Effect of Nickel Ions on the Physiological and Transcriptional Responses to Carbon and Nitrogen Metabolism in Tomato Roots under Low Nitrogen Levels
Abstract
:1. Introduction
2. Results
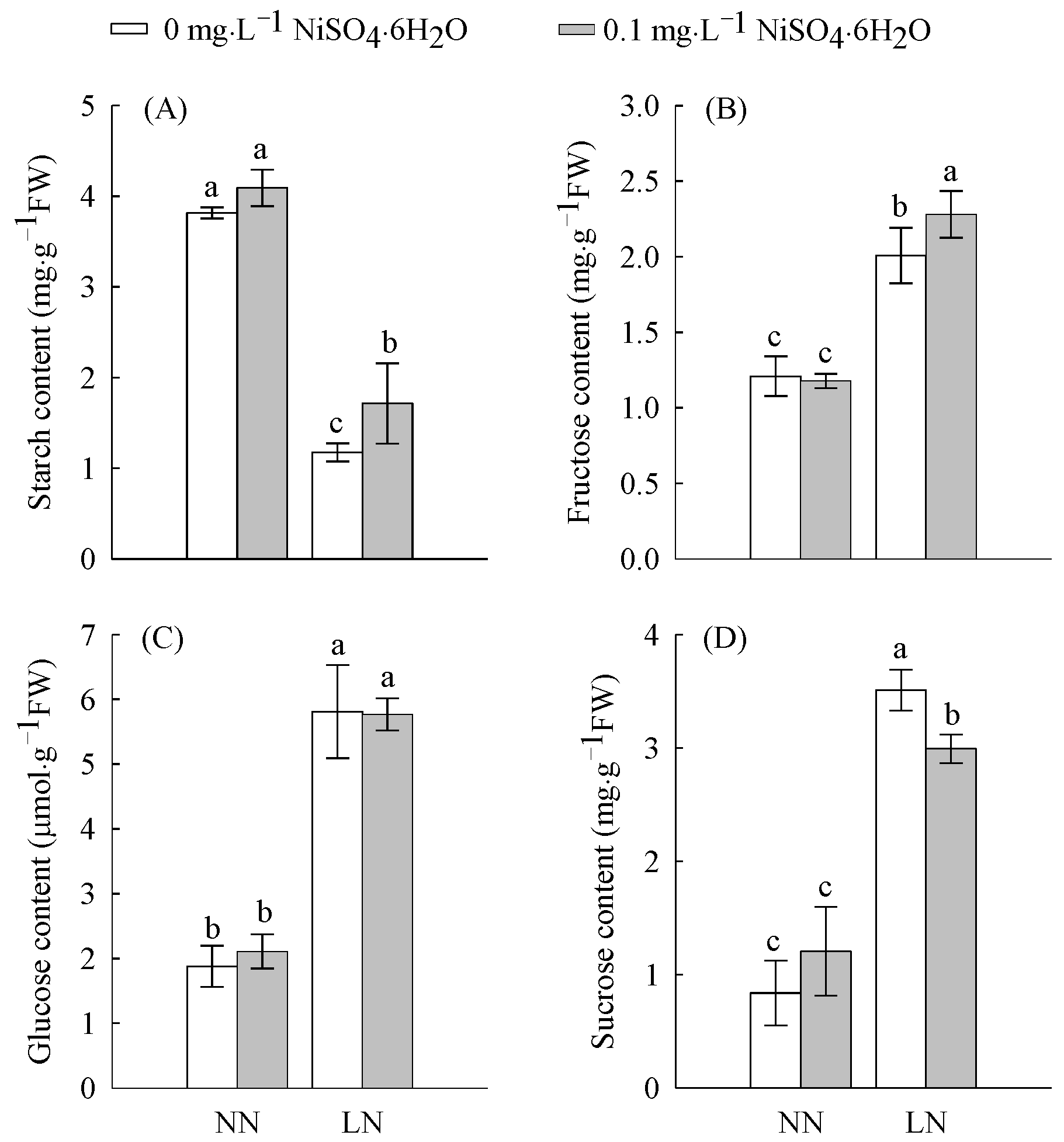
2.1. Carbohydrate Content
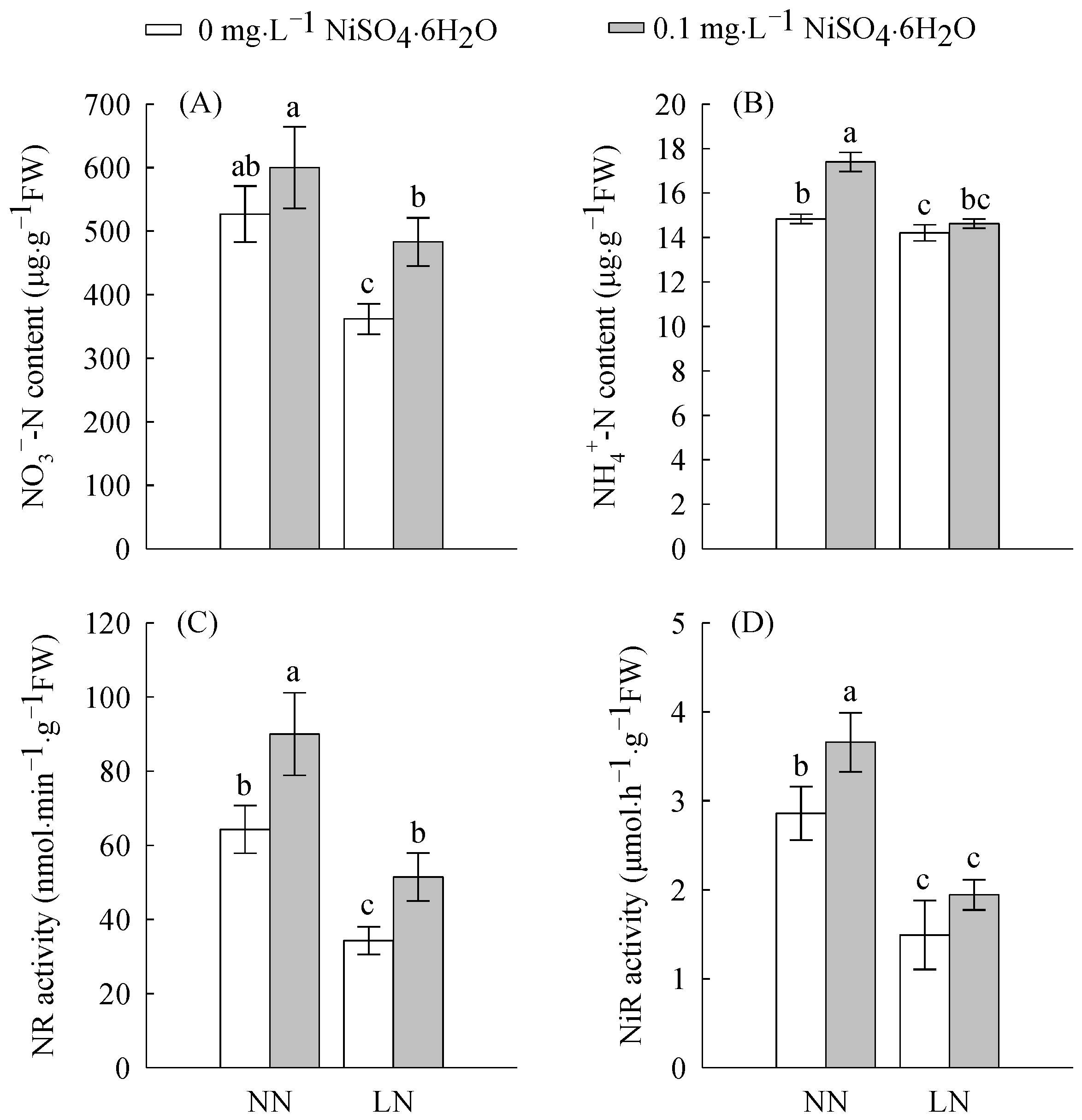
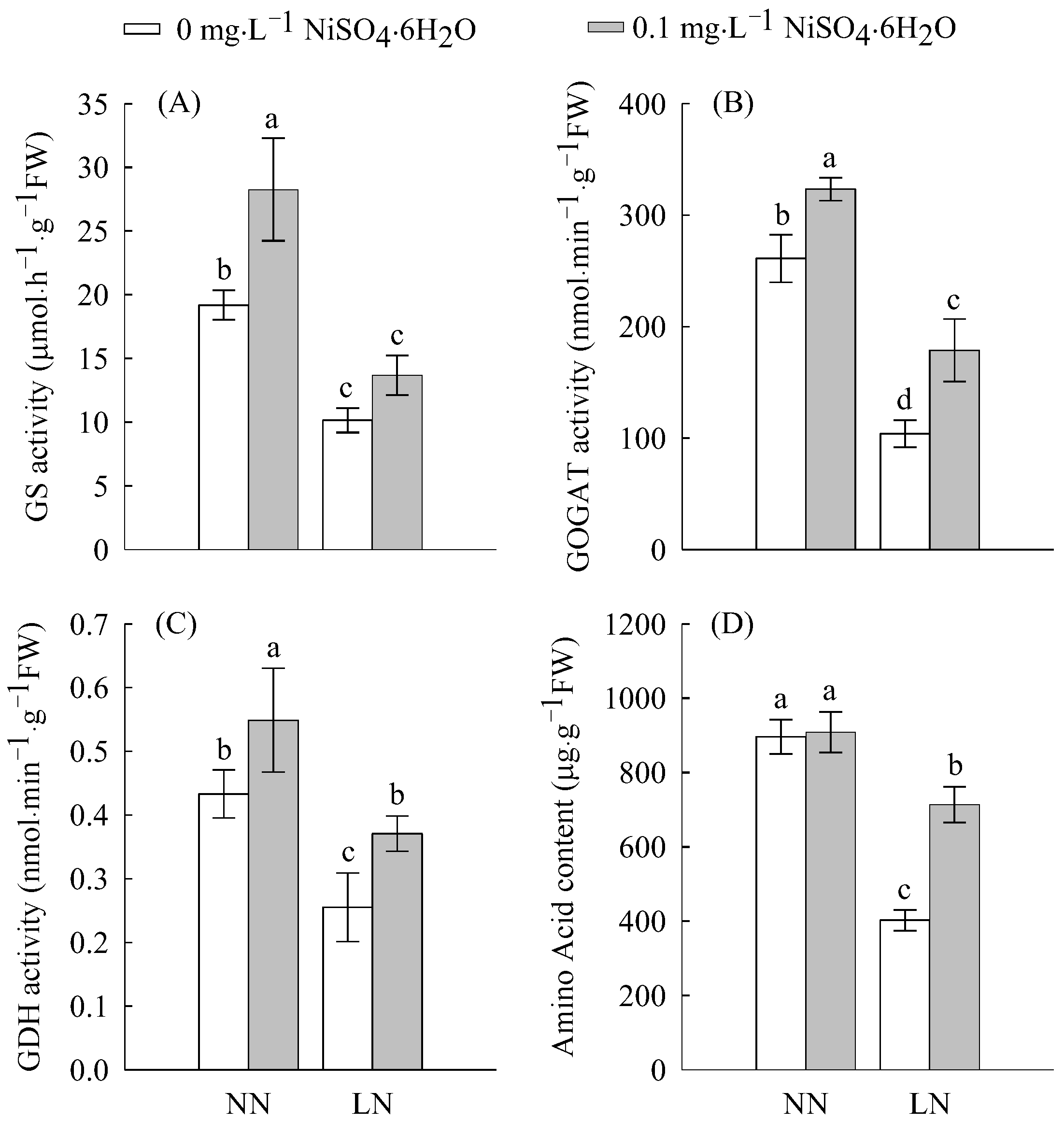
2.2. Enzymatic Activities and N Content
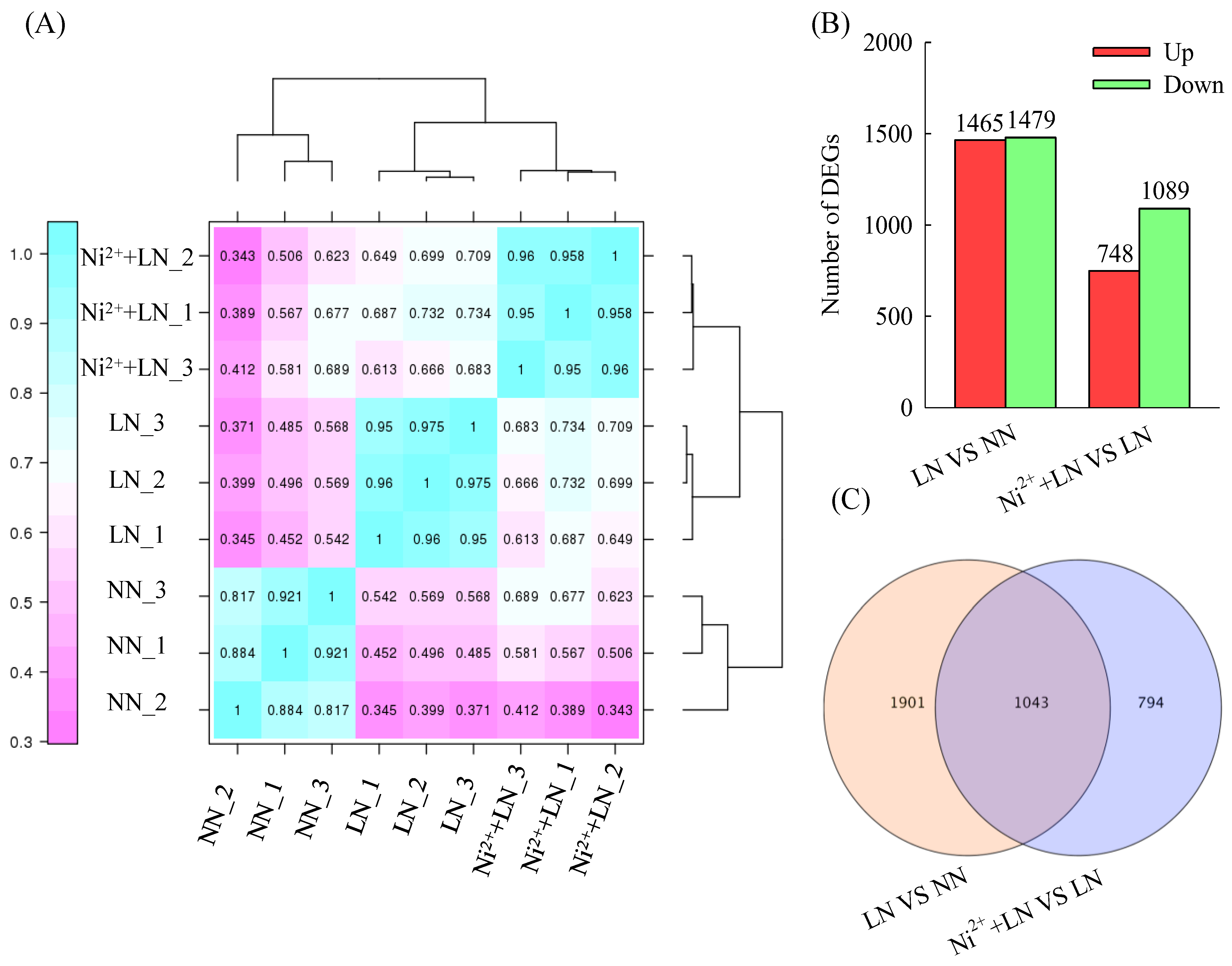
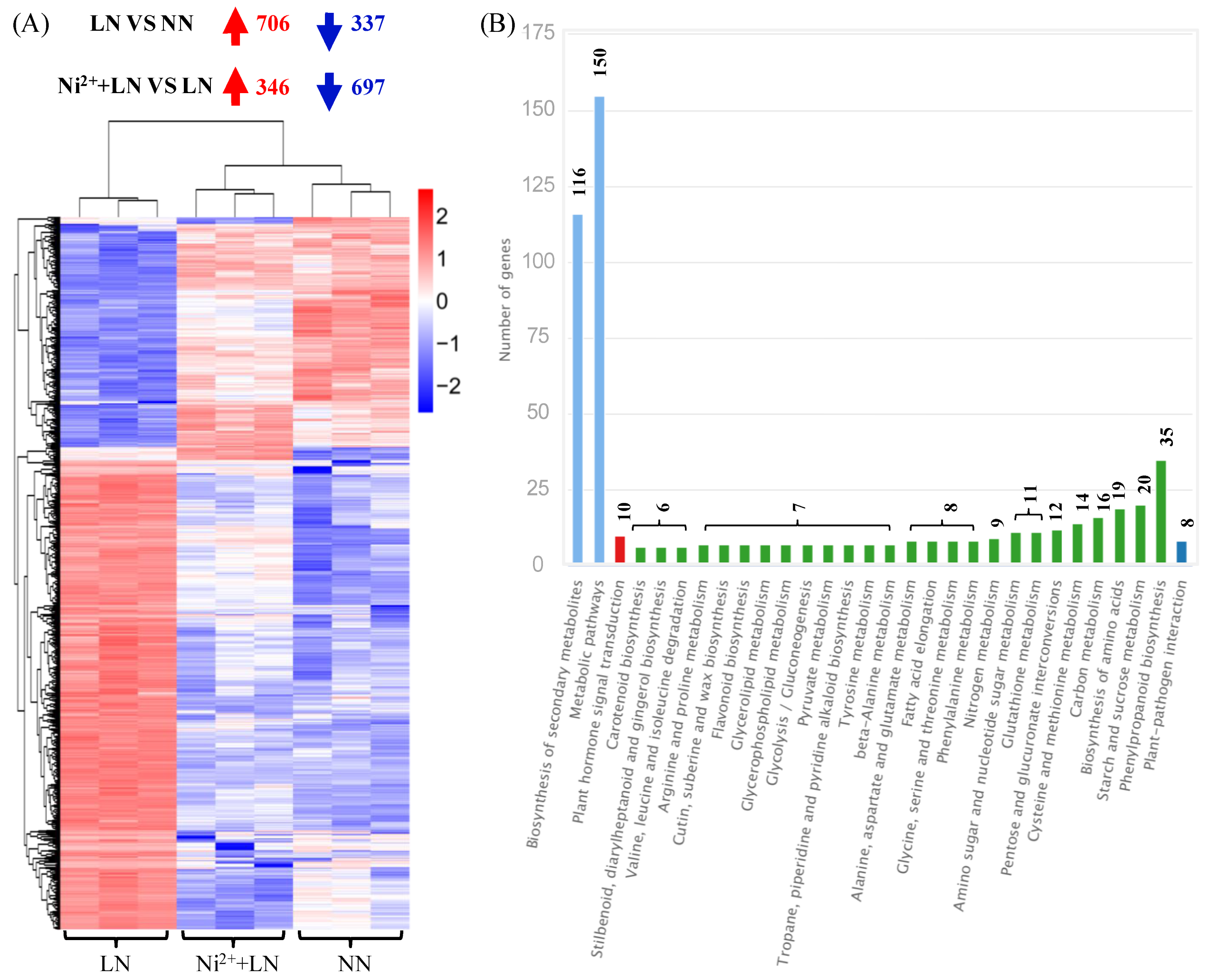
2.3. Analysis of Differentially Expressed Genes (DEGs)
2.4. Functional Classification of DEGs and Validation by Quantitative Real-Time PCR (RT-qPCR)
2.5. Metabolic Regulation of Enzymes Encoded by DEGs
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Treatments
4.2. Determination of Enzyme Activity in the C and N Metabolism
4.3. RNA-Seq and RT-qPCR Analysis
4.4. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
Appendix A.1. N Nutrient Solution Formula
| N Concentration (mmol L−1) | Reagent (mmol L−1) | ||||||
|---|---|---|---|---|---|---|---|
| Ca(NO3)2 4H2O | CaCl2 | KNO3 | NH4H2PO4 | KH2PO4 | KCl | MgSO4 7H2O | |
| 0.383 | 0.075 | 1.425 | 0.200 | 0.033 | 0.635 | 3.168 | 0.998 |
| 7.67 | 1.499 | 3.996 | 0.669 | 0.998 | |||
Appendix A.2. RNA Extraction and RNA-Seq Analysis
References
- Karki, S.; Poudel, N.S.; Bhusal, G.; Simkhada, S.; Regmi, B.R.; Adhikari, B.; Poudel, S. Growth parameter and yield attributes of rice (Oryza sativa) as influenced by different combination of nitrogen sources. World J. Agric. Res. 2018, 6, 58–64. [Google Scholar]
- Liang, Y.; Zhao, X.; Jones, A.M.; Gao, Y. G proteins sculp root architecture in response to nitrogen in rice and arabidopsis. Plant Sci. 2018, 274, 129–136. [Google Scholar] [CrossRef]
