Genome-Wide Identification of the A20/AN1 Zinc Finger Protein Family Genes in Ipomoea batatas and Its Two Relatives and Function Analysis of IbSAP16 in Salinity Tolerance
Abstract
:1. Introduction
2. Results
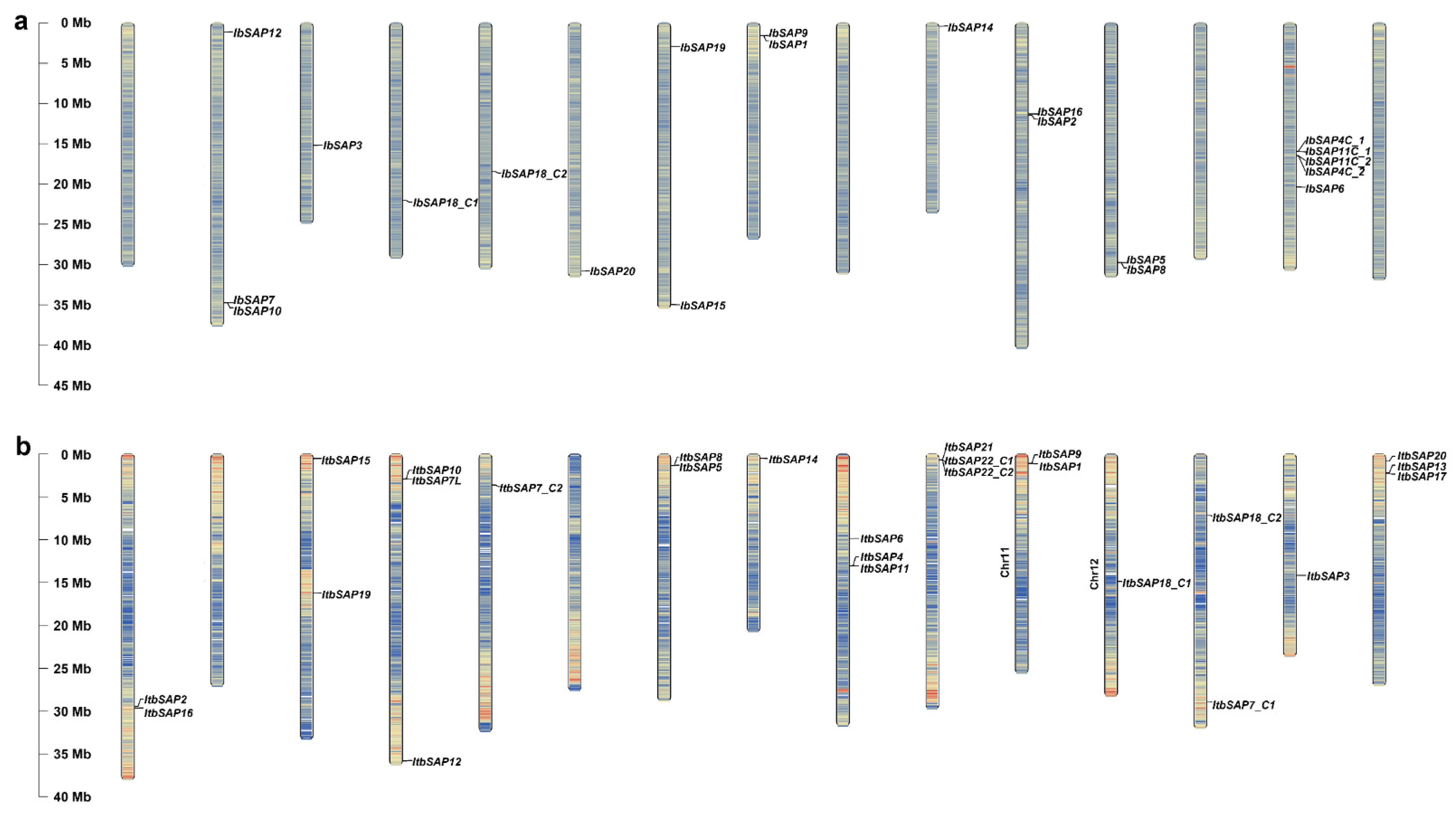
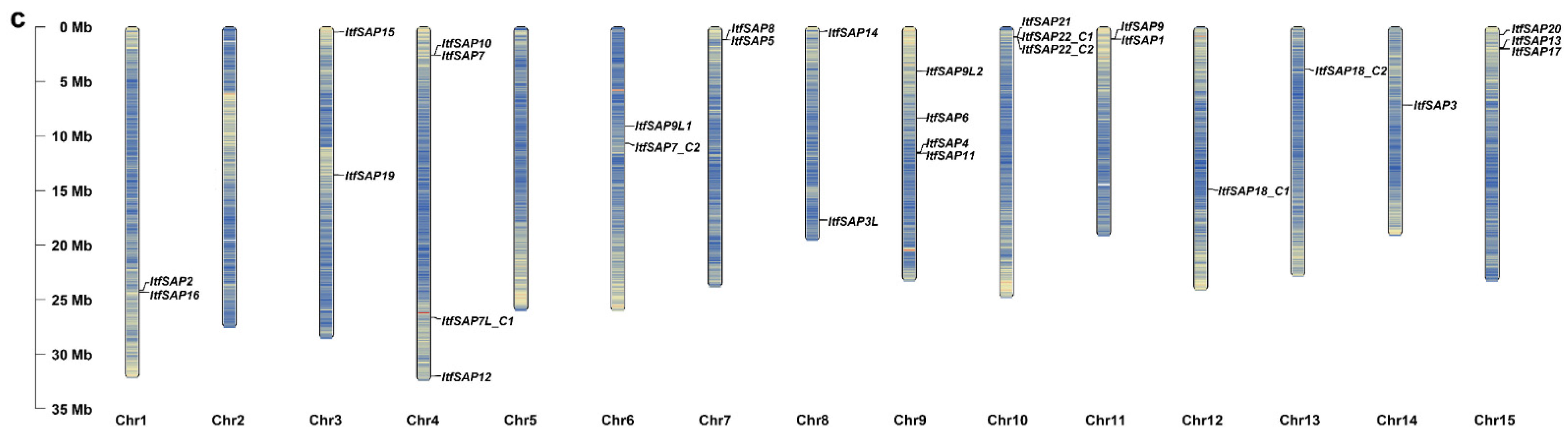
2.1. Identification of SAPs in Sweetpotato and Its Two Diploid Wild Relatives
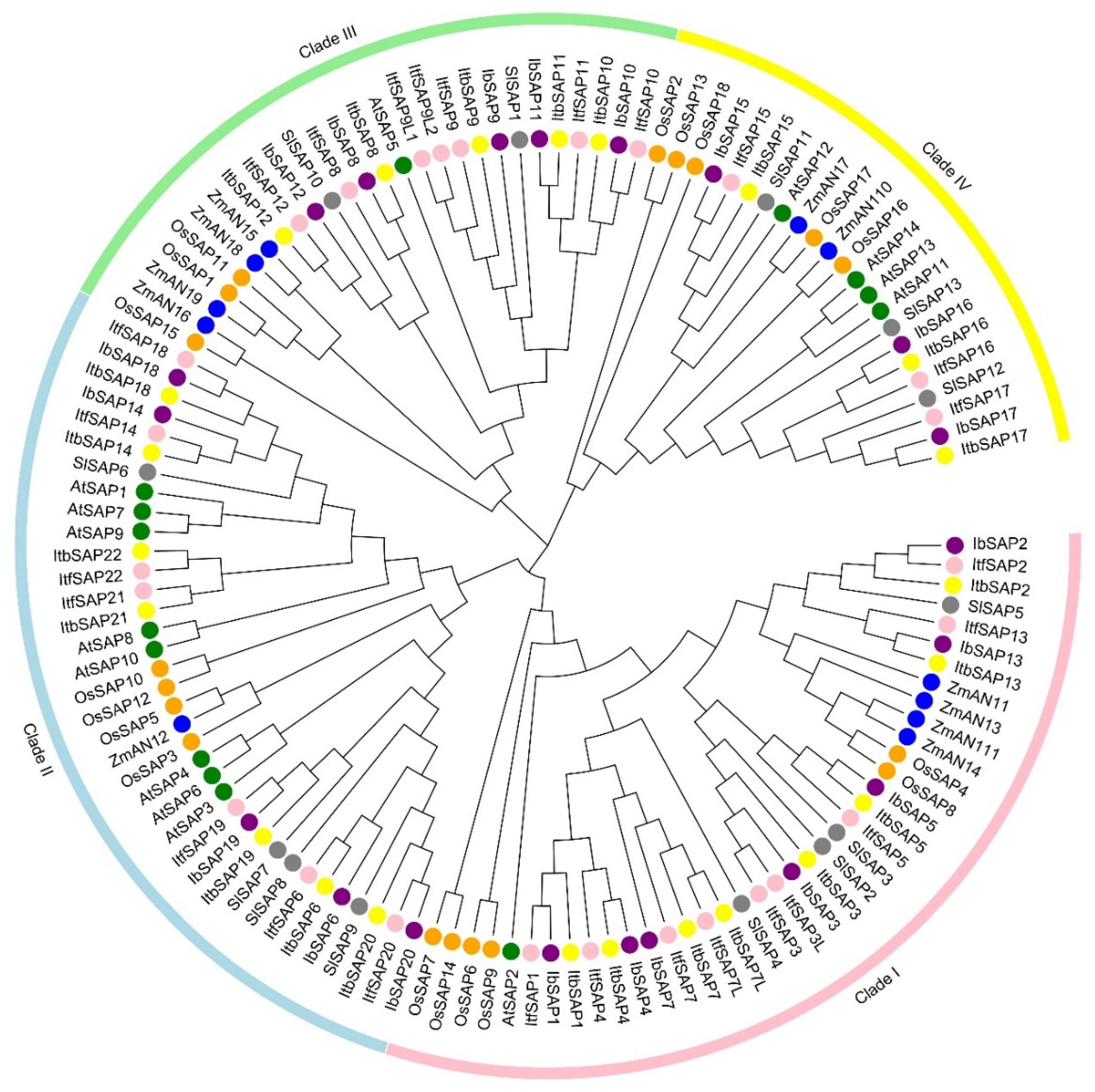
2.2. Phylogenetic Analysis of IbSAPs and SAPs from Some Other Plants
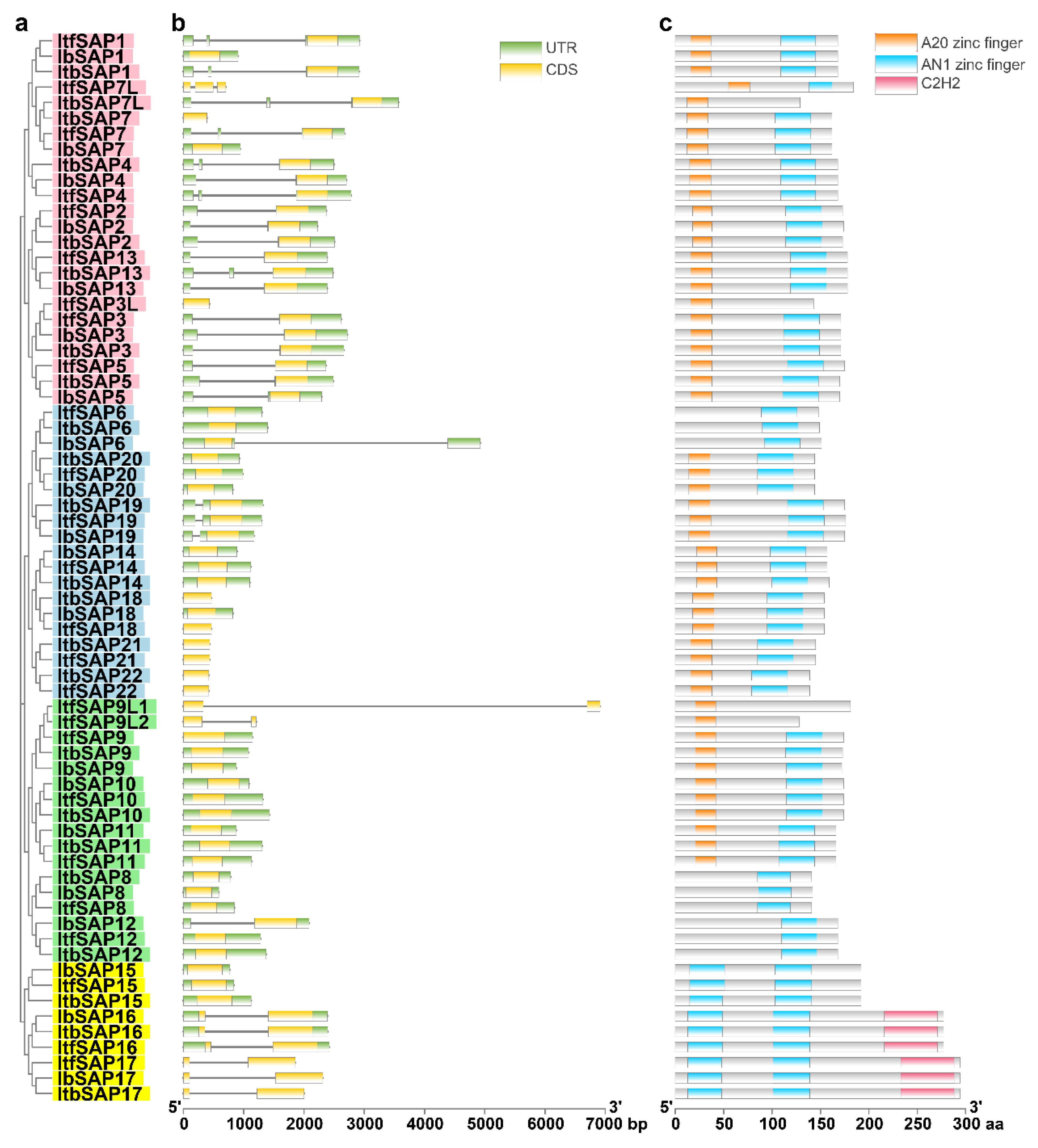
2.3. Gene and Protein Structures of IbSAPs, ItbSAPs, and ItfSAPs
2.4. Cis-Acting Elements in the Promoter of IbSAPs
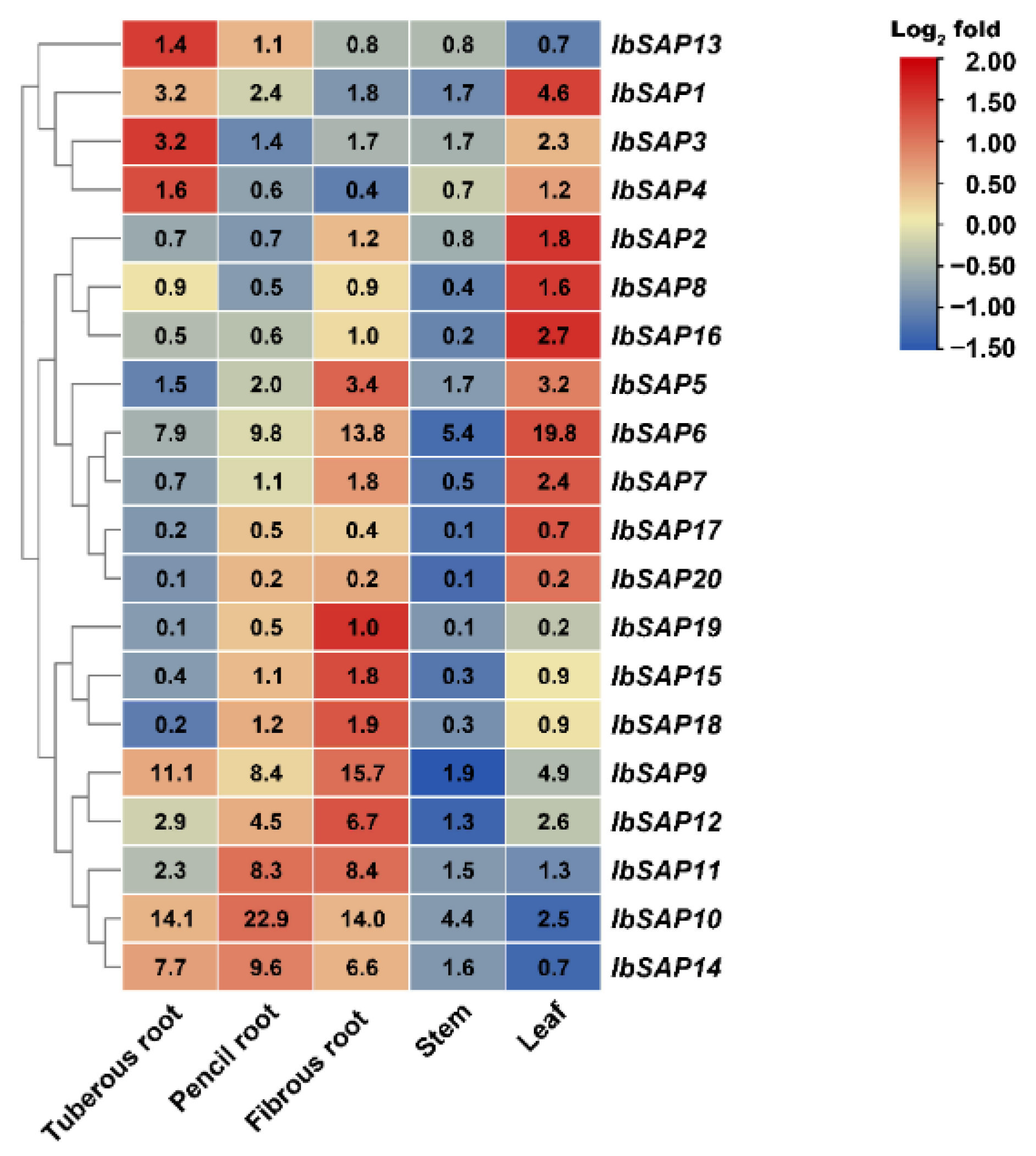
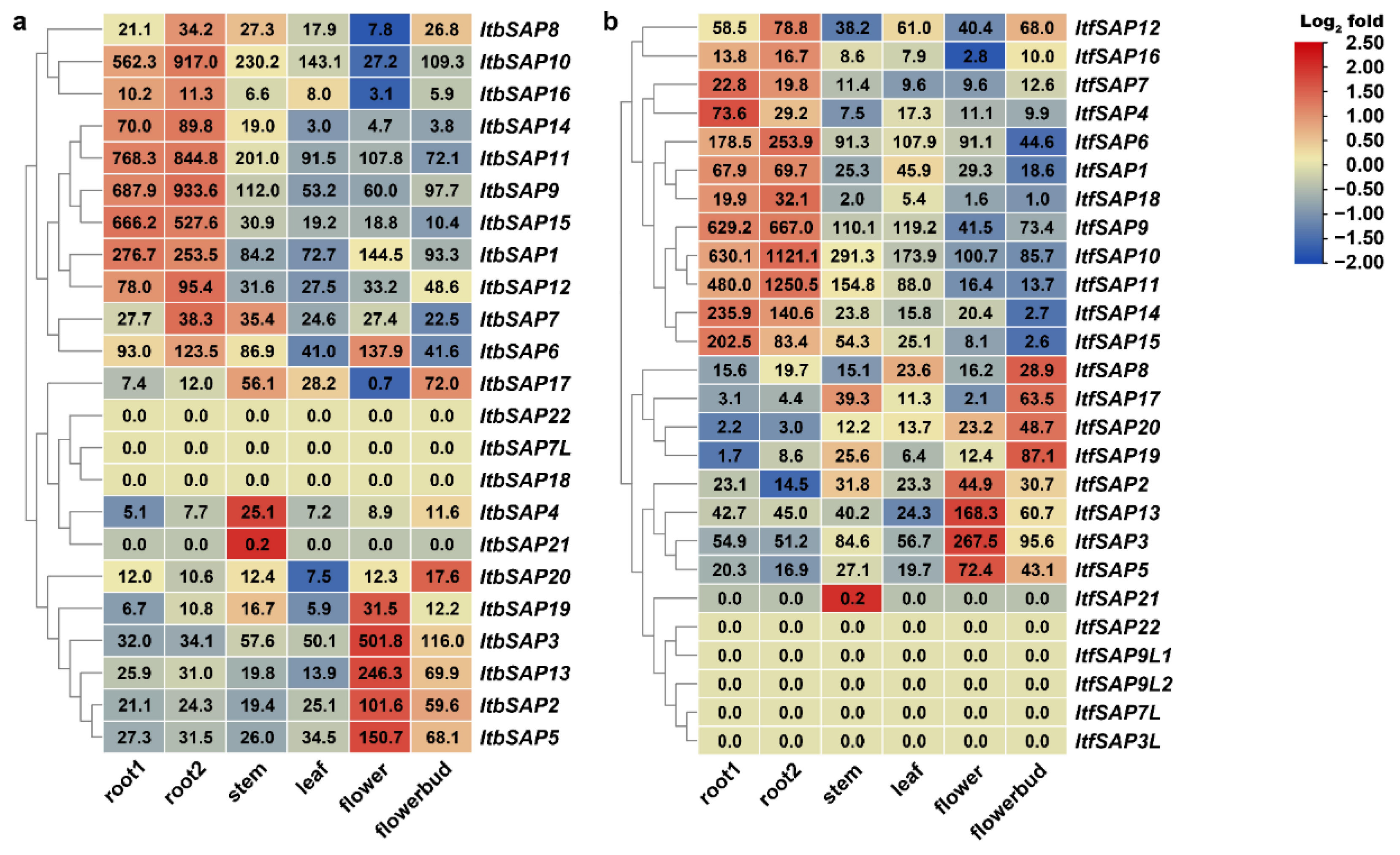
2.5. Expression Patterns in Different Organs of IbSAPs, ItbSAPs, and ItfSAPs
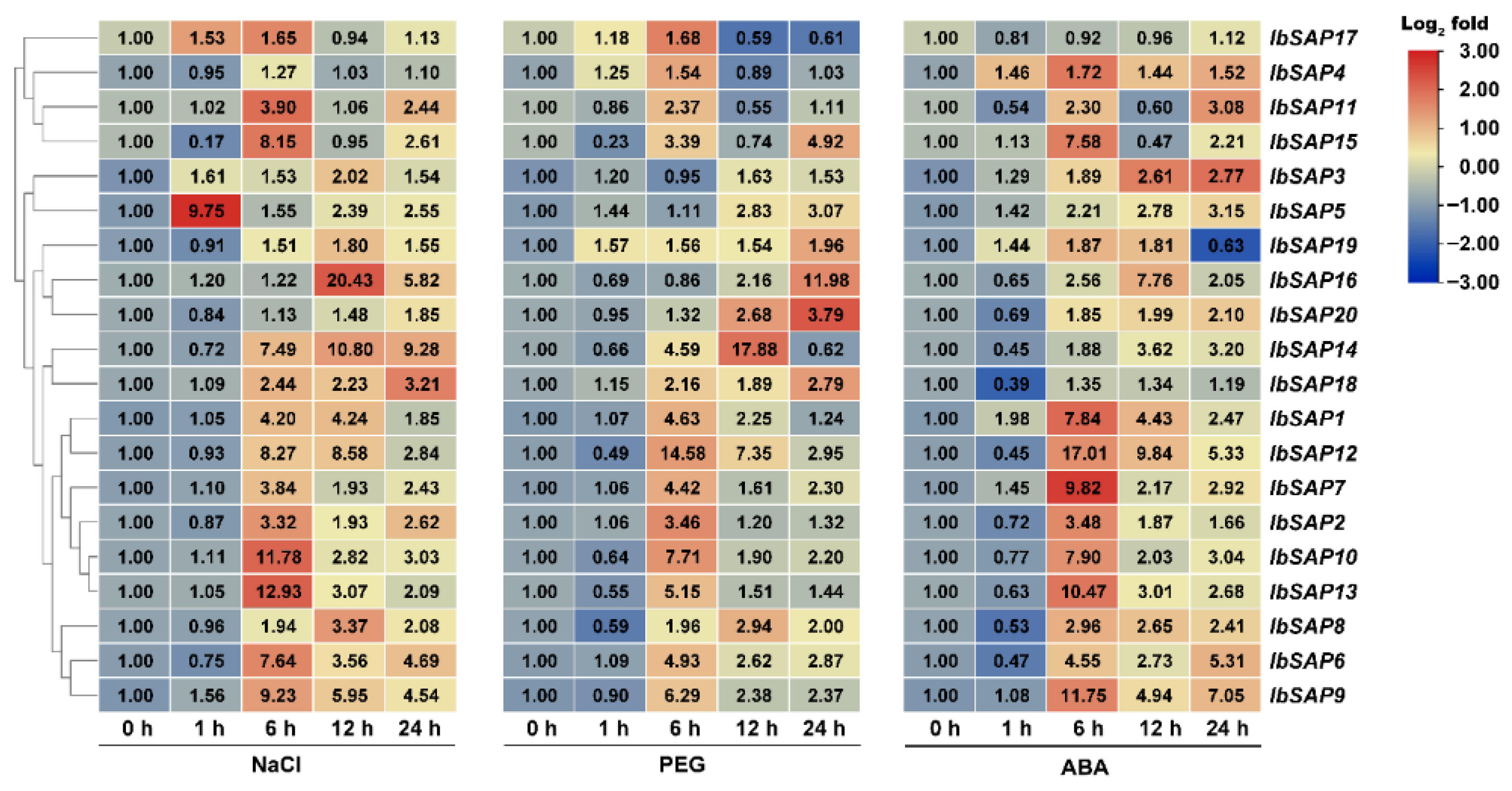
2.6. Expression Patterns of IbSAPs, ItbSAPs, and ItfSAPs in Response to Salinity, Drought, or ABA Treatments
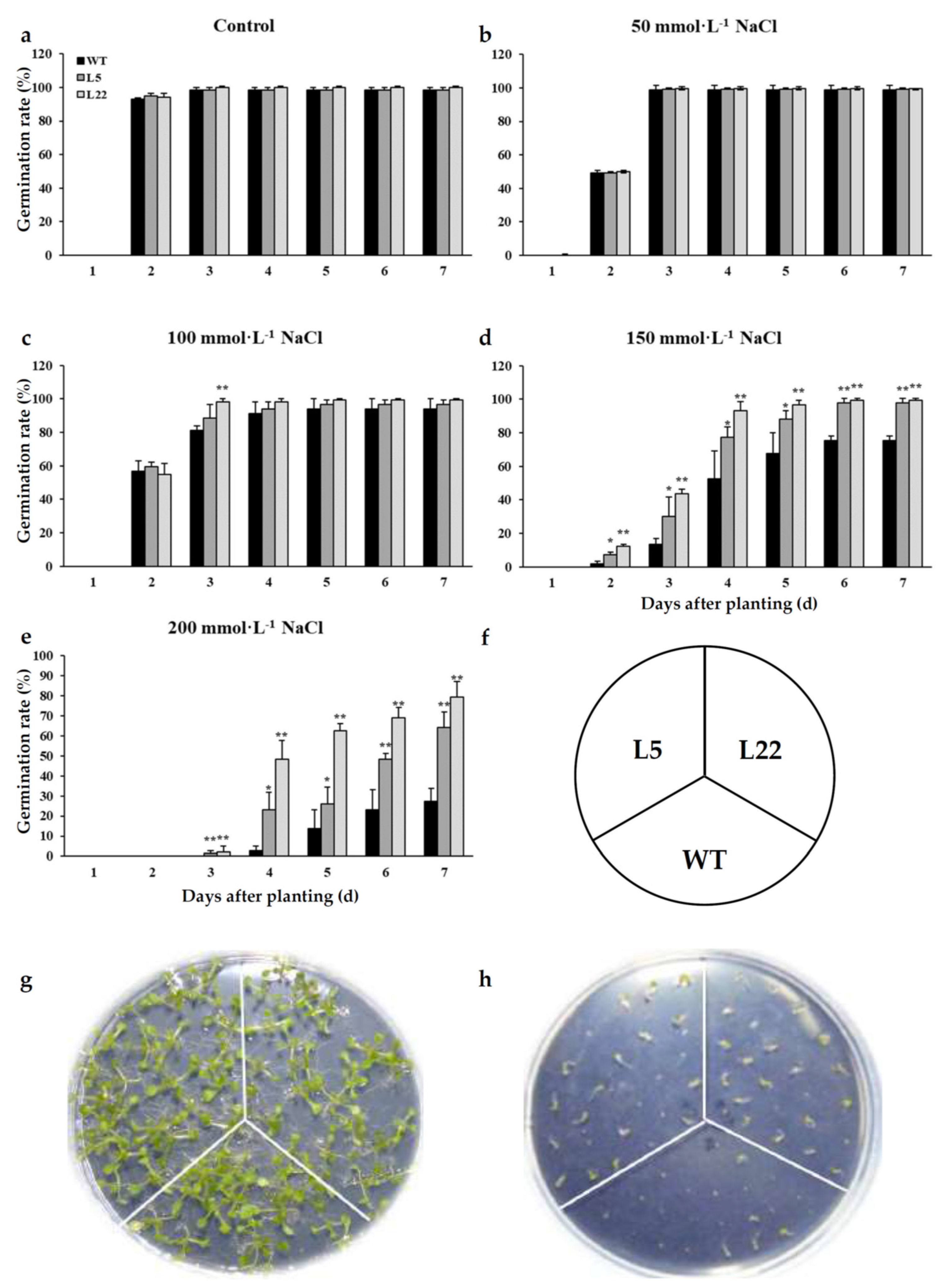
2.7. Ectopic Expression of IbSAP16 Enhanced Salinity Tolerance of Transgenic Arabidopsis Plants
3. Discussion
3.1. Evolution of the SAP Gene Family in Sweetpotato and Its Two Diploid Relatives
3.2. Different Functions of SAPs on Growth and Development between Sweetpotato and Its Two Diploid Relatives
3.3. Different Functions of SAPs in Multiple Abiotic Stress Responses between Sweetpotato and Its Two Diploid Relatives
3.4. AN1-AN1-C2H2-C2H2 Type SAPs Can Be Candidates to Improve Salinity Stress Tolerance of Plants
4. Materials and Methods
4.1. Characterization of SAP Members in Sweetpotato and Its Two Relative Species
4.2. Chromosomal Distribution of IbSAPs, ItbSAPs, and ItfSAPs
4.3. Phylogenetic Analysis of IbSAPs with SAPs from Six Other Plants
4.4. Analyses of Gene Structure and Conserved Domains of SAP Members
4.5. Cis-Acting Regulatory Elements Analyses of IbSAPs
4.6. Plant Materials
4.7. Stress Treatments of Sweetpotato Shoots
4.8. RNA Isolation and Quantitative Real-Time PCR Analysis
4.9. Transcriptome Analysis of ItbSAPs and ItfSAPs Expression Patterns
4.10. Plant Expression Vector Construction of IbSAP16 and Arabidopsis Transformation
4.11. Assay for Salinity Tolerance of Transgenic Arabidopsis
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Giri, J.; Dansana, P.K.; Kothari, K.S.; Sharma, G.; Vij, S.; Tyagi, A.K. SAPs as novel regulators of abiotic stress response in plants. Bioessays 2013, 35, 639–648. [Google Scholar] [CrossRef] [PubMed]
