Vesicular Glutamate Release from Feeder-FreehiPSC-Derived Neurons
Abstract
1. Introduction
2. Results
2.1. Generation and Characterization of iPSCs and Neuronal Differentiation
2.2. mRNA Expression Profile of hiPSC-Derived Neurons
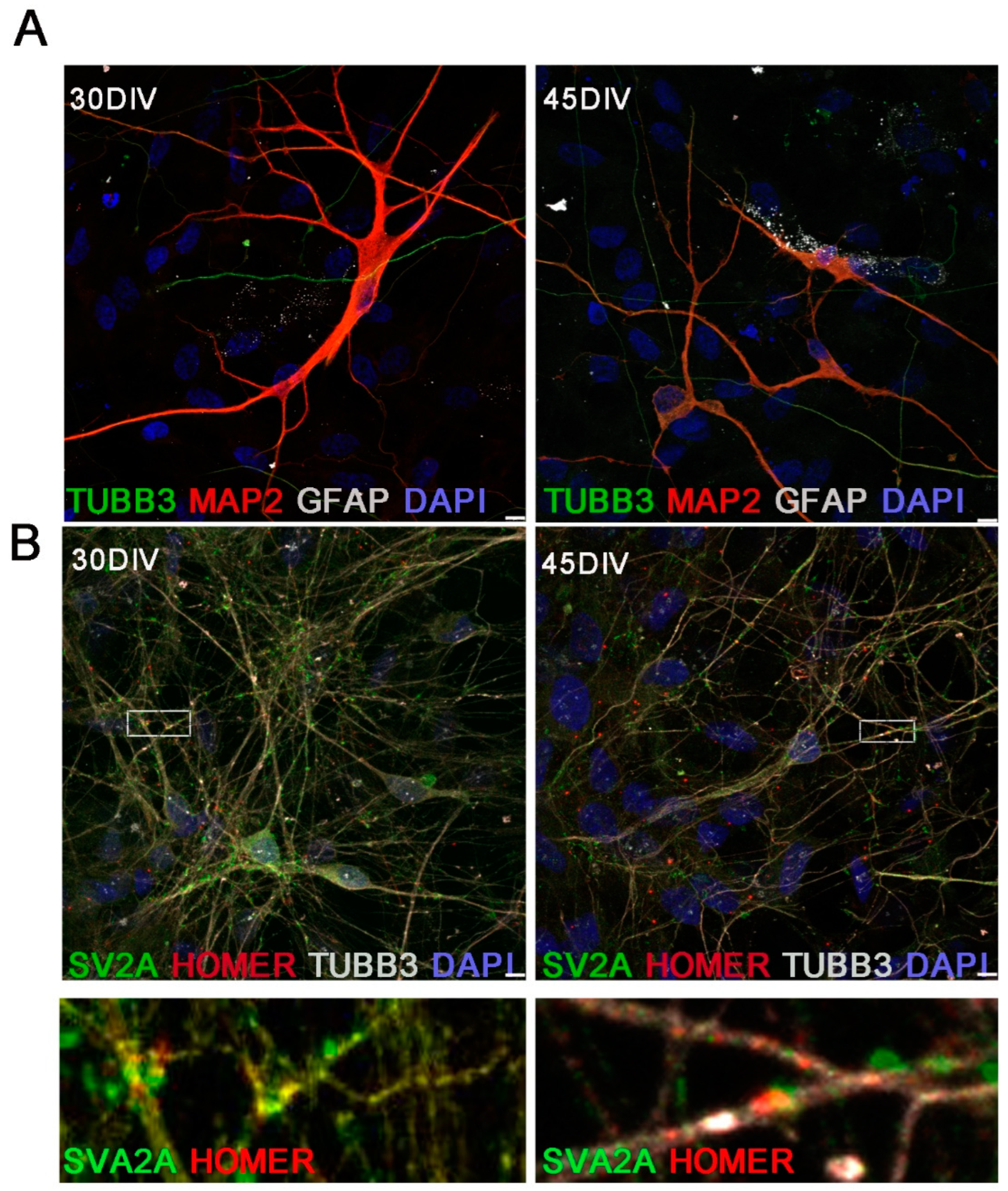
2.3. Morphological Analysis of hiPSC-DerivedNeurons
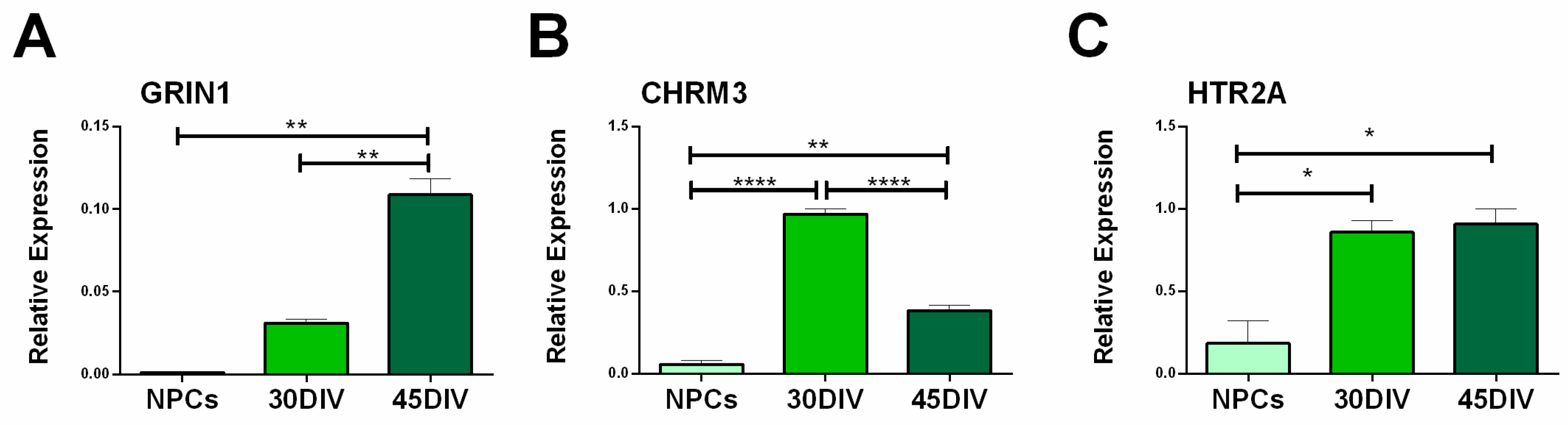
2.4. Analysis of NMDA, mAChRs and 5HT Receptors Expression in hiPSC-DerivedNeurons
2.5. Immunofluorescence Evaluation of the Synaptic Complex in hiPSC-DerivedNeurons
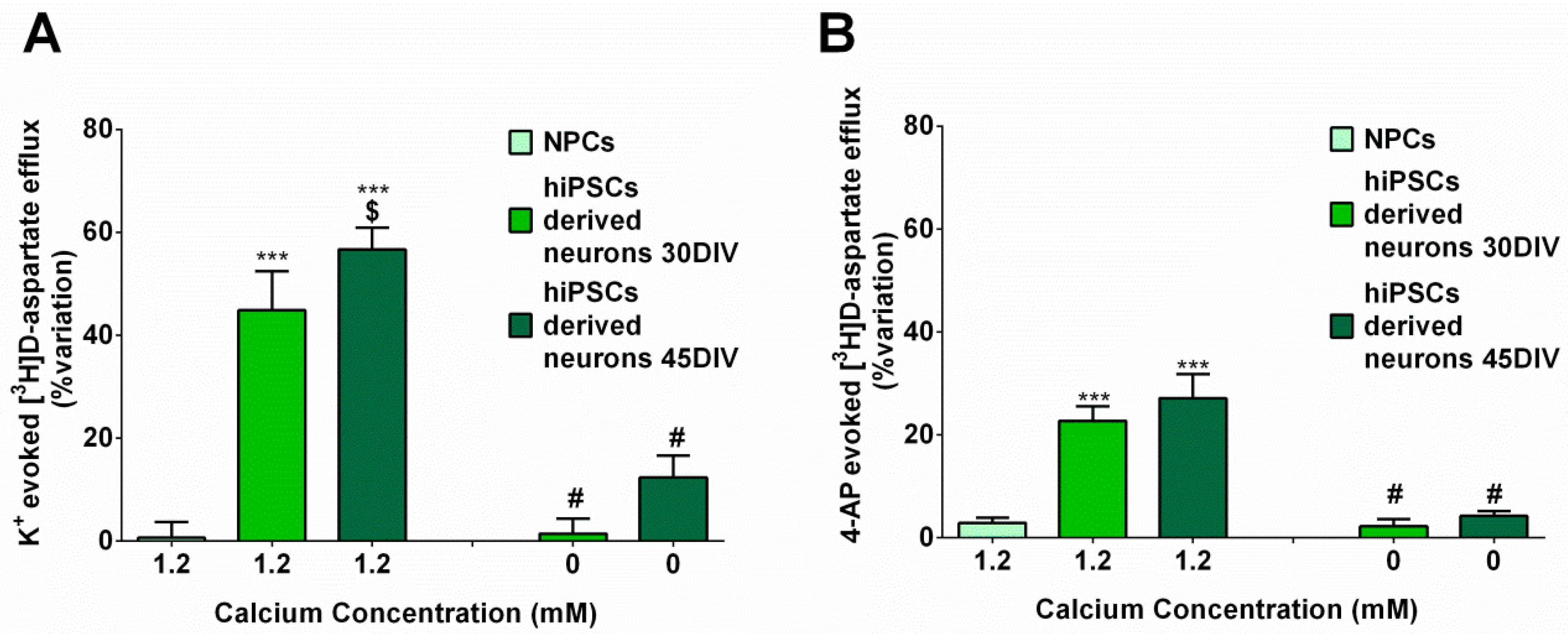
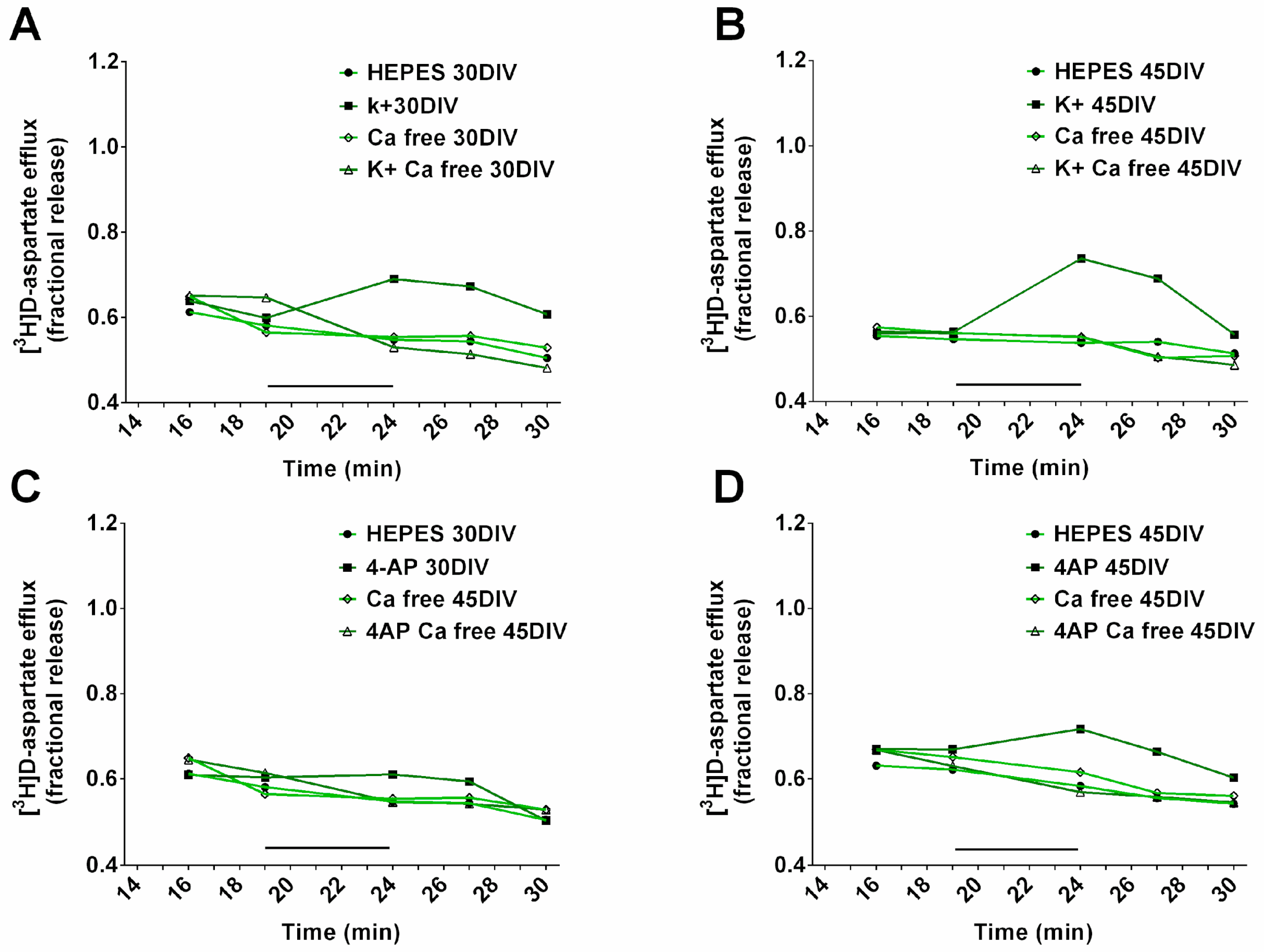
2.6. hiPSC-DerivedNeurons Release Glutamate in Response to Depolarization
3. Discussion
3.1. Feeder-Free hiPSC-DerivedNeurons Express Neuronal Markers and Morphology
3.2. hiPSC-DerivedNeurons Release Glutamate in Response to Depolarization
4. Materials and Methods
4.1. Generation and Maintenance of Human Induced Pluripotent Stem Cells (hiPSCs)
4.2. Array-CGH Assay
4.3. Feeder-Free Differentiation of hiPSC Clones into Neurons
4.4. qRT-PCR
4.5. Western Blot
4.6. Immunocytochemistry
4.7. Assessment of Glutamate Release
- Fx = fractional release in the fraction x
- Tx = tritium content in the fraction x
- Tcell = tritium content in the cells at the end of perfusion
- n = number of the fractions collected during perfusion
- 1 ≤ x ≤ n
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 4-AP | 4-Aminopyridine |
| AUC | Area under the curve |
| BDNF | Brain-Derived Neurotrophic Factor |
| bFGF | basic fibroblast growth factor |
| CACNA1 | Voltage-dependent calcium channels Alpha1 subunit |
| ChR2 | Channelrhodopsin-2 |
| CHRM3 | Cholinergic Receptor Muscarinic 3 |
| CNS | Central Nervous System |
| DAPI | 4′:6-diamidino-2-phenylindole |
| DIV | Days of differentiation in vitro |
| EBs | Embryoid bodies |
| FBS | Foetal bovine serum |
| GAPDH | Glyceraldehyde-3 phosphate dehydrogenase |
| GDNF | Glial cell-derived neurotrophic factor |
| GRIN1 | NMDA Type Subunit 1 |
| hiPSCs | Human induced pluripotent stem cells |
| HTR2A | 5-Hydroxytryptamine Receptor 2A |
| IF | Immunofluorescence |
| KLF4 | Kruppel Like Factor 4 |
| mAChR | muscarinic acetylcholine receptors |
| MAP2 | Microtubule-associated protein 2 |
| NeuN | Neuronal nuclear protein |
| NMDA | N-methyl-D-aspartate receptor |
| NPCs | Neuroprecursors cells |
| OCT4 | Octamer-binding transcription factor 4 |
| PAX6 | Paired box 6 |
| PBS | Phosphate Buffered Saline |
| PCR | Polymerase Chain Reaction |
| PPIA | Peptidylprolyl isomerase A |
| PSD95 | Discs Large MAGUK Scaffold Protein 4 |
| RA | Retinoic acid |
| RBS | Rat Brain Synaptosome |
| RPL13A | Ribosomal Protein L13A |
| SMAD | Small mother against decapentaplegic |
| SNAP25 | Synaptosome Associated Protein 25 |
| SOX1 and 2 | SRY-Box Transcription Factor 1 and 2 |
| SSEA1 | Stage specific embryonic antigen-1 |
| SV2A | Synaptic vesicle glycoprotein 2A |
| SYP | Synaptophysin |
| TGF-b | Transforming Growth Factor-β |
| Tra1-60 | T cell receptor alpha locus1-60 |
| TUBB3 | Tubulin Beta 3 Class III |
| VAMP2 | Vesicle Associated Membrane Protein 2 |
| VGLUT (2 and 3) | Vesicular glutamate transporters (type 2 and 3) |
References
- Bal-Price, A.; Pistollato, F.; Sachana, M.; Bopp, S.K.; Munn, S.; Worth, A. Strategies to improve the regulatory assessment of developmental neurotoxicity (DNT) using in vitro methods. Toxicol. Appl. Pharmacol. 2018, 354, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Krewski, D.; Acosta, D., Jr.; Andersen, M.; Anderson, H.; Bailar, J.C., 3rd; Boekelheide, K.; Brent, R.; Charnley, G.; Cheung, V.G.; Green, S., Jr.; et al. Toxicity testing in the 21st century: A vision and a strategy. J. Toxicol. Environ. Health B Crit. Rev. 2010, 13, 51–138. [Google Scholar] [CrossRef] [PubMed]
- Baumann, J.; Gassmann, K.; Masjosthusmann, S.; DeBoer, D.; Bendt, F.; Giersiefer, S.; Fritsche, E. Comparative human and rat neurospheres reveal species differences in chemical effects on neurodevelopmental key events. Arch. Toxicol. 2016, 90, 1415–1427. [Google Scholar] [CrossRef] [PubMed]