- Stein, L.Y.; Klotz, M.G. The nitrogen cycle. Curr. Biol. 2016, 26, 94–98. [Google Scholar] [CrossRef]
- Yang, X.L.; Lu, Y.L.; Ding, Y.; Yin, X.F.; Raza, S.; Tong, Y.A. Optimising nitrogen fertilisation: A key to improving nitrogen-use efficiency and minimising nitrate leaching losses in an intensive wheat/maize rotation (2008–2014). Field Crops Res. 2017, 206, 1–10. [Google Scholar] [CrossRef]
- Saleque, M.A.; Naher, U.A.; Islam, A.; Pathan, A.B.M.B.U.; Hossain, A.T.M.S.; Meisner, C.A. Inorganic and organic phosphorus fertilizer effects on the phosphorus fractionation in wetland rice soils. Soil Sci. Soc. Am. J. 2004, 68, 1635–1644. [Google Scholar] [CrossRef]
- Jensen, L.S.; Schjoerring, J.K.; Van der Hoek, K.W.; Poulsen, H.D.; Zevenbergen, J.F.; Pallière, C.; Lammel, J.; Brentrup, F.; Jongbloed, A.W.; Willems, J.; et al. Benefits of Nitrogen for Food, Fibre and Industrial Production; Cambridge University Press: Cambridge, UK, 2011; pp. 32–61. [Google Scholar]
- Rahman, H.; Sabreen, S.; Alam, S.; Kawai, S. Effects of nickel on growth and composition of metal micronutrients in barley plants grown in nutrient solution. J. Plant Nutr. 2005, 28, 393–404. [Google Scholar] [CrossRef]
- Wood, B.W.; Reilly, C.C.; Nyczepir, A.P. Mouse-ear of pecan: I. Symptomatology and occurrence. HortScience 2004, 39, 87–94. [Google Scholar] [CrossRef]
- Wood, B.W.; Reilly, C.C.; Nyczepir, A.P. Mouse-ear of pecan: II. Influence of nutrient applications. HortScience 2004, 39, 95–100. [Google Scholar] [CrossRef]
- Sirko, A.; Brodzik, R. Plant ureases: Roles and regulation. Acta Biochim. Pol. 2000, 47, 1189–1195. [Google Scholar] [CrossRef]