- Vij, S.; Tyagi, A.K. Genome-wide analysis of the stress associated protein (SAP) gene family containing A20/AN1 zinc-finger(s) in rice and their phylogenetic relationship with Arabidopsis. Mol. Genet. Genom. 2006, 276, 565–575. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, A.; Vij, S.; Tyagi, A.K. Overexpression of a zinc-finger protein gene from rice confers tolerance to cold, dehydration, and salt stress in transgenic tobacco. Proc. Natl. Acad. Sci. USA 2004, 101, 6309–6314. [Google Scholar] [CrossRef] [PubMed]
- Opipari, A.W., Jr.; Boguski, M.S.; Dixit, V.M. The A20 cDNA induced by tumor necrosis factor alpha encodes a novel type of zinc finger protein. J. Biol. Chem. 1990, 265, 14705–14708. [Google Scholar] [CrossRef]
- Linnen, J.M.; Bailey, C.P.; Weeks, D.L. Two related localized mRNAs from Xenopus laevis encode ubiquitin-like fusion proteins. Gene 1993, 128, 181–188. [Google Scholar] [CrossRef]
- Jin, Y.; Wang, M.; Fu, J.; Xuan, N.; Zhu, Y.; Lian, Y.; Jia, Z.; Zheng, J.; Wang, G. Phylogenetic and expression analysis of ZnF-AN1 genes in plants. Genomics 2007, 90, 265–275. [Google Scholar] [CrossRef]
- Giri, J.; Vij, S.; Dansana, P.K.; Tyagi, A.K. Rice A20/AN1 zinc-finger containing stress-associated proteins (SAP1/11) and a receptor-like cytoplasmic kinase (OsRLCK253) interact via A20 zinc-finger and confer abiotic stress tolerance in transgenic Arabidopsis plants. New Phytol. 2011, 191, 721–732. [Google Scholar] [CrossRef]
- Dansana, P.K.; Kothari, K.S.; Vij, S.; Tyagi, A.K. OsiSAP1 overexpression improves water-deficit stress tolerance in transgenic rice by affecting expression of endogenous stress-related genes. Plant Cell Rep. 2014, 33, 1425–1440. [Google Scholar] [CrossRef]
- Kim, G.-D.; Cho, Y.-H.; Yoo, S.-D. Regulatory functions of evolutionarily conserved AN1/A20-like Zinc finger family proteins in Arabidopsis stress responses under high temperature. Biochem. Biophys. Res. Commun. 2015, 457, 213–220. [Google Scholar] [CrossRef]
- Kang, M.; Fokar, M.; Abdelmageed, H.; Allen, R.D. Arabidopsis SAP5 functions as a positive regulator of stress responses and exhibits E3 ubiquitin ligase activity. Plant Mol. Biol. 2011, 75, 451–466. [Google Scholar] [CrossRef]
- Dixit, A.; Tomar, P.; Vaine, E.; Abdullah, H.; Hazen, S.; Dhankher, O.P. A stress-associated protein, AtSAP13, from Arabidopsis thaliana provides tolerance to multiple abiotic stresses. Plant Cell Environ. 2018, 41, 1171–1185. [Google Scholar] [CrossRef] [PubMed]
