An Integrative Analysis of Identified Schizophrenia-Associated Brain Cell Types and Gene Expression Changes
Abstract
:1. Introduction
2. Results
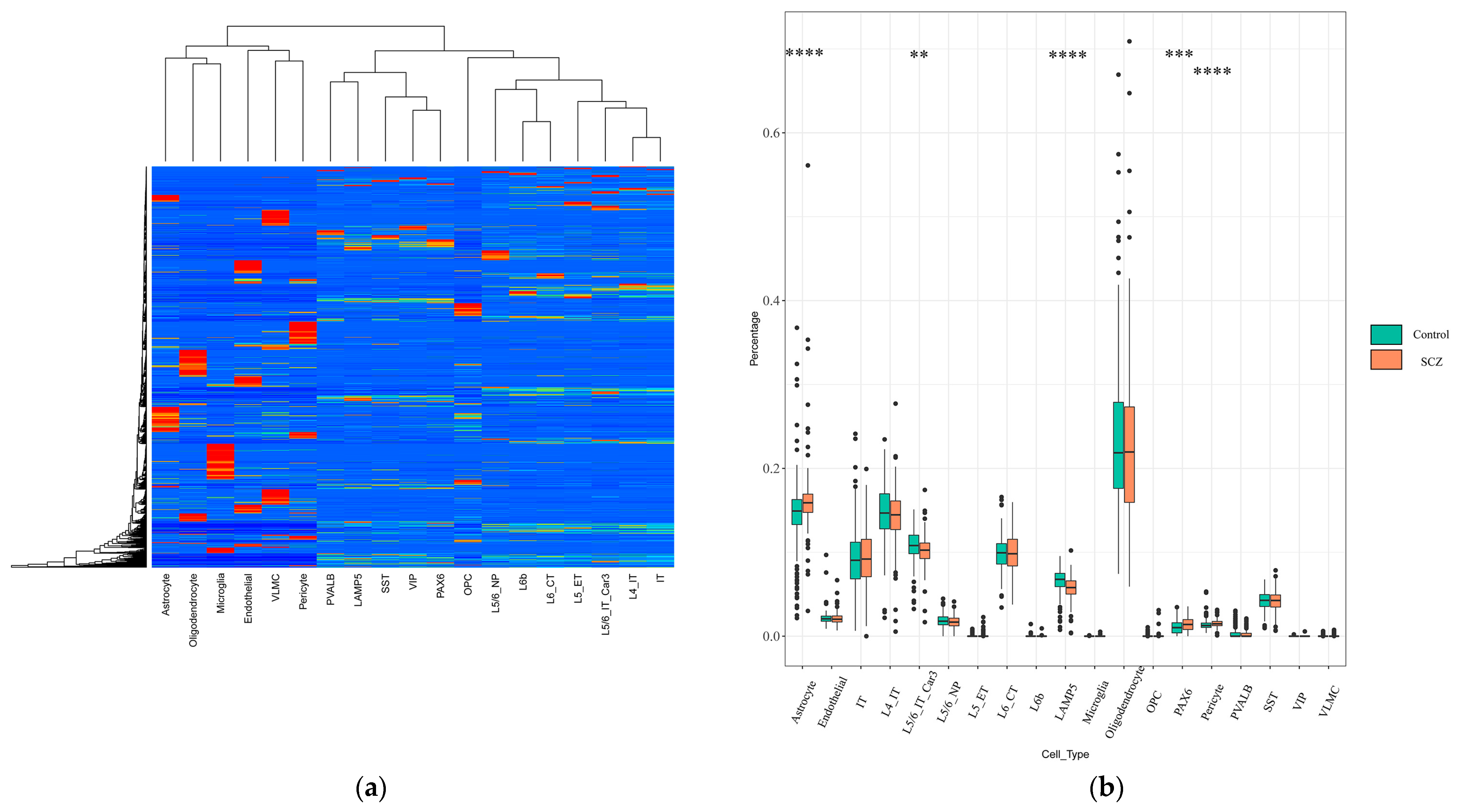