- Gerendas, J.; Sattelmacher, B. Significance of Ni supply for growth, urease activity and concentrations of urea, amino acids and mineral nutrients of urea-grown plants. Plant Soil 1997, 190, 153–162. [Google Scholar] [CrossRef]
- Gerendas, J.; Zhu, Z.; Sattelmacher, B. Influence of Ni and Ni supply on nitrogen metabolism and urease activity in rice (Oryzasativa L.). J. Exp. Bot. 1998, 49, 1545–1554. [Google Scholar] [CrossRef]
- Brown, P.H.; Welch, R.M.; Madison, J.T. Effect of nickel deficiency on soluble anion, amino acid, and nitrogen levels in barley. Plant Soil 1990, 125, 19–27. [Google Scholar] [CrossRef]
- Walker, C.D.; Graham, R.D.; Madison, J.T.; Cary, E.E.; Welch, R.M. Effects of ni deficiency on some nitrogen metabolites in cowpeas (Vigna unguiculata L. Walp). Plant Physiol. 1985, 79, 474–479. [Google Scholar] [CrossRef] [PubMed]
- Khoshgoftarmanesh, A.H.; Hosseini, F.; Afyuni, M. Nickel supplementation effect on the growth, urease activity and urea and nitrate concentrations in lettuce supplied with different nitrogen sources. Sci. Hortic. 2011, 130, 381–385. [Google Scholar] [CrossRef]
- Tan, X.W.; Ikeda, H.; Oda, M. Effects of nickel concentration in the nutrient solution on the nitrogen assimilation and growth of tomato seedlings in hydroponic culture supplied with urea or nitrate as the sole nitrogen source. Sci. Hortic. 2000, 84, 265–273. [Google Scholar] [CrossRef]
- Liu, M.Y.; Yan, Y.N.; Shang, C.Y.; Chen, L.; Li, J.M.; Zhong, F.L.; Lin, Y.Z. Effect of nickel ion on growth, physiology and nitrogen absorption in Lactuca sativa L. seedling. Acta Bot. Boreal.-Occident. Sin. 2018, 38, 2060–2071. [Google Scholar]
- Li, S.H.; Yang, D.Q.; Tian, J.; Wang, S.B.; Yan, Y.N.; He, X.L.; Du, Z.J.; Zhong, F.L. Physiological and transcriptional response of carbohydrate and nitrogen metabolism in tomato plant leaves to nickel ion and nitrogen levels. Sci. Hortic. 2022, 292, 110620. [Google Scholar] [CrossRef]