- Pistollato, F.; de Gyves, E.M.; Carpi, D.; Bopp, S.K.; Nunes, C.; Worth, A.; Bal-Price, A. Assessment of developmental neurotoxicity induced by chemical mixtures using an adverse outcome pathway concept. Environ. Health 2020, 19, 23. [Google Scholar] [CrossRef]
- Consiglio, E.D.; Pistollato, F.; Gyves, E.M.-D.; Bal-Price, A.; Testai, E. Integrating biokinetics and in vitro studies to evaluate developmental neurotoxicity induced by chlorpyrifos in human iPSC-derived neural stem cells undergoing differentiation towards neuronal and glial cells. Reprod. Toxicol. 2020, 98, 174–188. [Google Scholar] [CrossRef]
- Pistollato, F.; Carpi, D.; Gyves, E.M.-D.; Paini, A.; Bopp, S.K.; Worth, A.; Bal-Price, A. Combining in vitro assays and mathematical modelling to study developmental neurotoxicity induced by chemical mixtures. Reprod. Toxicol. 2021, 105, 101–119. [Google Scholar] [CrossRef]
- Mattis, V.B.; Tom, C.; Akimov, S.; Saeedian, J.; Østergaard, M.E.; Southwell, A.L.; Doty, C.N.; Ornelas, L.; Sahabian, A.; Lenaeus, L.; et al. HD iPSC-derived neural progenitors accumulate in culture and are susceptible to BDNF withdrawal due to glutamate toxicity. Hum. Mol. Genet. 2015, 24, 3257–3271. [Google Scholar] [CrossRef]
- Mertens, J.; Reid, D.; Lau, S.; Kim, Y.; Gage, F.H. Aging in a Dish: iPSC-Derived and Directly Induced Neurons for Studying Brain Aging and Age-Related Neurodegenerative Diseases. Annu. Rev. Genet. 2018, 52, 271–293. [Google Scholar] [CrossRef]
- Vasan, L.; Park, E.; David, L.A.; Fleming, T.; Schuurmans, C. Direct Neuronal Reprogramming: Bridging the Gap Between Basic Science and Clinical Application. Front. Cell Dev. Biol. 2021, 9, 681087. [Google Scholar] [CrossRef]
- Bonaventura, G.; Iemmolo, R.; Attaguile, G.A.; Cognata, V.L.; Pistone, B.S.; Raudino, G.; D’Agata, V.; Cantarella, G.; Barcellona, M.L.; Cavallaro, S. iPSCs: A Preclinical Drug Research Tool for Neurological Disorders. Int. J. Mol. Sci. 2021, 22, 4596. [Google Scholar] [CrossRef]
- Grunwald, L.M.; Stock, R.; Haag, K.; Buckenmaier, S.; Eberle, M.C.; Wildgruber, D.; Storchak, H.; Kriebel, M.; Weissgraeber, S.; Mathew, L.; et al. Comparative characterization of human induced pluripotent stem cells (hipsc) derived from patients with schizophrenia and autism. Transl. Psychiatry 2019, 9, 179. [Google Scholar] [CrossRef]
- Brennand, K.J.; Simone, A.; Jou, J.; Gelboin-Burkhart, C.; Tran, N.; Sangar, S.; Li, Y.; Mu, Y.; Chen, G.; Yu, D.; et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature 2011, 473, 221–225. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Jia, X.; Wu, H.; Xun, G.; Ou, J.; Zhang, Q.; Li, H.; Bai, T.; Hu, Z.; Zou, X.; et al. Genotype and phenotype correlations for shank3 de novo mutations in neurodevelopmental disorders. Am. J. Med. Genet. A 2018, 176, 2668–2676. [Google Scholar] [PubMed]
- Page, S.C.; Sripathy, S.R.; Farinelli, F.; Ye, Z.; Wang, Y.; Hiler, D.J.; Pattie, E.A.; Nguyen, C.V.; Tippani, M.; Moses, R.L.; et al. Electrophysiological measures from human iPSC-derived neurons are associated with schizophrenia clinical status and predict individual cognitive performance. Proc. Natl. Acad. Sci. USA 2022, 119, e2109395119. [Google Scholar] [CrossRef] [PubMed]
- Marchetto, M.C.; Belinson, H.; Tian, Y.; Freitas, B.C.; Fu, C.; Vadodaria, K.; Beltrao-Braga, P.; Trujillo, C.A.; Mendes, A.P.D.; Padmanabhan, K.; et al. Altered proliferation and networks in neural cells derived from idiopathic autistic individuals. Mol. Psychiatry 2017, 22, 820–835. [Google Scholar] [CrossRef] [PubMed]
- Heider, J.; Vogel, S.; Volkmer, H.; Breitmeyer, R. Human iPSC-Derived Glia as a Tool for Neuropsychiatric Research and Drug Development. Int. J. Mol. Sci. 2021, 22, 10254. [Google Scholar] [CrossRef] [PubMed]
- Lepski, G.; Maciaczyk, J.; Jannes, C.E.; Maciaczyk, D.; Bischofberger, J.; Nikkhah, G. Delayed Functional Maturation of Human Neuronal Progenitor Cells in Vitro. Mol. Cell Neurosci. 2011, 47, 36–44. [Google Scholar] [CrossRef]