2.1. Identification of Changes in Cell-Type Proportion Using Signature Matrix
2.2. Bulk RNA-Seq Revealed Abnormal Gene Expression and Immune-Related Pathways in SCZ Samples
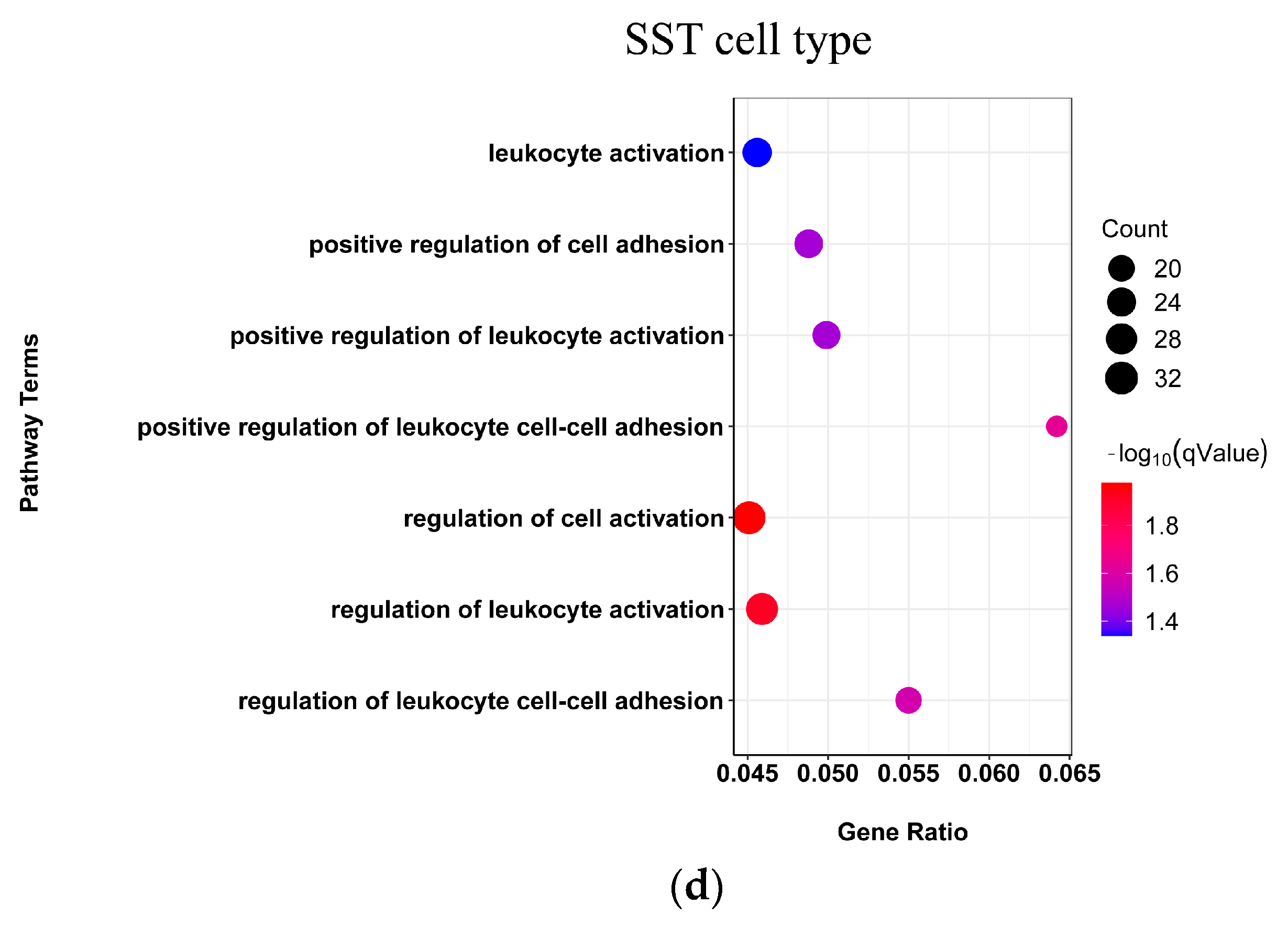
2.3. PVALB, SST, and LAMP5 Neurons Involved in Chemical Synaptic Transmission and Immune Function Dysregulation in SCZ
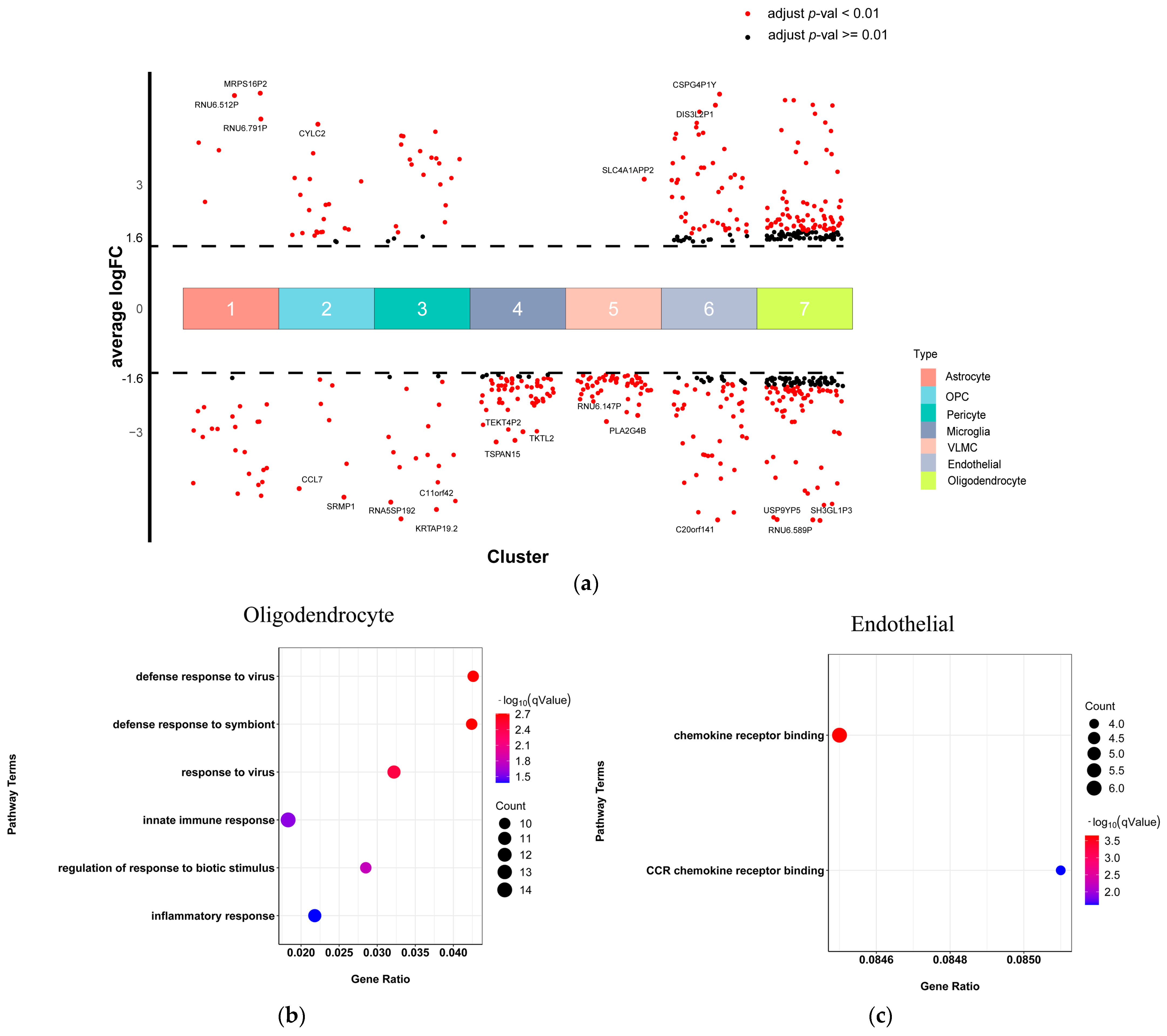