- Zhang, S.C.; Li, M.J.; Guo, J.K.; Shi, Z.L.; Fu, X.Y.; Di, R.Y.; Li, Y.M. Comparative transcriptome analysis of triticumaestivumin response to nitrogen stress. Russ. J. Plant Physl. 2016, 63, 365–374. [Google Scholar] [CrossRef]
- Stitt, M.; Müller, C.; Matt, P.; Gibon, Y.; Carillo, P.; Morcuende, R.; Scheible, W.; Krapp, A. Steps towards an integrated view of nitrogen metabolism. J. Exp. Bot. 2002, 53, 959–970. [Google Scholar] [CrossRef] [PubMed]
- Vidal, E.A.; Gutiérrez, R.A. A systems view of nitrogen nutrient and metabolite responses in arabidopsis. Curr. Opin. Plant Biol. 2008, 11, 521–529. [Google Scholar] [CrossRef]
- Sun, T.T.; Zhang, J.K.; Zhang, Q.; Li, X.L.; Li, M.J.; Yang, Y.Z.; Zhou, J.; Wei, Q.P.; Zhou, B.B. Integrative physiological, transcriptome, and metabolome analysis revealsthe effects of nitrogen sufficiency and deficiency conditions in apple leavesand roots. Environ. Exp. Bot. 2021, 192, 104633. [Google Scholar] [CrossRef]
- Xin, W.; Zhang, L.N.; Zhang, W.Z.; Gao, J.P.; Yi, J.; Zhen, X.X.; Du, M.; Zhao, Y.Z.; Chen, L.Q. An Integrated Analysis of the rice transcriptome and metabolome reveals root growth regulation mechanisms in response to nitrogen availability. Int. J. Mol. Sci. 2019, 20, 5893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernard, S.M.; Habash, D.Z. The importance of cytosolic glutamine synthetase in nitrogen assimilation and recycling. New Phytol. 2009, 182, 608–620. [Google Scholar] [CrossRef] [PubMed]