- Telias, M.; Segal, M.; Ben-Yosef, D. Electrical Maturation of Neurons Derived from Human Embryonic Stem Cells. F1000Resarch 2014, 3, 196. [Google Scholar] [CrossRef][Green Version]
- Belinsky, G.S.; Rich, M.T.; Sirois, C.L.; Short, S.M.; Pedrosa, E.; Lachman, H.M.; Antic, S.D. Patch-Clamp Recordings and Calcium Imaging Followed by Single-Cell Pcr Reveal the Developmental Profile of 13 Genes in Ipsc-Derived Human Neurons. Stem Cell Res. 2014, 12, 101–118. [Google Scholar] [CrossRef]
- Tofoli, F.A.; Chien, H.F.; Barbosa, E.R.; Pereira, L.V. Generation of 5 hiPSC lines derived from three unrelated idiopathic Parkinson disease patients and two unrelated healthy control individuals. Stem Cell Res. 2019, 41, 101640. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, K.; Zhou, L.; Gao, X.; Wang, J.; Yao, Y.; He, F.; Luo, Y.; Yu, Y.; Li, S.; et al. Coupled electrophysiological recording and single cell transcriptome analyses revealed molecular mechanisms underlying neuronal maturation. Protein Cell 2016, 7, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Kirwan, P.; Turner-Bridger, B.; Peter, M.; Momoh, A.; Arambepola, D.; Robinson, H.P.; Livesey, F.J. Development and Function of Human Cerebral Cortex Neural Networks from Pluripotent Stem Cells In Vitro. Development 2015, 142, 3178–3187. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, M.A.; Renner, M.; Martin, C.A.; Wenzel, D.; Bicknell, L.S.; Hurles, M.E.; Homfray, T.; Penninger, J.M.; Jackson, A.P.; Knoblich, J.A. Cerebral Organoids Model Human Brain Development and Microcephaly. Nature 2013, 501, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Hartfield, E.M.; Yamasaki-Mann, M.; Fernandes, H.J.R.; Vowles, J.; James, W.S.; Cowley, S.A.; Wade-Martins, R. Physiological Characterisation of Human Ips-Derived Dopaminergic Neurons. PLoS ONE 2014, 9, e87388. [Google Scholar] [CrossRef] [PubMed]
- Antonov, S.A.; Novosadova, E.V.; Kobylyansky, A.G.; Illarioshkin, S.N.; Tarantul, V.Z.; Grivennikov, I.A. Expression and Functional Properties of Nmda and Gaba(a) Receptors During Differentiation of Human Induced Pluripotent Stem Cells into Ventral Mesencephalic Neurons. Biochemistry 2019, 84, 310–320. [Google Scholar] [PubMed]
- Antonov, S.A.; Novosadova, E.V. Current State-of-the-Art and Unresolved Problems in Using Human Induced Pluripotent Stem Cell-Derived Dopamine Neurons for Parkinson’s Disease Drug Development. Int. J. Mol. Sci. 2021, 22, 3381. [Google Scholar] [CrossRef] [PubMed]
- Ruden, J.B.; Dixit, M.; Zepeda, J.C.; Grueter, B.A.; Dugan, L.L. Robust Expression of Functional NMDA Receptors in Human Induced Pluripotent Stem Cell-Derived Neuronal Cultures Using an Accelerated Protocol. Front. Mol. Neurosci. 2021, 14, 777049. [Google Scholar] [CrossRef]
- Cline, H.; Haas, K. The regulation of dendritic arbor development and plasticity by glutamatergic synaptic input: A review of the synaptotrophic hypothesis. J. Physiol. 2008, 586, 1509–1517. [Google Scholar] [CrossRef]
- Gambrill, A.C.; Barria, A. NMDA receptor subunit composition controls synaptogenesis and synapse stabilization. Proc. Natl. Acad. Sci. USA 2011, 108, 5855–5860. [Google Scholar] [CrossRef]
- Fedorova, V.; Vanova, T.; Elrefae, L.; Pospisil, J.; Petrasova, M.; Kolajova, V.; Hudacova, Z.; Baniariova, J.; Barak, M.; Peskova, L.; et al. Differentiation of neural rosettes from human pluripotent stem cells in vitro is sequentially regulated on a molecular level and accomplished by the mechanism reminiscent of secondary neurulation. Stem Cell Res. 2019, 40, 101563. [Google Scholar] [CrossRef]
- Engle, S.J.; Blaha, L.; Kleiman, R.J. Best Practices for Translational Disease Modeling Using Human iPSC-Derived Neurons. Neuron 2018, 100, 783–797. [Google Scholar] [CrossRef] [PubMed]
- Cox, W.G.; Hemmati-Brivanlou, A. Caudalization of neural fate by tissue recombination and bFGF. Development 1995, 121, 4349–4358. [Google Scholar] [CrossRef] [PubMed]
