Transcriptome Analysis Reveals an Essential Role of Exogenous Brassinolide on the Alkaloid Biosynthesis Pathway in Pinellia Ternata
Abstract
:1. Introduction
2. Results
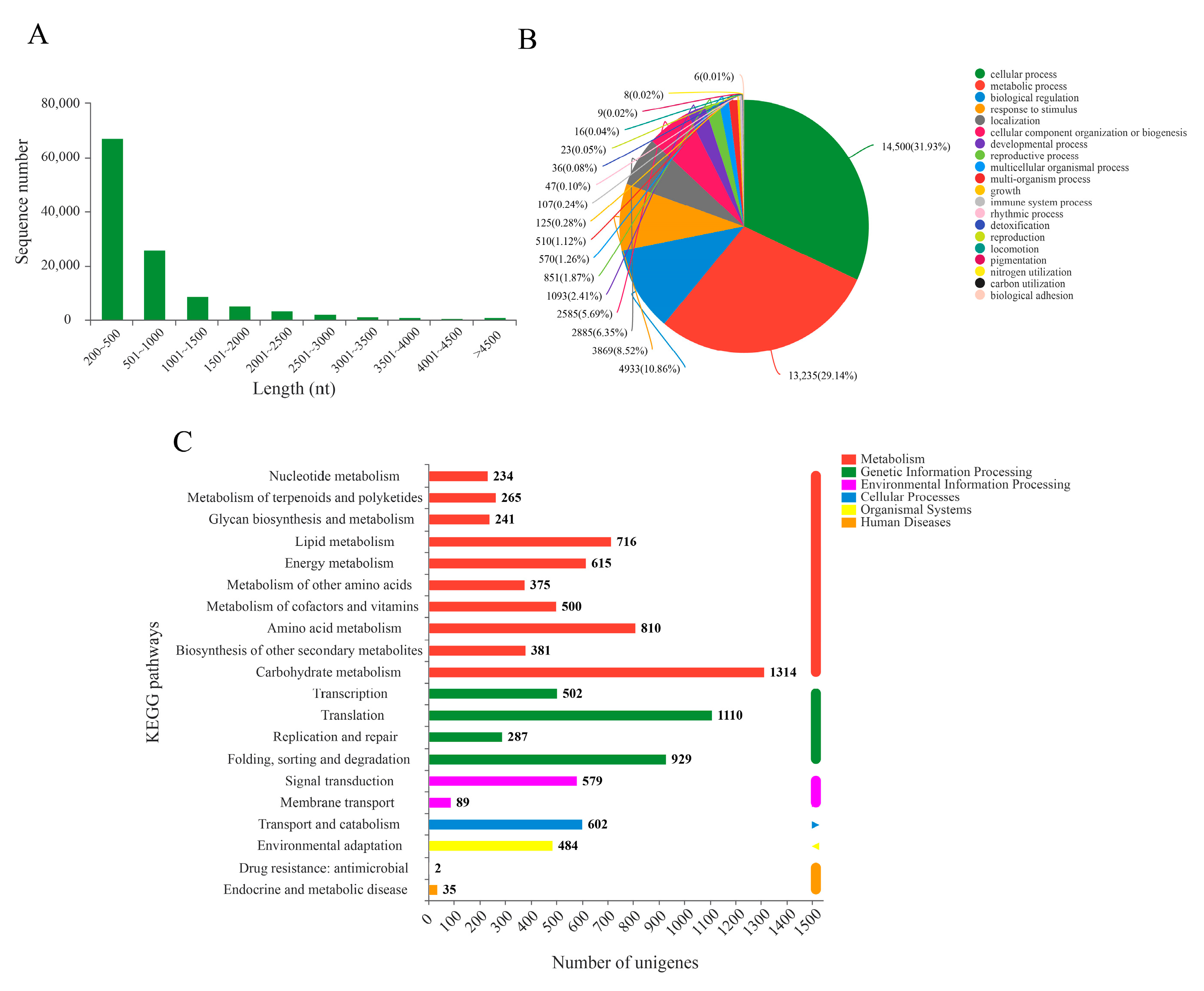
2.1. RNA-Seq Sequencing Analysis
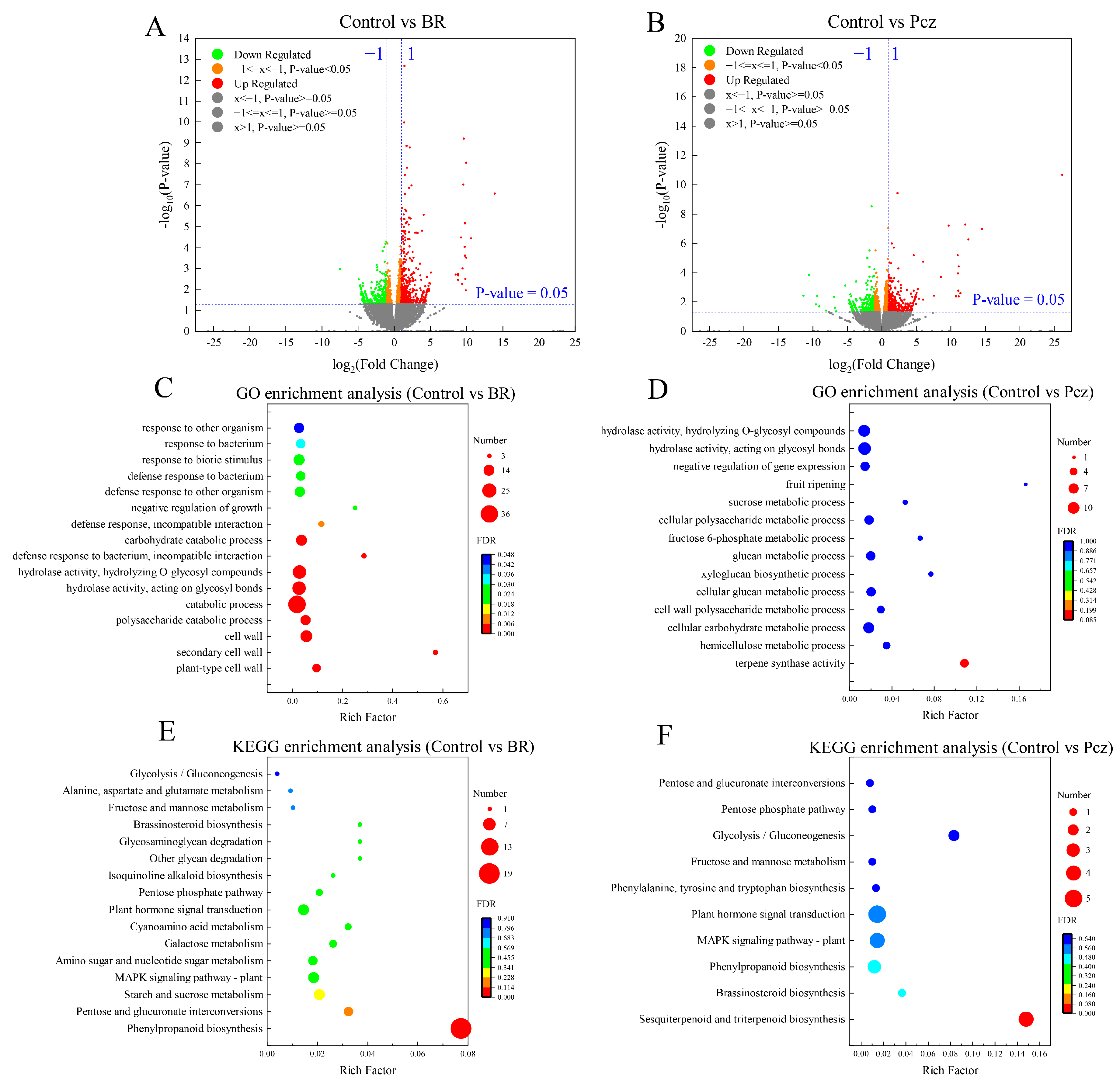
2.2. Comparative Analysis of DEGs
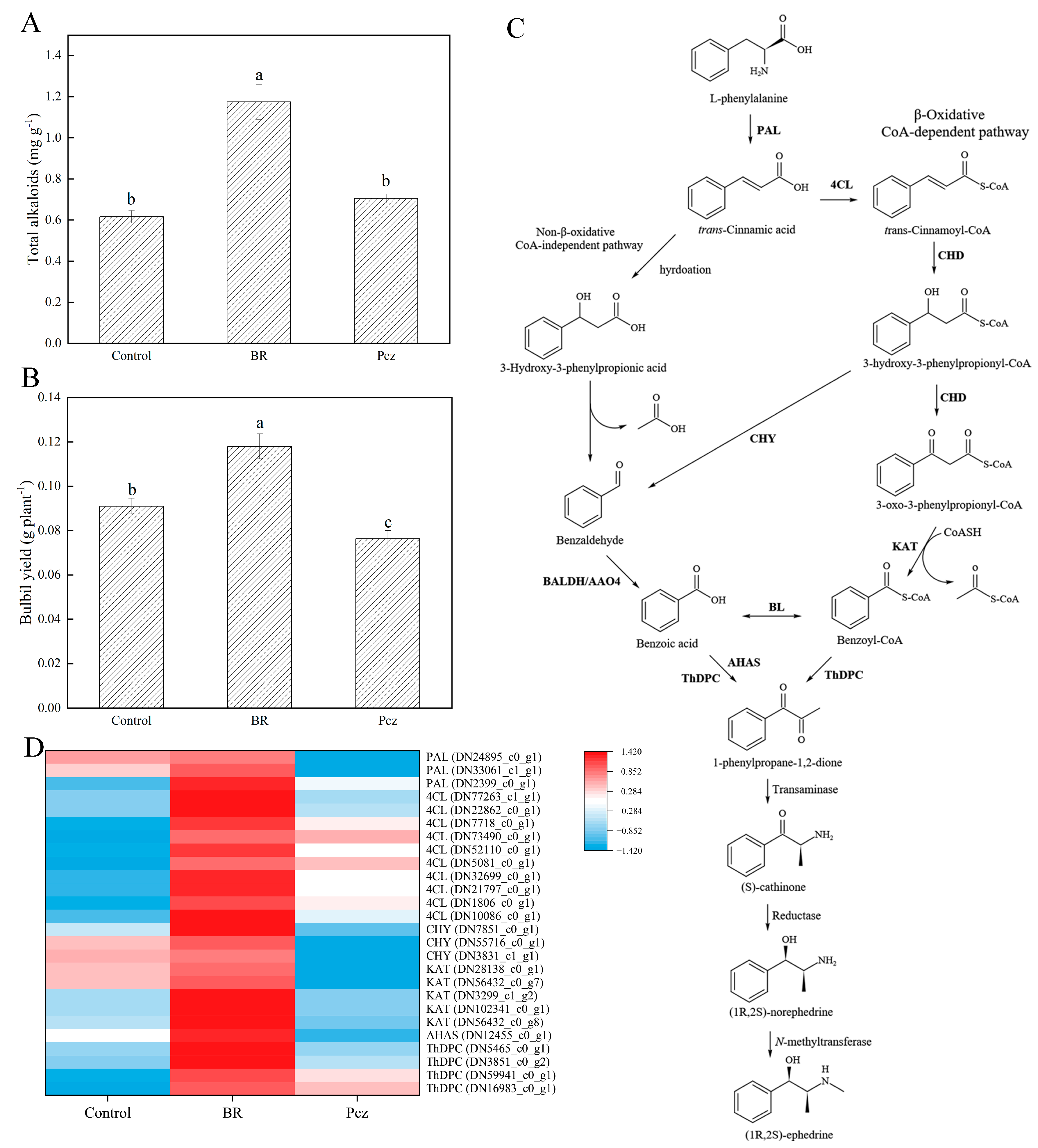
2.3. BR Treatment Improves Ephedrine Biosynthesis
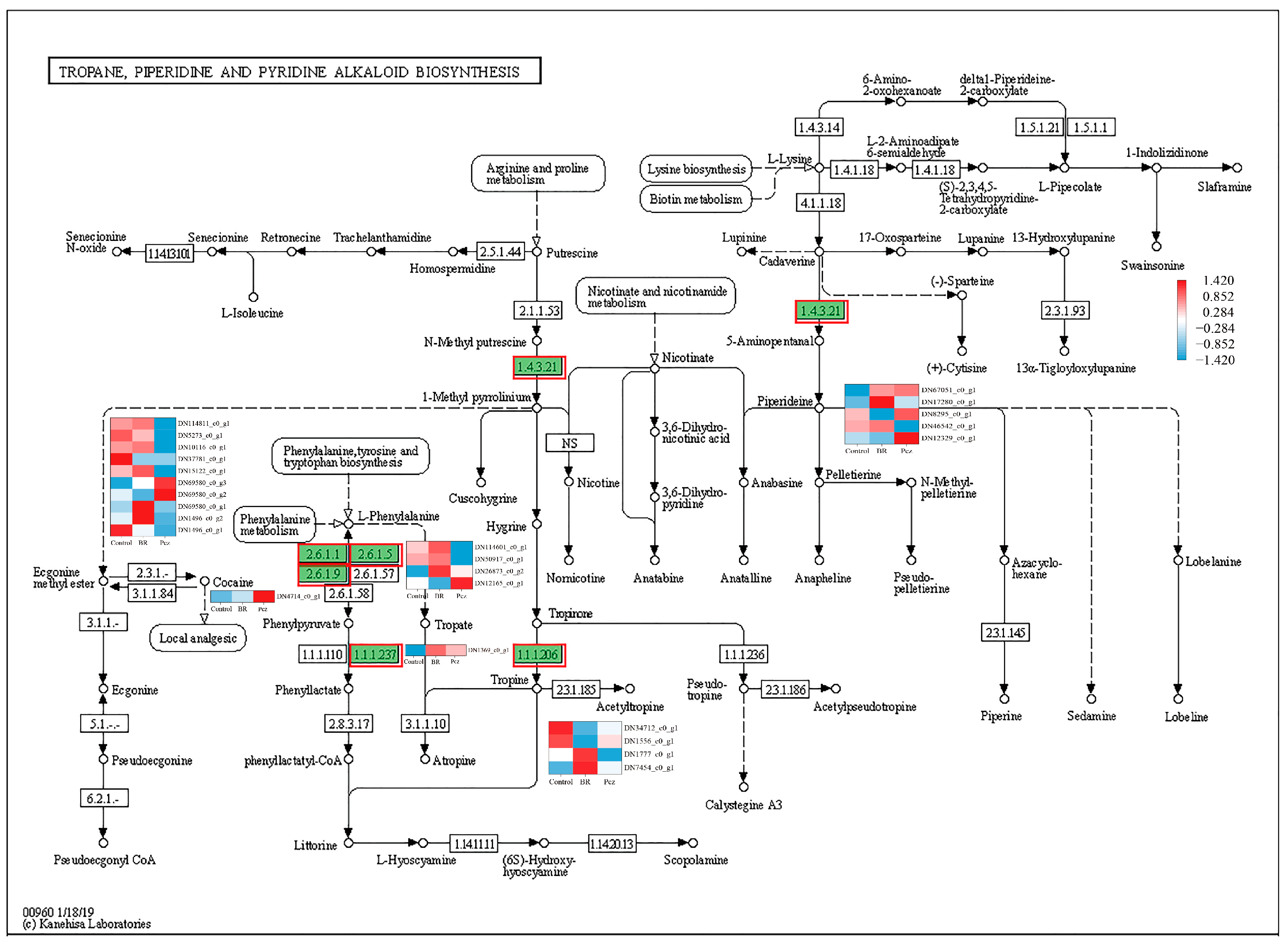
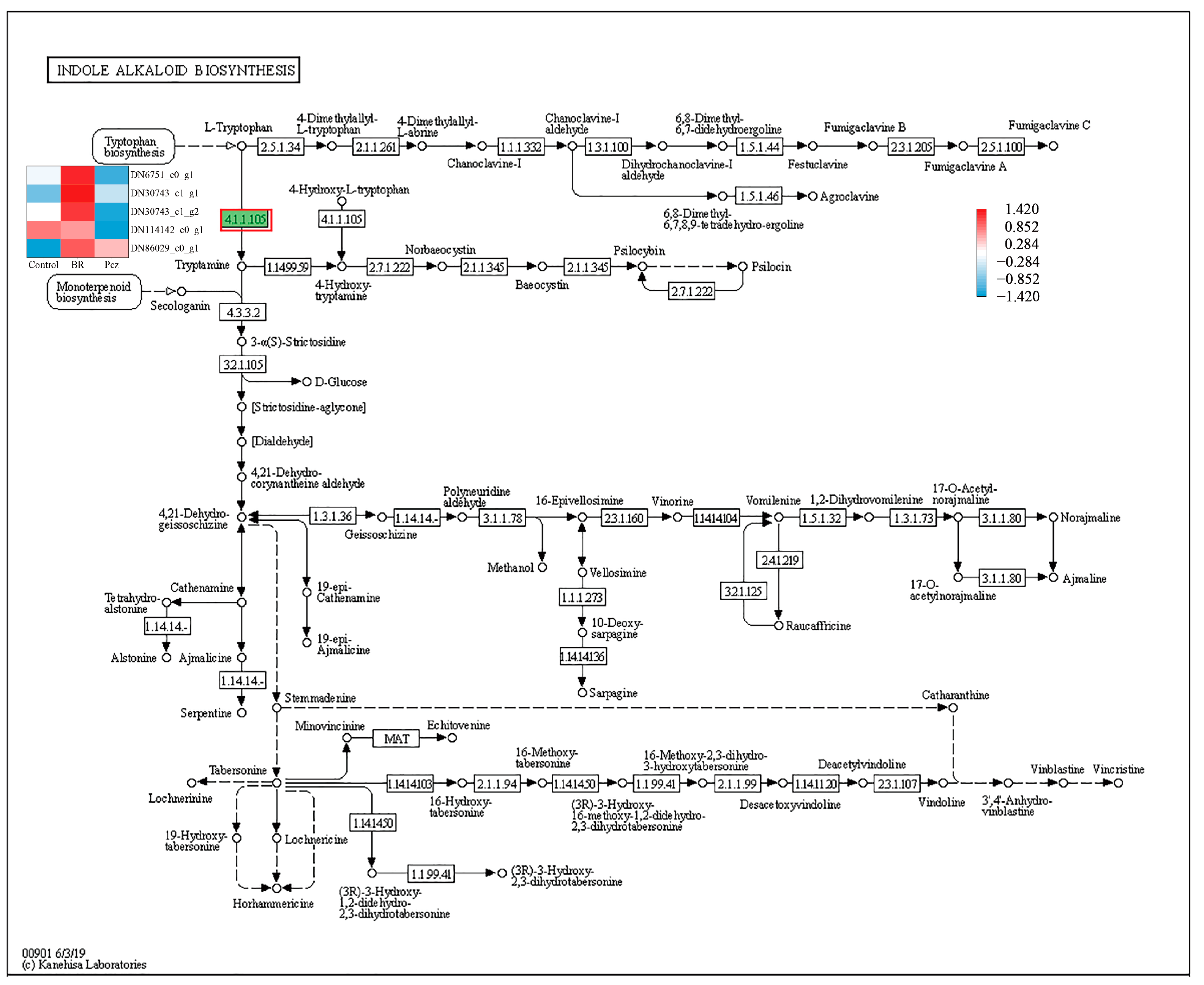
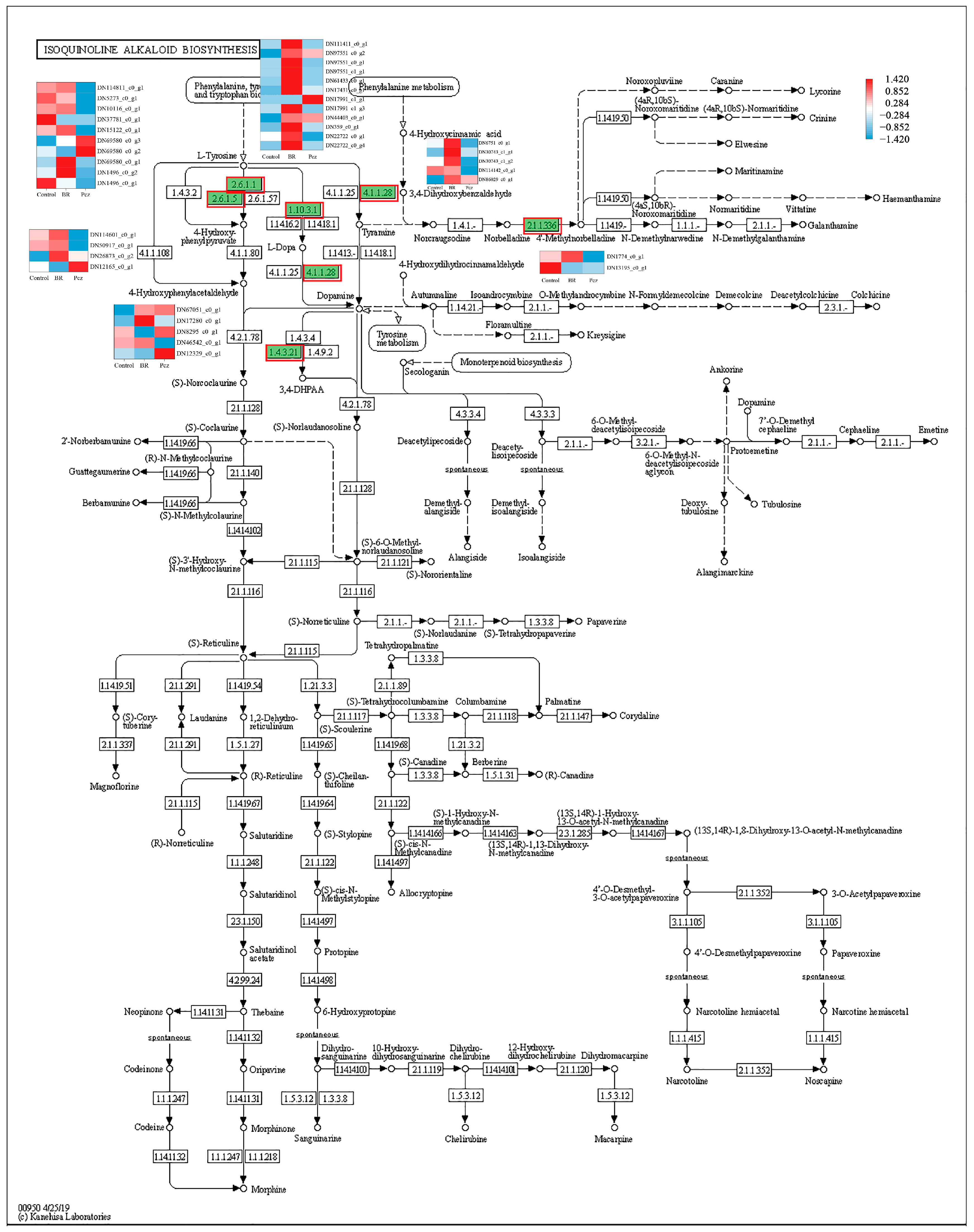
2.4. KEGG-Annotated Genes Involved in Alkaloid Biosynthetic Pathways
2.5. qRT-PCR Validation of the Sequencing Data
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Bulbil Yield and Total Alkaloid Content
4.3. Total RNA Isolation, cDNA Library Construction, and Sequencing
4.4. Transcriptome Assembly and Functional Annotation
4.5. Differentially Expressed Analysis
4.6. qRT-PCR Analysis
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ji, X.; Huang, B.; Wang, G.; Zhang, C. The ethnobotanical, phytochemical and pharmacological profile of the genus Pinellia. Fitoterapia 2014, 93, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.H.; Jiang, N.H.; Song, W.L.; Ma, C.H.; Yang, S.C.; Chen, J.W. De novo sequencing and transcriptome analysis of Pinellia ternata identify the candidate genes involved in the biosynthesis of benzoic acid and ephedrine. Front. Plant Sci. 2016, 7, 1209. [Google Scholar] [CrossRef] [PubMed]