- Camargo, E.L.O.; Nascimento, L.C.; Soler, M.; Salazar, M.M.; Lepikson-Neto, J.; Marques, W.L.; Alves, A.; Teixeira, P.J.P.L.; Carazzolle, M.F.; Martinez, Y.; et al. Contrasting nitrogen fertilization treatments impact xylem gene expression and secondary cell wall lignification in eucalyptus. BMC Plant Biol. 2014, 14, 256. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Meng, C.; Ji, J.; Li, M.H.; Zhang, X.M.; Wu, Y.Y.; Xie, T.T.; Du, C.J.; Sun, J.C.; Jiang, Z.P.; et al. Exogenous GABA promotes adaptation and growth by altering the carbon and nitrogen metabolic flux in poplar seedlings under low nitrogen conditions. Tree Physiol. 2020, 40, 1744–1761. [Google Scholar] [CrossRef]
- Cheng, L.; Yuan, H.Y.; Ren, R.; Zhao, S.Q.; Han, Y.P.; Zhou, Q.Y.; Ke, D.X.; Wang, Y.X.; Wang, L. Genome-wide identification, classification, and expression analysis of amino acid transporter gene family in glycine max. Front. Plant Sci. 2016, 7, 515. [Google Scholar] [CrossRef]
- Masdaux-Daubresse, C.; Daniel-Vedele, F.; Dechorgnat, J.; Chardon, F.; Gaufichon, L.; Suzuki, A. Nitrogen uptake, assimilation and remobilization in plants: Challenges for sustainable and productive agriculture. Ann. Bot. 2010, 105, 1141–1157. [Google Scholar] [CrossRef]
- Nawaz, M.A.; Chen, C.; Shireen, F.; Zheng, Z.H.; Sohail, H.; Afzal, M.; Ali, M.A.; Bie, Z.L.; Huang, Y. Genome-wide expression profiling of leaves and roots of watermelon in response to low nitrogen. BMC Genom. 2018, 19, 456. [Google Scholar] [CrossRef]
- Plett, D.C.; Holthama, L.R.; Okamotoa, M.; Garnett, T.P. Nitrate uptake and its regulation in relation to improving nitrogen use efficiency in cereals. Semin. Cell Dev. Biol. 2017, 74, 97–104. [Google Scholar] [CrossRef]
- Fan, X.R.; Tang, Z.; Tan, Y.W.; Zhang, Y.; Luo, B.B.; Yang, M.; Lian, X.M.; Shen, Q.R.; Miller, A.J.; Xu, G.H. Overexpression of a pH-sensitive nitrate transporter in rice increases crop yields. Proc. Natl. Acad. Sci. USA 2016, 113, 7118–7123. [Google Scholar] [CrossRef]