- Blumberg, B.; Bolado, J., Jr.; Moreno, T.A.; Kintner, C.; Evans, R.M.; Papalopulu, N. An essential role for retinoid signaling in anteroposterior neural patterning. Development 1997, 124, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Kiecker, C.; Niehrs, C. A morphogen gradient of Wnt/beta-catenin signalling regulates anteroposterior neural patterning in Xenopus. Development 2001, 128, 4189–4201. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Elkabetz, Y.; Panagiotakos, G.; Shamy, G.A.; Socci, N.D.; Tabar, V.; Studer, L. Human ES cell-derived neural rosettes reveal a functionally distinct early neural stem cell stage. Gene. Dev. 2008, 22, 152–165. [Google Scholar] [CrossRef] [PubMed]
- Wilson, P.G.; Stice, S.S. Development and differentiation of neural rosettes derived from human embryonic stem cells. Stem Cell Rev. 2006, 2, 67–77. [Google Scholar] [CrossRef]
- Zhang, S.C.; Wernig, M.; Duncan, I.D.; Brüstle, O.; Thomson, J.A. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat. Biotechnol. 2001, 19, 1129–1133. [Google Scholar] [CrossRef]
- O’Neill, K.M.; Akum, B.F.; Dhawan, S.T.; Kwon, M.; Langhammer, C.G.; Firestein, B.L. Assessing effects on dendritic arborization using novel Sholl analyses. Front. Cell Neurosci. 2015, 9, 285. [Google Scholar] [CrossRef]
- Gensel, J.C.; Schonberg, D.L.; Alexander, J.K.; McTigue, D.M.; Popovich, P.G. Semi-automated Sholl analysis for quantifying changes in growth and differentiation of neurons and glia. J. Neurosci. Methods 2010, 190, 71–79. [Google Scholar] [CrossRef]
- Bird, A.D.; Cuntz, H. Dissecting Sholl Analysis into Its Functional Components. Cell Rep. 2019, 27, 3081–3096. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wong, T.P.; Aarts, M.; Rooyakkers, A.; Liu, L.; Lai, T.W.; Wu, D.C.; Lu, J.; Tymianski, M.; Craig, A.M.; et al. NMDA receptor subunits have differential roles in mediating excitotoxic neuronal death both in vitro and in vivo. J. Neurosci. 2007, 27, 2846–2857. [Google Scholar] [CrossRef] [PubMed]
- Hunt, D.L.; Castillo, P.E. Synaptic plasticity of NMDA receptors: Mechanisms and functional implications. Curr. Opin. Neurobiol. 2012, 22, 496–508. [Google Scholar] [CrossRef] [PubMed]
- Luscher, C.; Malenka, R.C. NMDA receptor-dependent long-term potentiation and long-term depression (LTP/LTD). Cold Spring Harb. Perspect. Biol. 2012, 4, a0057. [Google Scholar] [CrossRef]
- Chakraborty, A.; Murphy, S.; Coleman, N. The role of NMDA receptors in neural stem cell proliferation and differentiation. Stem Cells Dev. 2017, 26, 798–807. [Google Scholar] [CrossRef]
- Anderson, N.C.; Chen, P.F.; Meganathan, K.; Saber, W.A.; Petersen, A.J.; Bhattacharyya, A.; Kroll, K.L.; Sahin, M. Cross-IDDRC Human Stem Cell Working Group. Balancing serendipity and reproducibility: Pluripotent stem cells as experimental systems for intellectual and developmental disorders. Stem Cell Rep. 2021, 16, 1446–1457. [Google Scholar] [CrossRef]
- Nakagawa, M.; Taniguchi, Y.; Senda, S.; Takizawa, N.; Ichisaka, T.; Asano, K.; Morizane, A.; Doi, D.; Takahashi, J.; Nishizawa, M.; et al. A novel efficient feeder-free culture system for the derivation of human induced pluripotent stem cells. Sci. Rep. 2014, 4, 3594. [Google Scholar] [CrossRef]
- Yuan, F.; Fang, K.H.; Cao, S.Y.; Qu, Z.Y.; Li, Q.; Krencik, R.; Xu, M.; Bhattacharyya, A.; Su, Y.W.; Zhu, D.Y.; et al. Efficient generation of region-specific forebrain neurons from human pluripotent stem cells under highly defined condition. Sci. Rep. 2015, 5, 18550. [Google Scholar] [CrossRef]
- Aprile, D.; Fruscione, F.; Baldassari, S.; Fadda, M.; Ferrante, D.; Falace, A.; Buhler, E.; Sartorelli, J.; Represa, A.; Baldelli, P.; et al. TBC1D24 regulates axonal outgrowth and membrane trafficking at the growth cone in rodent and human neurons. Cell Death Differ. 2019, 26, 2464–2478. [Google Scholar] [CrossRef]
- Anderson, S.; Vanderhaeghen, P. Cortical neurogenesis from pluripotent stem cells: Complexity emerging from simplicity. Curr. Opin. Neurobiol. 2014, 27, 151–157. [Google Scholar] [CrossRef]