- Xue, T.; Zhang, H.; Zhang, Y.; Wei, S.; Chao, Q.; Zhu, Y.; Teng, J.; Zhang, A.; Sheng, W.; Duan, Y.; et al. Full-length transcriptome analysis of shade-induced promotion of tuber production in Pinellia ternata. BMC Plant Biol. 2019, 19, 565. [Google Scholar] [CrossRef]
- Fang, C.; Huiling, D.; Yi, Z. Growth inhibition of human hepatocarcinoma cell strain Bel-7402 by Pinellia total alkaloids. China Pharm. 2011, 14, 1449–1451. [Google Scholar]
- Oshio, H.; Tsukui, M.; Matsuoka, T. Isolation of l-ephedrine from “pinelliae tuber”. Chem. Pharm. Bull. 1978, 26, 2096–2097. [Google Scholar] [CrossRef] [PubMed]
- Parsaeimehr, A.; Sargsyan, E. Ephedra Alkaloids-Alkaloids Derived by Amination Reaction: Phenylalanine Derived. In Natural Products; Springer: Berlin/Heidelberg, Germany, 2013; pp. 909–922. [Google Scholar]
- Krizevski, R.; Bar, E.; Shalit, O.; Sitrit, Y.; Ben-Shabat, S.; Lewinsohn, E. Composition and stereochemistry of ephedrine alkaloids accumulation in Ephedra sinica Stapf. Phytochemistry 2010, 71, 895–903. [Google Scholar] [CrossRef]
- Krizevski, R.; Dudai, N.; Bar, E.; Lewinsohn, E. Developmental patterns of phenylpropylamino alkaloids accumulation in khat (Catha edulis, Forsk.). J. Ethnopharmacol. 2007, 114, 432–438. [Google Scholar] [CrossRef]
- Unver, T.; Groves, R.A.; Hagel, J.M.; Zhang, Y.; Kilpatrick, K.; Levy, A.; Marsolais, F.; Lewinsohn, E.; Sensen, C.W.; Facchini, P.J. Transcriptome profiling of khat (Catha edulis) and ephedra sinica reveals gene candidates potentially involved in amphetamine-type alkaloid biosynthesis. PLoS ONE 2015, 10, e0119701. [Google Scholar]