- Sharma, G.; Giri, J.; Tyagi, A.K. Rice OsiSAP7 negatively regulates ABA stress signalling and imparts sensitivity to water-deficit stress in Arabidopsis. Plant Sci. 2015, 237, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.; Lee, S.; Abdelmageed, H.; Reichert, A.; Lee, H.-K.; Fokar, M.; Mysore, K.S.; Allen, R.D. Arabidopsis stress associated protein 9 mediates biotic and abiotic stress responsive ABA signaling via the proteasome pathway. Plant Cell Environ. 2017, 40, 702–716. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, H.; Jha, S.; Sharma, M.; Giri, J.; Tyagi, A.K. Rice SAPs are responsive to multiple biotic stresses and overexpression of OsSAP1, an A20/AN1 zinc-finger protein, enhances the basal resistance against pathogen infection in tobacco. Plant Sci. 2014, 225, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Chang, H.-H.; Chang, J.-C.; Lu, H.-C.; Wang, T.-T.; Hsu, D.-W.; Tzean, Y.; Cheng, A.-P.; Chiu, Y.-S.; Yeh, H.-H. Plant A20/AN1 protein serves as the important hub to mediate antiviral immunity. PLoS Pathog. 2018, 14, e1007288. [Google Scholar] [CrossRef]
- Liu, S.; Wang, J.; Jiang, S.; Wang, H.; Gao, Y.; Zhang, H.; Li, D.; Song, F. Tomato SlSAP3, a member of the stress-associated protein family, is a positive regulator of immunity against Pseudomonas syringae pv. tomato DC3000. Mol. Plant Pathol. 2019, 20, 815–830. [Google Scholar] [CrossRef]
- Solanke, A.U.; Sharma, M.K.; Tyagi, A.K.; Sharma, A.K. Characterization and phylogenetic analysis of environmental stress-responsive SAP gene family encoding A20/AN1 zinc finger proteins in tomato. Mol. Genet. Genom. 2009, 282, 153–164. [Google Scholar] [CrossRef]
- Gao, W.; Long, L.; Tian, X.; Jin, J.; Liu, H.; Zhang, H.; Xu, F.; Song, C. Genome-wide identification and expression analysis of stress-associated proteins (SAPs) containing A20/AN1 zinc finger in cotton. Mol. Genet. Genom. 2016, 291, 2199–2213. [Google Scholar] [CrossRef]
- Zhou, Y.; Zeng, L.; Chen, R.; Wang, Y.; Song, J. Genome-wide identification and characterization of stress-associated protein (SAP) gene family encoding A20/AN1 zinc-finger proteins in Medicago truncatula. Arch. Biol. Sci. 2018, 70, 87–98. [Google Scholar] [CrossRef]
- Dong, Q.; Duan, D.; Zhao, S.; Xu, B.; Luo, J.; Wang, Q.; Huang, D.; Liu, C.; Li, C.; Gong, X.; et al. Genome-wide analysis and cloning of the apple stress-associated protein gene family reveals MdSAP15, which confers tolerance to drought and osmotic stresses in transgenic Arabidopsis. Int. J. Mol. Sci. 2018, 19, 2478. [Google Scholar] [CrossRef] [Green Version]