- Gaspard, N.; Bouschet, T.; Hourez, R.; Dimidschstein, J.; Naeije, G.; van den Ameele, J.; Espuny-Camacho, I.; Herpoel, A.; Passante, L.; Schiffmann, S.N.; et al. An intrinsic mechanism of corticogenesis from embryonic stem cells. Nature 2008, 455, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Barak, M.; Fedorova, V.; Pospisilova, V.; Raska, J.; Vochyanova, S.; Sedmik, J.; Hribkova, H.; Klimova, H.; Vanova, T.; Bohaciakova, D. Human iPSC-Derived Neural Models for Studying Alzheimer’s Disease: From Neural Stem Cells to Cerebral Organoids. Stem Cell Rev. Rep. 2022, 18, 792–820. [Google Scholar] [CrossRef] [PubMed]
- Koch, P.; Opitz, T.; Steinbeck, J.A.; Ladewig, J.; Brustle, O. A rosette-type, self-renewing human ES cell-derived neural stem cell with potential for in vitro instruction and synaptic integration. Proc. Natl. Acad. Sci. USA 2009, 106, 3225–3230. [Google Scholar] [CrossRef] [PubMed]
- Grabiec, M.; Hříbková, H.; Vařecha, M.; Střítecká, D.; Hampl, A.; Dvořák, P.; Sun, Y.M. Stage-specific roles of FGF2 signaling in human neural development. Stem Cell Res. 2016, 17, 330–341. [Google Scholar] [CrossRef] [PubMed]
- Bohaciakova, D.; Hruska-Plochan, M.; Tsunemoto, R.; Gifford, W.D.; Driscoll, S.P.; Glenn, T.D.; Wu, S.; Marsala, S.; Navarro, M.; Tadokoro, T.; et al. A scalable solution for isolating human multipotent clinical-grade neural stem cells from ES precursors. Stem Cell Res. Ther. 2019, 10, 83. [Google Scholar] [CrossRef]
- Kim, Y.H.; Chung, J.-I.; Woo, H.G.; Jung, Y.S.; Lee, S.H.; Moon, C.-H.; Suh-Kim, H.; and Baik, E.J. Differential regulation of proliferation and differentiation in neural precursor cells by the Jak pathway. Stem Cells 2010, 28, 1816–1828. [Google Scholar] [CrossRef]
- Yin, X.; Xu, J.C.; Cho, G.S.; Kwon, C.; Dawson, T.M.; Dawson, V.L. Neurons Derived from Human Induced Pluripotent Stem Cells Integrate into Rat Brain Circuits and Maintain Both Excitatory and Inhibitory Synaptic Activities. eNeuro 2019, 6, 0148-19. [Google Scholar] [CrossRef]
- Fremeau, R.T., Jr.; Voglmaier, S.; Seal, R.P.; Edwards, R.H. VGLUTs define subsets of ex-citatory neurons and suggest novel roles for glutamate. Trends Neurosci. 2004, 27, 98–103. [Google Scholar] [CrossRef]
- D’Aiuto, L.; Zhi, Y.; Kumar Das, D.; Wilcox, M.R.; Johnson, J.W.; McClain, L.; MacDon-ald, M.L.; Di Maio, R.; Schurdak, M.E.; Piazza, P.; et al. Large-scale generation of human iPSC-derived neural stem cells/early neural progenitor cells and their neuronal differentiation. Organogenesis 2014, 10, 365–377. [Google Scholar] [CrossRef]
- Togo, K.; Fukusumi, H.; Shofuda, T.; Ohnishi, H.; Yamazaki, H.; Hayashi, M.K.; Kawasaki, N.; Takei, N.; Nakazawa, T.; Saito, Y.; et al. Postsynaptic structure formation of human iPS cell-derived neurons takes longer than presynaptic formation during neural differentiation in vitro. Mol. Brain 2021, 14, 149. [Google Scholar] [CrossRef]
- Setien, M.B.; Smith, K.R.; Howard, K.; Williams, K.; Suhr, S.T.; Purcell, E.K. Differentia-tion and characterization of neurons derived from rat iPSCs. J. Neurosci. Methods 2020, 338, 108693. [Google Scholar] [CrossRef] [PubMed]
- Dolphin, A.C. Functions of Presynaptic Voltage-gated Calcium Channels. Function 2021, 2, zqaa027. [Google Scholar] [CrossRef] [PubMed]
- Resende, R.R.; Adhikari, A. Cholinergic receptor pathways involved in apoptosis, cell proliferation and neuronal differentiation. Cell Commun. Signal 2009, 7, 20. [Google Scholar] [CrossRef] [PubMed]
- Abreu-Villaça, Y.; Filgueiras, C.C.; Manhães, A.C. Developmental aspects of the cholinergic system. Behav. Brain Res. 2011, 221, 367–378. [Google Scholar] [CrossRef]
- Azmitia, E.C. Modern views on an ancient chemical: Serotonin effects on cell proliferation, maturation, and apoptosis. Brain Res. Bull. 2001, 56, 413–424. [Google Scholar] [CrossRef]
- Xing, L.; Kalebic, N.; Namba, T.; Vaid, S.; Wimberger, P.; Huttner, W. Serotonin receptor 2A activation promotes evolutionarily relevant basal progenitor proliferation in the developing neocortex. Neuron 2020, 108, 1113–1129. [Google Scholar] [CrossRef]
- Weber, E.T.; Andrade, R. Htr2a Gene and 5-HT(2A) Receptor Expression in the Cerebral Cortex Studied Using Genetically Modified Mice. Front. Neurosci. 2010, 4, 36. [Google Scholar]