- Hagel, J.M.; Krizevski, R.; Marsolais, F.; Lewinsohn, E.; Facchini, P.J. Biosynthesis of amphetamine analogs in plants. Trends Plant Sci. 2012, 17, 404–412. [Google Scholar] [CrossRef]
- Duan, Y.; Zhang, H.; Meng, X.; Huang, M.; Zhang, Z.; Huang, C.; Zhao, F.; Xue, T.; Xue, J. Accumulation of salicylic acid-elicited alkaloid compounds in in vitro cultured Pinellia ternata microtubers and expression profiling of genes associated with benzoic acid-derived alkaloid biosynthesis. Plant Cell Tiss. Org. 2019, 139, 317–325. [Google Scholar] [CrossRef]
- Li, Q.; Xu, J.; Yang, L.; Sun, Y.; Zhou, X.; Zheng, Y.; Zhang, Y.; Cai, Y. LED light quality affect growth, alkaloids contents, and expressions of amaryllidaceae alkaloids biosynthetic pathway genes in Lycoris longituba. J. Plant Growth Regul. 2022, 41, 257–270. [Google Scholar] [CrossRef]
- Fraser, V.N.; Philmus, B.; Megraw, M. Metabolomics analysis reveals both plant variety and choice of hormone treatment modulate vinca alkaloid production in Catharanthus roseus. Plant Direct 2020, 4, e00267. [Google Scholar] [CrossRef] [PubMed]
- Ptak, A.; Simlat, M.; Morańska, E.; Skrzypek, E.; Warchoł, M.; Tarakemeh, A.; Laurain-Mattar, D. Exogenous melatonin stimulated Amaryllidaceae alkaloid biosynthesis in in vitro cultures of Leucojum aestivum L. Ind. Crops Prod. 2019, 138, 111458. [Google Scholar] [CrossRef]