- He, X.; Xie, S.; Xie, P.; Yao, M.; Liu, W.; Qin, L.; Liu, Z.; Zheng, M.; Liu, H.; Guan, M.; et al. Genome-wide identification of stress-associated proteins (SAP) with A20/AN1 zinc finger domains associated with abiotic stresses responses in Brassica napus. Environ. Exp. Bot. 2019, 165, 108–119. [Google Scholar] [CrossRef]
- Zhang, X.-Z.; Zheng, W.-J.; Cao, X.-Y.; Cui, X.-Y.; Zhao, S.-P.; Yu, T.-F.; Chen, J.; Zhou, Y.-B.; Chen, M.; Chai, S.-C.; et al. Genomic analysis of stress associated proteins in Soybean and the role of GmSAP16 in abiotic stress responses in Arabidopsis and soybean. Front. Plant Sci. 2019, 10, 1453. [Google Scholar] [CrossRef] [PubMed]
- Lai, W.; Zhou, Y.; Pan, R.; Liao, L.; He, J.; Liu, H.; Yang, Y.; Liu, S. Identification and expression analysis of stress-associated proteins (SAPs) containing A20/AN1 zinc finger in Cucumber. Plants 2020, 9, 400. [Google Scholar] [CrossRef]
- Liu, Q. Improvement for agronomically important traits by gene engineering in sweetpotato. Breed. Sci. 2017, 67, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Qiu, X.; Yang, Y.; Kim, H.S.; Jia, X.; Yu, H.; Kwak, S.-S. Sweetpotato bZIP transcription factor IbABF4 confers tolerance to multiple abiotic stresses. Front. Plant Sci. 2019, 10, 630. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Xie, Y.; Sun, H.; Bian, X.; Ke, Q.; Kim, H.S.; Ji, C.Y.; Jin, R.; Wang, W.; Zhang, C.; et al. IbINH positively regulates drought stress tolerance in sweetpotato. Plant Physiol. Biochem. 2020, 146, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Liu, H.; Wang, H.; Zhang, P.; Shi, C. A novel sucrose transporter gene IbSUT4 involves in plant growth and response to abiotic stress through the ABF-dependent ABA signaling pathway in Sweetpotato. BMC Plant Biol. 2020, 20, 157. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, Z.; Li, X.; Gao, X.; Dai, Z.; Cui, Y.; Zhi, Y.; Liu, Q.; Zhai, H.; Gao, S.; et al. The IbBBX24–IbTOE3–IbPRX17 module enhances abiotic stress tolerance by scavenging reactive oxygen species in sweet potato. New Phytol. 2022, 233, 1133–1152. [Google Scholar] [CrossRef]
- Yang, J.; Moeinzadeh, M.-H.; Kuhl, H.; Helmuth, J.; Xiao, P.; Haas, S.; Liu, G.; Zheng, J.; Sun, Z.; Fan, W.; et al. Haplotype-resolved sweet potato genome traces back its hexaploidization history. Nat. Plants 2017, 3, 696–703. [Google Scholar] [CrossRef]
- Wu, S.; Lau, K.H.; Cao, Q.H.; Hamilton, J.P.; Sun, H.H.; Zhou, C.X.; Eserman, L.; Gemenet, D.C.; Olukolu, B.A.; Wang, H.Y.; et al. Genome sequences of two diploid wild relatives of cultivated sweetpotato reveal targets for genetic improvement. Nat. Commun. 2018, 9, 4580. [Google Scholar] [CrossRef] [Green Version]