- Tu, J.C.; Xiao, B.; Naisbitt, S.; Yuan, J.P.; Petralia, R.S.; Brakeman, P.; Doan, A.; Aakalu, V.K.; Lanahan, A.A.; Sheng, M.; et al. Coupling of mGluR/Homer and PSD-95 complexes by the Shank family of postsynaptic density proteins. Neuron 1999, 23, 583–592. [Google Scholar] [CrossRef]
- Cervetto, C.; Maura, G.; Marcoli, M. Inhibition of presynaptic release-facilitatory kainate autoreceptors by extracellular cyclic GMP. J. Pharmacol. Exp. Ther. 2010, 332, 210–219. [Google Scholar] [CrossRef]
- Caiazzo, M.; Dell’Anno, M.T.; Dvoretskova, E.; Lazarevic, D.; Taverna, S.; Leo, D.; Sotnikova, T.D.; Menegon, A.; Roncaglia, P.; Colciago, G.; et al. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature 2011, 476, 224–227. [Google Scholar] [CrossRef]
- Hook, V.; Brennand, K.J.; Kim, Y.; Toneff, T.; Funkelstein, L.; Lee, K.C.; Ziegler, M.; Gage, F.H. Human iPSC neurons display activity-dependent neurotransmitter secretion: Aberrant catecholamine levels in schizophrenia neurons. Stem Cell Rep. 2014, 3, 531–538. [Google Scholar] [CrossRef] [PubMed]
- Ohta, E.; Nihira, T.; Uchino, A.; Imaizumi, Y.; Okada, Y.; Akamatsu, W.; Takahashi, K.; Hayakawa, H.; Nagai, M.; Ohyama, M.; et al. I2020T mutant LRRK2 iPSC-derived neurons in the Sagamihara family exhibit increased Tau phosphorylation through the AKT/GSK-3β signaling pathway. Hum. Mol. Genet. 2015, 24, 4879–4900. [Google Scholar] [CrossRef] [PubMed]
- Han, F.; Liu, Y.; Huang, J.; Zhang, X.; Wei, C. Current Approaches and Molecular Mechanisms for Directly Reprogramming Fibroblasts Into Neurons and Dopamine Neurons. Front. Aging Neurosci. 2021, 13, 738529. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Chen, R.; Wu, X.; Zhao, Y.; Fan, Y.; Xiao, Z.; Han, J.; Sun, L.; Wang, X.; Dai, J. Rapid and Efficient Conversion of Human Fibroblasts into Functional Neurons by Small Molecules. Stem Cell Rep. 2019, 3, 862–876. [Google Scholar] [CrossRef] [PubMed]
- Autar, K.; Guo, X.; Rumsey, J.W.; Long, C.J.; Akanda, N.; Jackson, M.; Narasimhan, N.S.; Caneus, J.; Morgan, D.; Hickman, J.J. A functional hiPSC-cortical neuron differentiation and maturation model and its application to neurological disorders. Stem Cell Rep. 2022, 17, 96–109. [Google Scholar] [CrossRef]
- Wen, Z.; Nguyen, H.N.; Guo, Z.; Lalli, M.A.; Wang, X.; Su, Y.; Kim, N.S.; Yoon, K.J.; Shin, J.; Zhang, C.; et al. Synaptic dysregulation in a human iPS cell model of mental disorders. Nature 2014, 515, 414–418. [Google Scholar] [CrossRef]
- Cao, S.Y.; Hu, Y.; Chen, C.; Yuan, F.; Xu, M.; Li, Q.; Fang, K.H.; Chen, Y.; Liu, Y. Enhanced derivation of human pluripotent stem cell-derived cortical glutamatergic neurons by a small molecule. Sci. Rep. 2017, 7, 3282. [Google Scholar] [CrossRef]
- Dong, Y.; Xiong, M.; Chen, Y.; Tao, Y.; Li, X.; Bhattacharyya, A.; Zhang, S.C. Plasticity of Synaptic Transmission in Human Stem Cell-Derived Neural Networks. iScience 2020, 23, 100829. [Google Scholar] [CrossRef]
- Meldrum, B.S. Glutamate as a neurotransmitter in the brain: Review of physiology and pathology. J. Nutr. 2000, 130, 1007S–1015S. [Google Scholar] [CrossRef]
- Cheung, G.; Bataveljic, D.; Visser, J.; Kumar, N.; Moulard, J.; Dallérac, G.; Mozheiko, D.; Rollenhagen, A.; Ezan, P.; Mongin, C.; et al. Physiological synaptic activity and recognition memory require astroglial glutamine. Nat. Commun. 2022, 13, 753. [Google Scholar] [CrossRef]
- Lewerenz, J.; Maher, P. Chronic Glutamate Toxicity in Neurodegenerative Diseases-What is the Evidence? Front. Neurosci. 2015, 9, 469. [Google Scholar] [CrossRef] [PubMed]
- Bal-Price, A.; Meek, M.E. Adverse outcome pathways: Application to enhance mechanistic understanding of neurotoxicity. Pharmacol. Ther. 2017, 179, 84–95. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y. Induced Pluripotent Stem Cell-Derived Human Glutamatergic Neurons as a Platform for Mechanistic Assessment of Inducible Excitotoxicity in Drug Discovery. In Neurotoxins; McDuffie, J.E., Ed.; IntechOpen: London, UK, 2018. [Google Scholar]