- Nolan, T.M.; Vukasinovic, N.; Liu, D.; Russinova, E.; Yin, Y. Brassinosteroids: Multidimensional regulators of plant growth, development, and stress responses. Plant Cell 2020, 32, 295–318. [Google Scholar] [CrossRef] [PubMed]
- Basit, F.; Bhat, J.A.; Dong, Z.; Mou, Q.; Zhu, X.; Wang, Y.; Hu, J.; Jan, B.L.; Shakoor, A.; Guan, Y.; et al. Chromium toxicity induced oxidative damage in two rice cultivars and its mitigation through external supplementation of brassinosteroids and spermine. Chemosphere 2022, 302, 134423. [Google Scholar] [CrossRef]
- Faizan, M.; Bhat, J.A.; Noureldeen, A.; Ahmad, P.; Yu, F. Zinc oxide nanoparticles and 24-epibrassinolide alleviates Cu toxicity in tomato by regulating ROS scavenging, stomatal movement and photosynthesis. Ecotox. Environ. Saf. 2021, 218, 112293. [Google Scholar] [CrossRef]
- Ghassemi-Golezani, K.; Hassanzadeh, N.; Shakiba, M.-R.; Esmaeilpour, B. Exogenous salicylic acid and 24-epi-brassinolide improve antioxidant capacity and secondary metabolites of Brassica nigra. Biocatal. Agric. Biotechnol. 2020, 26, 101636. [Google Scholar] [CrossRef]
- Swamy, K.; Rao, S.S.R. Effect of brassinosteroids on the performance of coleus (Coleus forskohlii). J. Herbs, Spices Med. Plants 2011, 17, 12–20. [Google Scholar] [CrossRef]
- Guo, C.; Shen, Y.; Li, M.; Chen, Y.; Xu, X.; Chu, J.; Yao, X. Principal component analysis to assess the changes of yield and quality of two Pinellia ternata cultivars after brassinolide treatments. J. Plant Growth Regul. 2022, 41, 2185–2197. [Google Scholar] [CrossRef]