- Hishiya, A.; Iemura, S.-I.; Natsume, T.; Takayama, S.; Ikeda, K.; Watanabe, K. A novel ubiquitin-binding protein ZNF216 functioning in muscle atrophy. EMBO J. 2006, 25, 554–564. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Teng, L.; Li, L.; Liu, T.; Li, L.; Chen, D.; Xu, L.-G.; Zhai, Z.; Shu, H.-B. ZNF216 is an A20-like and IκB Kinase γ-interacting inhibitor of NFκB activation. J. Biol. Chem. 2004, 279, 16847–16853. [Google Scholar] [CrossRef] [PubMed]
- Evans, P.C.; Ovaa, H.; Hamon, M.; Kilshaw, P.J.; Hamm, S.; Bauer, S.; Ploegh, H.L.; Smith, T.S. Zinc-finger protein A20, a regulator of inflammation and cell survival, has de-ubiquitinating activity. Biochem. J. 2004, 378, 727–734. [Google Scholar] [CrossRef] [PubMed]
- Opipari, A.W., Jr.; Hu, H.M.; Yabkowitz, R.; Dixit, V.M. The A20 zinc finger protein protects cells from tumor necrosis factor cytotoxicity. J. Biol. Chem. 1992, 267, 12424–12427. [Google Scholar] [CrossRef]
- Eserman, L.A.; Tiley, G.P.; Jarret, R.L.; Leebens-Mack, J.H.; Miller, R.E. Phylogenetics and diversification of morning glories (tribe Ipomoeeae, Convolvulaceae) based on whole plastome sequences. Am. J. Bot. 2014, 101, 92–103. [Google Scholar] [CrossRef]
- Rajapakse, S.; Nilmalgoda, S.D.; Molnar, M.; Ballard, R.E.; Austin, D.F.; Bohac, J.R. Phylogenetic relationships of the sweetpotato in Ipomoea series Batatas (Convolvulaceae) based on nuclear β-amylase gene sequences. Mol. Phylogenet. Evol. 2004, 30, 623–632. [Google Scholar] [CrossRef]
- Zhang, Y.; Lan, H.; Shao, Q.; Wang, R.; Chen, H.; Tang, H.; Zhang, H.; Huang, J. An A20/AN1-type zinc finger protein modulates gibberellins and abscisic acid contents and increases sensitivity to abiotic stress in rice (Oryza sativa). J. Exp. Bot. 2015, 67, 315–326. [Google Scholar] [CrossRef]
- Kang, M.; Abdelmageed, H.; Lee, S.; Reichert, A.; Mysore, K.S.; Allen, R.D. AtMBP-1, an alternative translation product of LOS2, affects abscisic acid responses and is modulated by the E3 ubiquitin ligase AtSAP5. Plant J. 2013, 76, 481–493. [Google Scholar] [CrossRef]
- Ströher, E.; Wang, X.-J.; Roloff, N.; Klein, P.; Husemann, A.; Dietz, K.-J. Redox-dependent regulation of the stress-induced zinc-finger protein SAP12 in Arabidopsis thaliana. Mol. Plant 2009, 2, 357–367. [Google Scholar] [CrossRef]
- Sreedharan, S.; Shekhawat, U.K.S.; Ganapathi, T.R. MusaSAP1, a A20/AN1 zinc finger gene from banana functions as a positive regulator in different stress responses. Plant Mol. Biol. 2012, 80, 503–517. [Google Scholar] [CrossRef]