- Shirakawa, T.; Suzuki, I. Approach to Neurotoxicity using Human iPSC Neurons: Consortium for Safety Assessment using Human iPS Cells. Curr. Pharm. Biotechnol. 2020, 21, 780–786. [Google Scholar] [CrossRef]
- Qian, L.; TCW, J. Human iPSC-Based Modeling of Central Nerve System Disorders for Drug Discovery. Int. J. Mol. Sci. 2021, 22, 1203. [Google Scholar] [CrossRef]
- Eriksen, J.; Li, F.; Edwards, R.H. The mechanism and regulation of vesicular glutamate transport: Coordination with the synaptic vesicle cycle. Biochim. Biophys. Acta BBA Biomembr. 2020, 1862, 183259. [Google Scholar] [CrossRef] [PubMed]
- Phillis, J.W.; Ren, J.; O’Regan, M.H. Transporter reversal as a mechanism of glutamate release from the ischemic rat cerebral cortex: Studies with DL-threo-beta-benzyloxyaspartate. Brain Res. 2000, 868, 105–112. [Google Scholar] [CrossRef]
- Bridges, R.J.; Natale, N.R.; Patel, S.A. System xc− cystine/glutamate antiporter: An update on molecular pharmacology and roles within the CNS. Br. J. Pharmacol. 2012, 165, 20–34. [Google Scholar] [CrossRef]
- Cervetto, C.; Alloisio, S.; Frattaroli, D.; Mazzotta, M.C.; Milanese, M.; Gavazzo, P.; Passalacqua, M.; Nobile, M.; Maura, G.; Marcoli, M. The P2X7 receptor as a route for non-exocytotic glutamate release: Dependence on the carboxyl tail. J. Neurochem. 2013, 124, 821–831. [Google Scholar] [CrossRef]
- Mannelli, L.D.C.; Marcoli, M.; Micheli, L.; Zanardelli, M.; Maura, G.; Ghelardini, C.; Cervetto, C. Oxaliplatin evokes P2X7-dependent glutamate release in the cerebral cortex: A pain mechanism mediated by Pannexin 1. Neuropharmacology 2015, 97, 133–141. [Google Scholar] [CrossRef]
- Naito, S.; Ueda, T. Characterization of glutamate uptake into synaptic vesicles. J. Neurochem. 1985, 44, 99–109. [Google Scholar] [CrossRef]
- Nicholls, D.; Attwell, D. The release and uptake of excitatory amino acids. Trends Pharmacol. Sci. 1990, 11, 462–468. [Google Scholar] [CrossRef]
- Gundersen, V.; Chaudhry, F.A.; Bjaalie, J.G.; Fonnum, F.; Ottersen, O.P.; Storm-Mathisen, J. Synaptic vesicular localization and exocytosis of L-aspartate in excitatory nerve terminals: A quantitative immunogold analysis in rat hippocampus. J. Neurosci. 1998, 18, 6059–6070. [Google Scholar] [CrossRef] [PubMed]
- Morland, C.; Nordengen, K.; Larsson, M.; Prolo, L.M.; Farzampour, Z.; Reimer, R.J.; Gundersen, V. Vesicular uptake and exocytosis of L-aspartate is independent of sialin. FASEB J. 2013, 27, 1264–1274. [Google Scholar] [CrossRef] [PubMed]
- Herring, B.E.; Silm, K.; Edwards, R.H.; Nicoll, R.A. Is aspartate an excitatory neurotransmitter? J. Neurosci. 2015, 35, 10168–10171. [Google Scholar] [CrossRef]
- Gras, C.; Amilhon, B.; Lepicard, E.M.; Poirel, O.; Vinatier, J.; Herbin, M.; Dumas, S.; Tzavara, E.T.; Wade, M.R.; Nomikos, G.G.; et al. The vesicular glutamate transporter VGLUT3 synergizes striatal acetylcholine tone. Nat. Neurosci. 2008, 11, 292–300. [Google Scholar] [CrossRef] [PubMed]
- Amilhon, B.; Lepicard, E.; Renoir, T.; Mongeau, R.; Popa, D.; Poirel, O.; Miot, S.; Gras, C.; Gardier, A.M.; Gallego, J.; et al. VGLUT3 (vesicular glutamate transporter type 3) contribution to the regulation of serotonergic transmission and anxiety. J. Neurosci. 2010, 30, 2198–2210. [Google Scholar] [CrossRef] [PubMed]
- Vigneault, É.; Poirel, O.; Riad, M.; Prud’homme, J.; Dumas, S.; Turecki, G.; Fasano, C.; Mechawar, N.; El Mestikawy, S. Distribution of vesicular glutamate transporters in the human brain. Front. Neuroanat. 2015, 9, 23. [Google Scholar] [CrossRef]
- Stensrud, M.J.; Sogn, C.J.; Gundersen, V. Immunogold characteristics of VGLUT3-positive GABAergic nerve terminals suggest corelease of glutamate. J. Comp. Neurol. 2015, 523, 2698–2713. [Google Scholar] [CrossRef]
- Gajera, C.R.; Fernandez, R.; Postupna, N.; Montine, K.S.; Fox, E.J.; Tebaykin, D.; Angelo, M.; Bendall, S.C.; Keene, C.D.; Montine, T.J. Mass synaptometry: High-dimensional multi parametric assay for single synapses. J. Neurosci. Methods 2019, 312, 73–83. [Google Scholar] [CrossRef]
