK, encoding integrin-linked kinase, are associated with arrhythmogenic cardiomyopathy. Transl. Res. 2019, 208, 15–29. [Google Scholar] [CrossRef] [PubMed]
- Huttner, I.G.; Wang, L.W.; Santiago, C.F.; Horvat, C.; Johnson, R.; Cheng, D.; Von Frieling-Salewsky, M.; Hillcoat, K.; Bemand, T.J.; Trivedi, G.; et al. A-Band Titin Truncation in Zebrafish Causes Dilated Cardiomyopathy and Hemodynamic Stress Intolerance. Circ. Genom. Precis. Med. 2018, 11, e002135. [Google Scholar] [CrossRef] [PubMed]
- Kirchhof, P.; Fabritz, L.; Zwiener, M.; Witt, H.; Schäfers, M.; Zellerhoff, S.; Paul, M.; Athai, T.; Hiller, K.-H.; Baba, H.A.; et al. Age- and training-dependent development of arrhythmogenic right ventricular cardiomyopathy in heterozygous plakoglobin-deficient mice. Circulation 2006, 114, 1799–1806. [Google Scholar] [CrossRef] [PubMed]
- James, C.A.; Bhonsale, A.; Tichnell, C.; Murray, B.; Russell, S.D.; Tandri, H.; Tedford, R.J.; Judge, D.P.; Calkins, H. Exercise increases age-related penetrance and arrhythmic risk in arrhythmogenic right ventricular dysplasia/cardiomyopathy-associated desmosomal mutation carriers. J. Am. Coll. Cardiol. 2013, 62, 1290–1297. [Google Scholar] [CrossRef] [PubMed]
- Sawant, A.C.; Bhonsale, A.; te Riele, A.S.; Tichnell, C.; Murray, B.; Russell, S.D.; Tandri, H.; Tedford, R.J.; Judge, D.P.; Calkins, H.; et al. Exercise has a disproportionate role in the pathogenesis of arrhythmogenic right ventricular dysplasia/cardiomyopathy in patients without desmosomal mutations. J. Am. Heart Assoc. 2014, 3, e001471. [Google Scholar] [CrossRef]
- Spinelli, J.B.; Haigis, M.C. The multifaceted contributions of mitochondria to cellular metabolism. Nat. Cell Biol. 2018, 20, 745–754. [Google Scholar] [CrossRef]
- Galloway, C.A.; Yoon, Y. Mitochondrial morphology in metabolic diseases. Antioxid. Redox Signal 2013, 19, 415–430. [Google Scholar] [CrossRef]
- Schaper, J.; Froede, R.; Hein, S.; Buck, A.; Hashizume, H.; Speiser, B.; Friedl, A.; Bleese, N. Impairment of the myocardial ultrastructure and changes of the cytoskeleton in dilated cardiomyopathy. Circulation 1991, 83, 504–514. [Google Scholar] [CrossRef]
- Chaanine, A.H.; Joyce, L.D.; Stulak, J.M.; Maltais, S.; Joyce, D.L.; Dearani, J.A.; Klaus, K.; Nair, K.S.; Hajjar, R.J.; Redfield, M.M. Mitochondrial Morphology, Dynamics, and Function in Human Pressure Overload or Ischemic Heart Disease with Preserved or Reduced Ejection Fraction. Circ. Heart Fail. 2019, 12, e005131. [Google Scholar] [CrossRef]
- Kwan, K.M.; Fujimoto, E.; Grabher, C.; Mangum, B.D.; Hardy, M.E.; Campbell, D.S.; Parant, J.M.; Yost, H.J.; Kanki, J.P.; Chien, C.-B. The Tol2kit: A multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 2007, 236, 3088–3099. [Google Scholar] [CrossRef]
- Mackey, A.S.; Redd, P.S.; DeLaurier, A.; Hancock, C.N.; Draper, B. Codon optimized Tol2 transposase results in increased transient expression of a crystallin-GFP transgene in zebrafish. microPubl. Biol. 2020, 2020. [Google Scholar] [CrossRef]
- Jao, L.E.; Wente, S.R.; Chen, W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci. USA 2013, 110, 13904–13909. [Google Scholar] [CrossRef] [PubMed]
- Thisse, C.; Thisse, B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 2008, 3, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Xu, X. Immunostaining of dissected zebrafish embryonic heart. J. Vis. Exp. 2012, 59, e3510. [Google Scholar] [CrossRef] [PubMed]
- Sullivan-Brown, J.; Bisher, M.E.; Burdine, R.D. Embedding, serial sectioning and staining of zebrafish embryos using JB-4 resin. Nat. Protoc. 2011, 6, 46–55. [Google Scholar] [CrossRef]
- Ellman, D.G.; Slaiman, I.; Mathiesen, S.; Andersen, K.; Hofmeister, W.; Ober, E.; Andersen, D. Apex Resection in Zebrafish (Danio rerio) as a Model of Heart Regeneration: A Video-Assisted Guide. Int. J. Mol. Sci. 2021, 22, 5865. [Google Scholar] [CrossRef]
- Helmprobst, F.; Frank, M.; Stigloher, C. Presynaptic architecture of the larval zebrafish neuromuscular junction. J. Comp. Neurol. 2015, 523, 1984–1997. [Google Scholar] [CrossRef]
- Martin, M. CUTADAPT removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef] [PubMed]
- Madeira, F.; Pearce, M.; Tivey, A.R.N.; Basutkar, P.; Lee, J.; Edbali, O.; Madhusoodanan, N.; Kolesnikov, A.; Lopez, R. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 2022, 50, W276–W279. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Feng, B.J. PERCH: A Unified Framework for Disease Gene Prioritization. Hum. Mutat. 2017, 38, 243–251. [Google Scholar] [CrossRef]
- Niroula, A.; Urolagin, S.; Vihinen, M. PON-P2: Prediction method for fast and reliable identification of harmful variants. PLoS ONE 2015, 10, e0117380. [Google Scholar]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zink, M.; Seewald, A.; Rohrbach, M.; Brodehl, A.; Liedtke, D.; Williams, T.; Childs, S.J.; Gerull, B. Altered Expression of TMEM43 Causes Abnormal Cardiac Structure and Function in Zebrafish. Int. J. Mol. Sci. 2022, 23, 9530. https://doi.org/10.3390/ijms23179530
Zink M, Seewald A, Rohrbach M, Brodehl A, Liedtke D, Williams T, Childs SJ, Gerull B. Altered Expression of TMEM43 Causes Abnormal Cardiac Structure and Function in Zebrafish. International Journal of Molecular Sciences. 2022; 23(17):9530. https://doi.org/10.3390/ijms23179530
Chicago/Turabian StyleZink, Miriam, Anne Seewald, Mareike Rohrbach, Andreas Brodehl, Daniel Liedtke, Tatjana Williams, Sarah J. Childs, and Brenda Gerull. 2022. "Altered Expression of TMEM43 Causes Abnormal Cardiac Structure and Function in Zebrafish" International Journal of Molecular Sciences 23, no. 17: 9530. https://doi.org/10.3390/ijms23179530
APA StyleZink, M., Seewald, A., Rohrbach, M., Brodehl, A., Liedtke, D., Williams, T., Childs, S. J., & Gerull, B. (2022). Altered Expression of TMEM43 Causes Abnormal Cardiac Structure and Function in Zebrafish. International Journal of Molecular Sciences, 23(17), 9530. https://doi.org/10.3390/ijms23179530