- Song, L.; Chen, W.; Yao, Q.; Guo, B.; Valliyodan, B.; Wang, Z.; Nguyen, H.T. Genome-wide transcriptional profiling for elucidating the effects of brassinosteroids on Glycine max during early vegetative development. Sci. Rep. 2019, 9, 16085. [Google Scholar] [CrossRef]
- Hartwig, T.; Corvalan, C.; Best, N.B.; Budka, J.S.; Zhu, J.Y.; Choe, S.; Schulz, B. Propiconazole is a specific and accessible brassinosteroid (BR) biosynthesis inhibitor for Arabidopsis and maize. PLoS ONE 2012, 7, e36625. [Google Scholar] [CrossRef] [PubMed]
- Sekimata, K.; Han, S.-Y.; Yoneyama, K.; Takeuchi, Y.; Yoshida, S.; Asami, T. A specific and potent inhibitor of brassinosteroid biosynthesis possessing a dioxolane ring. J. Agr. Food Chem. 2002, 50, 3486–3490. [Google Scholar] [CrossRef] [PubMed]
- Sridhara, S.; Ramesh, N.; Gopakkali, P.; Paramesh, V.; Tamam, N.; Abdelbacki, A.M.; Elansary, H.O.; El-Sabrout, A.M.; Abdelmohsen, S.A. Application of homobrassinolide enhances growth, yield and quality of tomato. Saudi J. Biol. Sci. 2021, 28, 4800–4806. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Liang, X.-G.; Zhang, L.; Lin, S.; Zhao, X.; Zhou, L.-L.; Shen, S.; Zhou, S.-L. Spraying exogenous 6-benzyladenine and brassinolide at tasseling increases maize yield by enhancing source and sink capacity. Field Crops Res. 2017, 211, 1–9. [Google Scholar] [CrossRef]