- Malagoli, P.Q.; Laine, P.; Deunff, E.L.; Rossato, L.; Ney, B.; Ourry, A. Modeling nitrogen uptake in oilseed rape cv capitol during a growth cycle using influx kinetics of root nitrate transport systems and field experimental data. Plant Physiol. 2004, 134, 388–400. [Google Scholar] [CrossRef] [PubMed]
- Gu, R.; Duan, F.; An, X.; Zhang, F.; Von Wirén, N.; Yuan, L. Characterization of AMT-mediated high-affinity ammonium uptake in roots of maize (Zea mays L.). Plant Cell Physiol. 2013, 54, 1515–1524. [Google Scholar] [CrossRef] [PubMed]
- Reich, P.B.; Hobbie, S.E.; Lee, T.; Ellsworth, D.S.; West, J.B.; Tilman, D.; Knops, J.M.H.; Naeem, S.; Trost, J. Nitrogen limitation constrains sustainability of ecosystem response to CO2. Nature 2006, 440, 922–925. [Google Scholar] [CrossRef] [PubMed]
- Li, M.X.; Xu, J.Y.; Wang, X.X.; Fu, H.; Zhao, M.L.; Wang, H.; Shi, L.X. Photosynthetic characteristics and metabolic analyses of two soybean genotypes revealed adaptive strategies to low-nitrogen stress. J. Plant Physiol. 2018, 229, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Quan, X.Y.; Qian, Q.F.; Ye, Z.L.; Zeng, J.B.; Han, Z.G.; Zhang, G.P. Metabolic analysis of two contrasting wild barley genotypes grown hydroponically reveals adaptive strategies in response to low nitrogen stress. J. Plant Physiol. 2016, 206, 59–67. [Google Scholar] [CrossRef]

| Sample Name | Clean Reads | Mapped Reads | Mapping Rate (%) | Uniq Mapped Reads | Uniq Mapping Rate (%) | Q30 (%) |
|---|---|---|---|---|---|---|
| NN_1 | 40,944,522 | 37,175,166 | 90.79 | 36,353,079 | 88.79 | 91.62 |