- Gajera, C.R.; Fernandez, R.; Postupna, N.; Montine, K.S.; Keene, C.D.; Bendall, S.C.; Montine, T.J. Mass Synaptometry: Applying Mass Cytometry to Single Synapse Analysis. Methods Mol. Biol. 2022, 2417, 69–88. [Google Scholar]
- Amaroli, A.; Marcoli, M.; Venturini, A.; Passalacqua, M.; Agnati, L.F.; Signore, A.; Raffetto, M.; Maura, G.; Benedicenti, S.; Cervetto, C. Near-infrared laser photons induce glutamate release from cerebrocortical nerve terminals. J. Biophotonics 2018, 11, e201800102. [Google Scholar] [CrossRef] [PubMed]
- Cervetto, C.; Vergani, L.; Passalacqua, M.; Ragazzoni, M.; Venturini, A.; Cecconi, F.; Berretta, N.; Mercuri, N.; D’Amelio, M.; Maura, G.; et al. Astrocyte-Dependent Vulnerability to Excitotoxicity in Spermine Oxidase-Overexpressing Mouse. Neuromol. Med. 2016, 18, 50–68. [Google Scholar] [CrossRef]
- Phongpreecha, T.; Gajera, C.R.; Liu, C.C.; Vijayaragavan, K.; Chang, A.L.; Becker, M.; Fallahzadeh, R.; Fernandez, R.; Postupna, N.; Sherfield, E.; et al. Single-synapse analyses of Alzheimer’s disease implicate pathologic tau, DJ1, CD47, and ApoE. Sci. Adv. 2021, 7, eabk0473. [Google Scholar] [CrossRef]
- Zhang, X.; Hu, D.; Shang, Y.; Qi, X. Using induced pluripotent stem cell neuronal models to study neurodegenerative diseases. Biochim. Biophys. Acta BBA Mol. Basis Dis. 2020, 1866, 165431. [Google Scholar] [CrossRef] [PubMed]
- Fruscione, F.; Valente, P.; Sterlini, B.; Romei, A.; Baldassari, S.; Fadda, M.; Prestigio, C.; Giansante, G.; Sartorelli, J.; Rossi, P.; et al. PRRT2 controls neuronal excitability by negatively modulating Na+ channel 1.2/1.6 activity. Brain 2018, 141, 1000–1016. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Schmuck, M.R.; Keil, K.P.; Sethi, S.; Morgan, R.K.; Lein, P.J. Automated high content image analysis of dendritic arborization in primary mouse hippocampal and rat cortical neurons in culture. J. Neurosci. Methods 2020, 341, 108793. [Google Scholar] [CrossRef] [PubMed]
- Benfenati, V.; Caprini, M.; Nicchia, G.P.; Rossi., A.; Dovizio, M.; Cervetto, C.; Nobile, M.; Ferroni, S. Carbenoxolone inhibits volume-regulated anion conductance in cultured rat cortical astroglia. Channels 2009, 3, 323–336. [Google Scholar] [CrossRef]
- Pietropaoli, S.; Leonetti, A.; Cervetto, C.; Venturini, A.; Mastrantonio, R.; Baroli, G.; Persichini, T.; Colasanti, M.; Maura, G.; Marcoli, M.; et al. Glutamate Excitotoxicity Linked to Spermine Oxidase Overexpression. Mol. Neurobiol. 2018, 55, 7259–7270. [Google Scholar] [CrossRef]
- Marcoli, M.; Cervetto, C.; Paluzzi, P.; Guarnieri, S.; Raiteri, M.; Maura, G. Nitric oxide-evoked glutamate release and cGMP production in cerebellar slices: Control by presynaptic 5-HT1D receptors. Neurochem. Int. 2006, 49, 12–19. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baldassari, S.; Cervetto, C.; Amato, S.; Fruscione, F.; Balagura, G.; Pelassa, S.; Musante, I.; Iacomino, M.; Traverso, M.; Corradi, A.; et al. Vesicular Glutamate Release from Feeder-FreehiPSC-Derived Neurons. Int. J. Mol. Sci. 2022, 23, 10545. https://doi.org/10.3390/ijms231810545
Baldassari S, Cervetto C, Amato S, Fruscione F, Balagura G, Pelassa S, Musante I, Iacomino M, Traverso M, Corradi A, et al. Vesicular Glutamate Release from Feeder-FreehiPSC-Derived Neurons. International Journal of Molecular Sciences. 2022; 23(18):10545. https://doi.org/10.3390/ijms231810545
Chicago/Turabian StyleBaldassari, Simona, Chiara Cervetto, Sarah Amato, Floriana Fruscione, Ganna Balagura, Simone Pelassa, Ilaria Musante, Michele Iacomino, Monica Traverso, Anna Corradi, and et al. 2022. "Vesicular Glutamate Release from Feeder-FreehiPSC-Derived Neurons" International Journal of Molecular Sciences 23, no. 18: 10545. https://doi.org/10.3390/ijms231810545
APA StyleBaldassari, S., Cervetto, C., Amato, S., Fruscione, F., Balagura, G., Pelassa, S., Musante, I., Iacomino, M., Traverso, M., Corradi, A., Scudieri, P., Maura, G., Marcoli, M., & Zara, F. (2022). Vesicular Glutamate Release from Feeder-FreehiPSC-Derived Neurons. International Journal of Molecular Sciences, 23(18), 10545. https://doi.org/10.3390/ijms231810545