- Maia, C.F.; da Silva, B.R.S.; da Silva Lobato, A.K. Brassinosteroids positively modulate growth: Physiological, biochemical and anatomical evidence using two tomato genotypes contrasting to dwarfism. J. Plant Growth Regul. 2018, 37, 1099–1112. [Google Scholar] [CrossRef]
- Ahmad, P.; Ahanger, M.A.; Egamberdieva, D.; Alam, P.; Alyemeni, M.N.; Ashraf, M. Modification of osmolytes and antioxidant enzymes by 24-Epibrassinolide in chickpea seedlings under mercury (Hg) toxicity. J. Plant Growth Regul. 2017, 37, 309–322. [Google Scholar] [CrossRef]
- Wang, F.; Zhi, J.; Zhang, Z.; Wang, L.; Suo, Y.; Xie, C.; Li, M.; Zhang, B.; Du, J.; Gu, L.; et al. Transcriptome analysis of salicylic acid treatment in Rehmannia glutinosa hairy roots using RNA-seq technique for identification of genes involved in acteoside biosynthesis. Front. Plant Sci. 2017, 8, 787. [Google Scholar] [CrossRef]
- Klempien, A.; Kaminaga, Y.; Qualley, A.; Nagegowda, D.A.; Widhalm, J.R.; Orlova, I.; Shasany, A.K.; Taguchi, G.; Kish, C.M.; Cooper, B.R.; et al. Contribution of CoA ligases to benzenoid biosynthesis in petunia flowers. Plant Cell 2012, 24, 2015–2030. [Google Scholar] [CrossRef]
- Van Moerkercke, A.; Schauvinhold, I.; Pichersky, E.; Haring, M.A.; Schuurink, R.C. A plant thiolase involved in benzoic acid biosynthesis and volatile benzenoid production. Plant J. 2009, 60, 292–302. [Google Scholar] [CrossRef]