| NN_2 | 42,183,292 | 36,617,683 | 86.81 | 35,772,363 | 84.80 | 91.17 |
| NN_3 | 40,683,494 | 36,883,568 | 90.66 | 36,079,625 | 88.68 | 91.42 |
| LN_1 | 54,418,492 | 49,722,687 | 91.37 | 48,893,527 | 89.85 | 91.45 |
| LN_2 | 51,221,772 | 46,191,761 | 90.18 | 45,404,214 | 88.64 | 91.95 |
| LN_3 | 44,687,904 | 38,817,524 | 86.86 | 38,152,833 | 85.38 | 91.91 |
| Ni2+ + LN_1 | 40,918,376 | 37,467,413 | 91.57 | 36,774,788 | 89.87 | 91.85 |
| Ni2+ + LN_2 | 43,462,968 | 39,298,045 | 90.42 | 38,608,365 | 88.83 | 91.85 |
| Ni2+ + LN_3 | 41,466,278 | 37,055,020 | 89.36 | 36,343,722 | 87.65 | 91.76 |
| Gene | Gene ID | LN vs. NN | Ni2+ + LN vs. LN | ||
|---|---|---|---|---|---|
| RNA-Seq | RT-qPCR | RNA-Seq | RT-qPCR | ||
| NR | Solyc11g013810.2 | −3.35 | −0.08 | 4.32 | 1.40 |
| NiR | Solyc10g050890.2 | −3.57 | −0.53 | 3.58 | 1.40 |
| GS | Solyc04g014510.3 | −2.43 | −0.24 | 2.00 | 0.26 |
| GOGAT | Solyc03g083440.3 | −2.70 | −1.36 | 2.54 | 0.35 |
| CA | Solyc09g010970.3 | 1.08 | 0.34 | −1.16 | −0.92 |
| PK | Solyc11g007690.2 | −1.05 | −0.10 | 1.15 | 0.58 |
| INV | Solyc08g079080.4 | 3.09 | 1.72 | −1.43 | −1.17 |
| NiSO4 6H2O Concentration (mg L−1) | N Concentration (mmol L−1) |
|---|---|
| 0 | 0.383 |
| 7.66 | |
| 0.1 | 0.383 |
| 7.66 |
| Gene | Gene ID | Primer Sequences (Forward/Reverse) |
|---|---|---|
| Actin | LOC101253675 | F 5′-GTCCTCTTCCAGCCATCCAT-3′ R 5′-ACCACTGAGCACAATGTTACCG-3′ |
| NR | Solyc11g013810.2 | F 5′-GCAACTTCCCTCCTTCATCCAACC-3′ R 5′-CGTCATCGTCATCCTCGTCTTCAC-3′ |
| NiR | Solyc10g050890.2 | F 5′-TGCTTGTGGGTGGATTCTTCAGTC-3′ R 5′-TTCTGCCTGTTCCCTCGGGTAC-3′ |
| GS | Solyc04g014510.3 | F 5′-CAACGGAGAAGTGATGCCTGGAC-3′ R 5′-GCCCACAACTCGTCACCTGATG-3′ |
| GOGAT | Solyc03g083440.3 | F 5′-GGCTGGTATGAGTGGTGGTGTTG-3′ R 5′-ACGCTGGTGTTGCTGTATCATCATC-3′ |