- Dixit, A.R.; Dhankher, O.P. A novel stress-associated protein ‘AtSAP10′ from Arabidopsis thaliana confers tolerance to nickel, manganese, zinc, and high temperature stress. PLoS ONE 2011, 6, e20921. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xu, Y.; Xiao, J.; Ma, Q.; Li, D.; Xue, Z.; Chong, K. OsDOG, a gibberellin-induced A20/AN1 zinc-finger protein, negatively regulates gibberellin-mediated cell elongation in rice. J. Plant Physiol. 2011, 168, 1098–1105. [Google Scholar] [CrossRef] [PubMed]
- Dong, F.; Zhang, D.; Zhang, C. Research progress and prospect on prevention and control measures of sweet potato virus disease. Agric. Biotechnol. 2018, 7, 54–59. [Google Scholar]
- Shu, X.; Ding, L.; Gu, B.; Zhang, H.; Guan, P.; Zhang, J. A stress associated protein from Chinese wild Vitis amurensis, VaSAP15, enhances the cold tolerance of transgenic grapes. Sci. Hortic. 2021, 285, 110147. [Google Scholar] [CrossRef]
- Xu, Q.-F.; Mao, X.-G.; Wang, Y.-X.; Wang, J.-Y.; Xi, Y.-J.; Jing, R.-L. A wheat gene TaSAP17-D encoding an AN1/AN1 zinc finger protein improves salt stress tolerance in transgenic Arabidopsis. J. Integr. Agric. 2018, 17, 507–516. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar]
- Goldberg, T.; Hecht, M.; Hamp, T.; Karl, T.; Yachdav, G.; Ahmed, N.; Altermann, U.; Angerer, P.; Ansorge, S.; Balasz, K.; et al. LocTree3 prediction of localization. Nucleic Acids Res. 2014, 42, W350–W355. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, B.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.-H. Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 2019, 47, W270–W275. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Nijveen, H.; Rao, X.; Bisseling, T.; Geurts, R.; Leunissen, J.A. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007, 35, W71–W74. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-C.; Kim, Y.-H.; Ji, C.Y.; Park, S.; Jeong, J.C.; Lee, H.-S.; Kwak, S.-S. Stable internal reference genes for the normalization of real-time PCR in different sweetpotato cultivars subjected to abiotic stress conditions. PLoS ONE 2012, 7, e51502. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]


| Gene ID | Locus 1 | CDS Length (bp) | Protein Length (aa) | pI | Mw (kDa) | GRAVY | Predicted Subcellular Localization |
|---|---|---|---|---|---|---|---|
| IbSAP1 | g30531 | 507 | 168 | 7.98 | 17.87 | −0.504 | cytoplasm |
| IbSAP2 | g43065 | 525 | 174 | 7.98 | 18.32 | −0.346 | cytoplasm |