- Fernandez, F.J.; Vega, M.C.; Lehmann, F.; Sandmeier, E.; Gehring, H.; Christen, P.; Wilmanns, M. Structural studies of the catalytic reaction pathway of a hyperthermophilic histidinol-phosphate aminotransferase. J. Biol. Chem. 2004, 279, 21478–21488. [Google Scholar] [CrossRef]
- Zhou, Y.; Cai, H.; Xiao, J.; Li, X.; Zhang, Q.; Lian, X. Over-expression of aspartate aminotransferase genes in rice resulted in altered nitrogen metabolism and increased amino acid content in seeds. Theor. Appl. Genet. 2009, 118, 1381–1390. [Google Scholar] [CrossRef] [PubMed]
- Bedewitz, M.A.; Gongora-Castillo, E.; Uebler, J.B.; Gonzales-Vigil, E.; Wiegert-Rininger, K.E.; Childs, K.L.; Hamilton, J.P.; Vaillancourt, B.; Yeo, Y.S.; Chappell, J.; et al. A root-expressed L-phenylalanine: 4-hydroxyphenylpyruvate aminotransferase is required for tropane alkaloid biosynthesis in Atropa belladonna. Plant Cell 2014, 26, 3745–3762. [Google Scholar] [CrossRef] [Green Version]
- Qiu, F.; Yang, C.; Yuan, L.; Xiang, D.; Lan, X.; Chen, M.; Liao, Z. A phenylpyruvic acid reductase is required for biosynthesis of tropane alkaloids. Org. Lett. 2018, 20, 7807–7810. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Zeng, J.; Zhao, T.; Zhang, H.; Qiu, F.; Yang, C.; Zeng, L.; Liu, X.; Chen, M.; Lan, X.; et al. Enhancing tropane alkaloid production based on the functional identification of tropine-forming reductase in Scopolia lurida, a tibetan medicinal plant. Front. Plant Sci. 2017, 8, 1745. [Google Scholar] [CrossRef]