| CA | Solyc09g010970.3 | F 5′-TGGTGCCTCCTTATGGAGCTGATCC-3′ R 5′-CGAATGCCTCCACAGCGACTATG-3′ |
| PK | Solyc11g007690.2 | F 5′-TGCCTTGAATCGGGAATGTCTGTG-3′ R 5′-TAGTTCAGGACCACCAGTGTCTAGC-3′ |
| INV | Solyc08g079080.4 | F 5′-ACGGTAACAACGACGGTACTGATG-3′ R 5′-TCCTCATGGTGGTTAACGGCATTAG-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, K.; Li, S.; Xu, Y.; Zhou, Y.; Ran, S.; Zhao, H.; Huang, W.; Xu, R.; Zhong, F. Effect of Nickel Ions on the Physiological and Transcriptional Responses to Carbon and Nitrogen Metabolism in Tomato Roots under Low Nitrogen Levels. Int. J. Mol. Sci. 2022, 23, 11398. https://doi.org/10.3390/ijms231911398
Zhang K, Li S, Xu Y, Zhou Y, Ran S, Zhao H, Huang W, Xu R, Zhong F. Effect of Nickel Ions on the Physiological and Transcriptional Responses to Carbon and Nitrogen Metabolism in Tomato Roots under Low Nitrogen Levels. International Journal of Molecular Sciences. 2022; 23(19):11398. https://doi.org/10.3390/ijms231911398
Chicago/Turabian StyleZhang, Kun, Shuhao Li, Yang Xu, Yuqi Zhou, Shengxiang Ran, Huanhuan Zhao, Weiqun Huang, Ru Xu, and Fenglin Zhong. 2022. "Effect of Nickel Ions on the Physiological and Transcriptional Responses to Carbon and Nitrogen Metabolism in Tomato Roots under Low Nitrogen Levels" International Journal of Molecular Sciences 23, no. 19: 11398. https://doi.org/10.3390/ijms231911398
APA StyleZhang, K., Li, S., Xu, Y., Zhou, Y., Ran, S., Zhao, H., Huang, W., Xu, R., & Zhong, F. (2022). Effect of Nickel Ions on the Physiological and Transcriptional Responses to Carbon and Nitrogen Metabolism in Tomato Roots under Low Nitrogen Levels. International Journal of Molecular Sciences, 23(19), 11398. https://doi.org/10.3390/ijms231911398