| IbSAP3 | g11483 | 516 | 171 | 7.51 | 18.28 | −0.488 | cytoplasm |
| IbSAP4 | g57575, g57655 | 507 | 168 | 7.49 | 18.11 | −0.388 | cytoplasm |
| IbSAP5 | g50881 | 513 | 170 | 7.49 | 18.07 | −0.379 | cytoplasm |
| IbSAP6 | g58165 | 456 | 151 | 9.05 | 16.32 | −0.418 | cytoplasm |
| IbSAP7 | g8952 | 489 | 162 | 8.52 | 17.29 | −0.430 | cytoplasm |
| IbSAP8 | g50882 | 429 | 142 | 8.67 | 15.59 | −0.601 | cytoplasm |
| IbSAP9 | g30530 | 519 | 172 | 9.06 | 19.12 | −0.935 | cytoplasm |
| IbSAP10 | g8953 | 525 | 174 | 9.14 | 19.07 | −0.726 | cytoplasm |
| IbSAP11 | g57577, g57652 | 501 | 166 | 9.36 | 18.18 | −0.713 | cytoplasm |
| IbSAP12 | g4341 | 507 | 168 | 8.72 | 18.20 | −0.813 | cytoplasm |
| IbSAP13 | / | 537 | 178 | 8.81 | 18.63 | −0.365 | cytoplasm |
| IbSAP14 | g38216 | 474 | 157 | 9.48 | 16.89 | −0.492 | cytoplasm |
| IbSAP15 | g30138 | 579 | 192 | 9.06 | 20.96 | −0.588 | endoplasmic reticulum |
| IbSAP16 | g43042 | 834 | 277 | 8.63 | 30.59 | −0.577 | endoplasmic reticulum |
| IbSAP17 | / | 885 | 294 | 8.63 | 32.56 | −0.640 | nucleus |
| IbSAP18 | g15701, g19161 | 465 | 154 | 8.65 | 16.71 | −0.501 | cytoplasm |
| IbSAP19 | g25751 | 528 | 175 | 8.79 | 18.48 | −0.608 | cytoplasm |
| IbSAP20 | g25129 | 435 | 144 | 9.21 | 15.40 | −0.363 | cytoplasm |
| Cis-Acting Elements | Functions | Sequences | Genes |
|---|---|---|---|
| Hormone response cis-acting elements | |||
| ABRE | cis-acting element involved in the abscisic acid responsiveness | ACGTG | IbSAP2–5, 8–12, 14–16, 19 |
| as-1 | involved in the response to auxin, salicylic acid, and methyl jasmonate | TGACG | IbSAP1, 3–5, 7–12, 14, 15, 18, 19 |
| ERE | ethene responsive element | ATTTCATA/ ATTTTAAA | IbSAP2, 5, 6, 8–12, 15, 16, 18–20 |
| GARE-motif | gibberellin-responsive element | TCTGTTG | IbSAP1, 7, 8, 12, 16 |
| P-box | gibberellin-responsive element | CCTTTTG | IbSAP1, 2, 4, 11, 12, 14, 15, 20 |
| TCA-element | cis-acting element involved in salicylic acid responsiveness | TCATCTTCAT/ CCATCTTTTT/ TCAGAAGAGG | IbSAP3, 4, 5, 7, 8, 10, 15, 16, 19 |
| TGA-box | auxin-responsive element | TGACGTAA/ AACGAC | IbSAP1, 7–11, 14, 16, 19 |
| Stress response cis-acting elements | |||
| DRE | drought responsive element | GCCGAC/ ACCGAGA | IbSAP3, 5, 8, 15 |
| LTR | cis-acting element involved in low-temperature responsiveness | CCGAAA | IbSAP2, 3, 5, 7, 8, 11, 15, 18, 19 |
| MBS | MYB binding site involved in drought-inducibility | CAACTG | IbSAP2, 7, 8–10, 12, 15, 16 |
| MYB | MYB binding site | CAACGG/CAACAG/ CAACCA/TAACCA/ TAACTG/CAACTG/ CCGTTG | IbSAP1-IbSAP12, 14–16, 18–20 |