- Peng, X.; Luo, Y.; Wang, J.; Ji, T.; Yuan, L.; Kai, G. Integrated analysis of the transcriptome, metabolome and analgesic effect provide insight into potential applications of different parts of Lindera aggregata. Food Res. Int. 2020, 138, 109799. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Qiao, C.; Pang, J.; Zhang, G.; Luo, Y. The versatile O-methyltransferase LrOMT catalyzes multiple O-methylation reactions in amaryllidaceae alkaloids biosynthesis. Int. J. Biol. Macromol. 2019, 141, 680–692. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, C.; Chen, Y.; Wu, D.; Du, Y.; Wang, M.; Liu, C.; Chu, J.; Yao, X. Transcriptome Analysis Reveals an Essential Role of Exogenous Brassinolide on the Alkaloid Biosynthesis Pathway in Pinellia Ternata. Int. J. Mol. Sci. 2022, 23, 10898. https://doi.org/10.3390/ijms231810898
Guo C, Chen Y, Wu D, Du Y, Wang M, Liu C, Chu J, Yao X. Transcriptome Analysis Reveals an Essential Role of Exogenous Brassinolide on the Alkaloid Biosynthesis Pathway in Pinellia Ternata. International Journal of Molecular Sciences. 2022; 23(18):10898. https://doi.org/10.3390/ijms231810898
Chicago/Turabian StyleGuo, Chenchen, Ying Chen, Dengyun Wu, Yu Du, Mengyue Wang, Cunqi Liu, Jianzhou Chu, and Xiaoqin Yao. 2022. "Transcriptome Analysis Reveals an Essential Role of Exogenous Brassinolide on the Alkaloid Biosynthesis Pathway in Pinellia Ternata" International Journal of Molecular Sciences 23, no. 18: 10898. https://doi.org/10.3390/ijms231810898