| STRE | stress-responsive elements | AGGGG | IbSAP2, 3, 5–11, 15, 16, 18, 20 |
| TC-rich repeats | cis-acting element involved in defense and stress responsiveness | ATTCTCTAAC | IbSAP1, 2, 5, 8 |
| W-box | cis-acting element involved in sugar metabolism and plant defense signaling | TTGACC | IbSAP1, 2, 4–6, 9, 10, 12, 14–16, 18, 20 |
| WRE3 | wound-responsive element | CCACCT | IbSAP2–5, 7, 11, 15, 18–20 |
| WUN-motif | wound-responsive element | AAATTACT/ AAATTTCTT/ TTATTACAT/ CAATTACAT/ AAATTTCCT | IbSAP1, 5, 6, 8, 10, 12, 14, 16, 18–20 |
| Light signal response cis-acting elements | |||
| Box 4 | part of a conserved DNA module involved in light responsiveness | ATTAAT | IbSAP2–6, 8–12, 14, 15, 18–20 |
| TCT-motif | part of a light responsive element | TCTTAC | IbSAP3, 5–8, 11, 14, 15, 18, 19 |
| Sp1 | light responsive element | GGGCGG | IbSAP3, 9, 11, 12 |
| chs-CMA1a/2a | part of a light responsive element | TTACTTAA/ TCACTTGA | IbSAP3–10 |
| GATA-motif | part of a light responsive element | AAGATAAGATT/ AAGGATAAGG/ GATAGGA/ GATAGGG | IbSAP1, 4, 6, 8–11, 14–16, 18, 20 |
| G-box | cis-acting regulatory element involved in light responsiveness | TACGTG/ CACGTG/CACGTC /CACGTT/CACGAC | IbSAP2–12, 14–16, 19 |
| GT1-motif | light responsive element | GGTTAA | IbSAP3, 6, 7, 9, 12, 14, 19, 20 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, H.; Yang, Q.; Wang, X.; Schläppi, M.R.; Yan, H.; Kou, M.; Tang, W.; Wang, X.; Zhang, Y.; Li, Q.; et al. Genome-Wide Identification of the A20/AN1 Zinc Finger Protein Family Genes in Ipomoea batatas and Its Two Relatives and Function Analysis of IbSAP16 in Salinity Tolerance. Int. J. Mol. Sci. 2022, 23, 11551. https://doi.org/10.3390/ijms231911551
Xie H, Yang Q, Wang X, Schläppi MR, Yan H, Kou M, Tang W, Wang X, Zhang Y, Li Q, et al. Genome-Wide Identification of the A20/AN1 Zinc Finger Protein Family Genes in Ipomoea batatas and Its Two Relatives and Function Analysis of IbSAP16 in Salinity Tolerance. International Journal of Molecular Sciences. 2022; 23(19):11551. https://doi.org/10.3390/ijms231911551
Chicago/Turabian StyleXie, Hao, Qiangqiang Yang, Xiaoxiao Wang, Michael R. Schläppi, Hui Yan, Meng Kou, Wei Tang, Xin Wang, Yungang Zhang, Qiang Li, and et al. 2022. "Genome-Wide Identification of the A20/AN1 Zinc Finger Protein Family Genes in Ipomoea batatas and Its Two Relatives and Function Analysis of IbSAP16 in Salinity Tolerance" International Journal of Molecular Sciences 23, no. 19: 11551. https://doi.org/10.3390/ijms231911551