2.4. Oligodendrocyte and Endothelial Cells Involved in Immune Function Dysregulation in SCZ
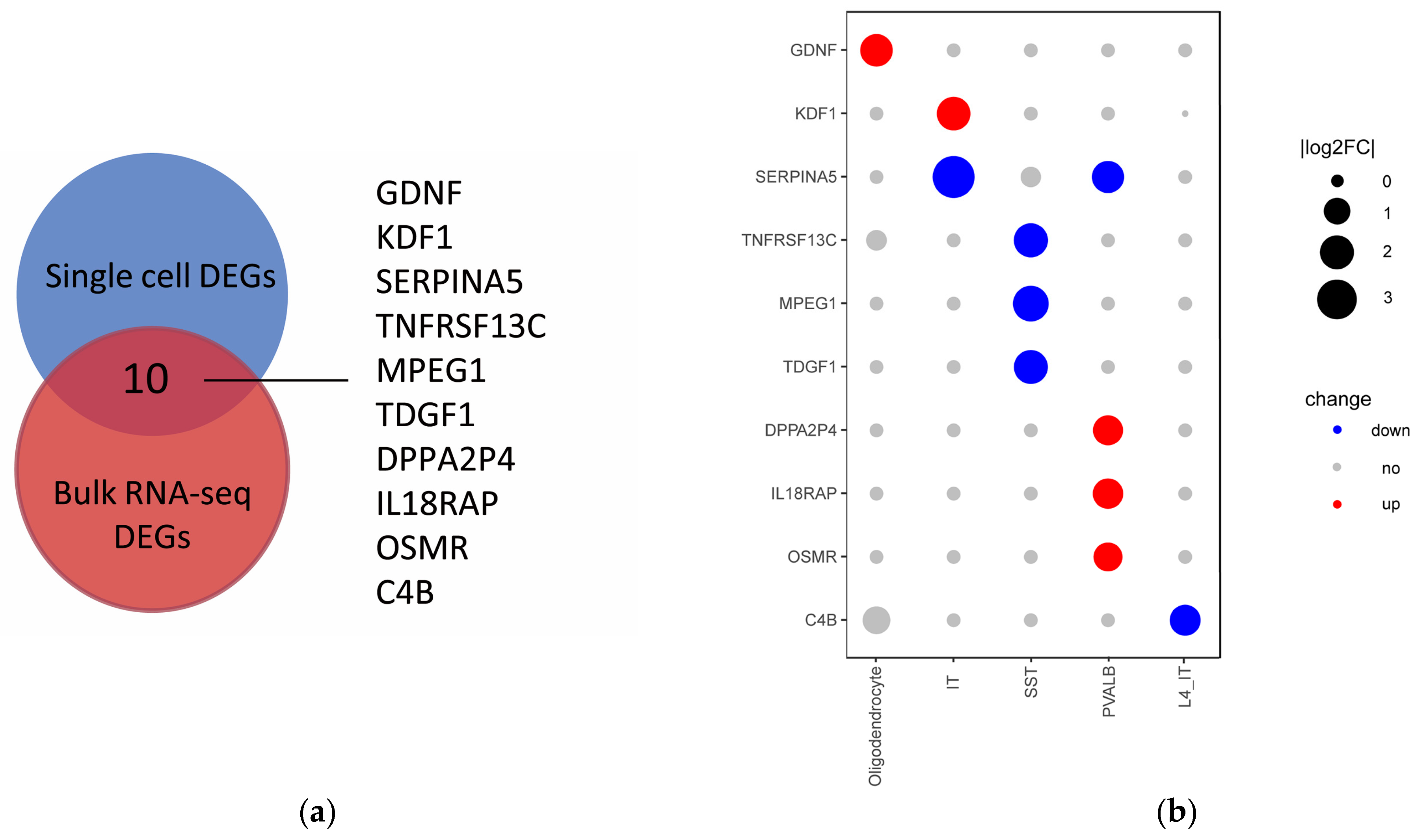
2.5. Bulk RNA-Seq DEGs Expression Changes at the Cellular Layer
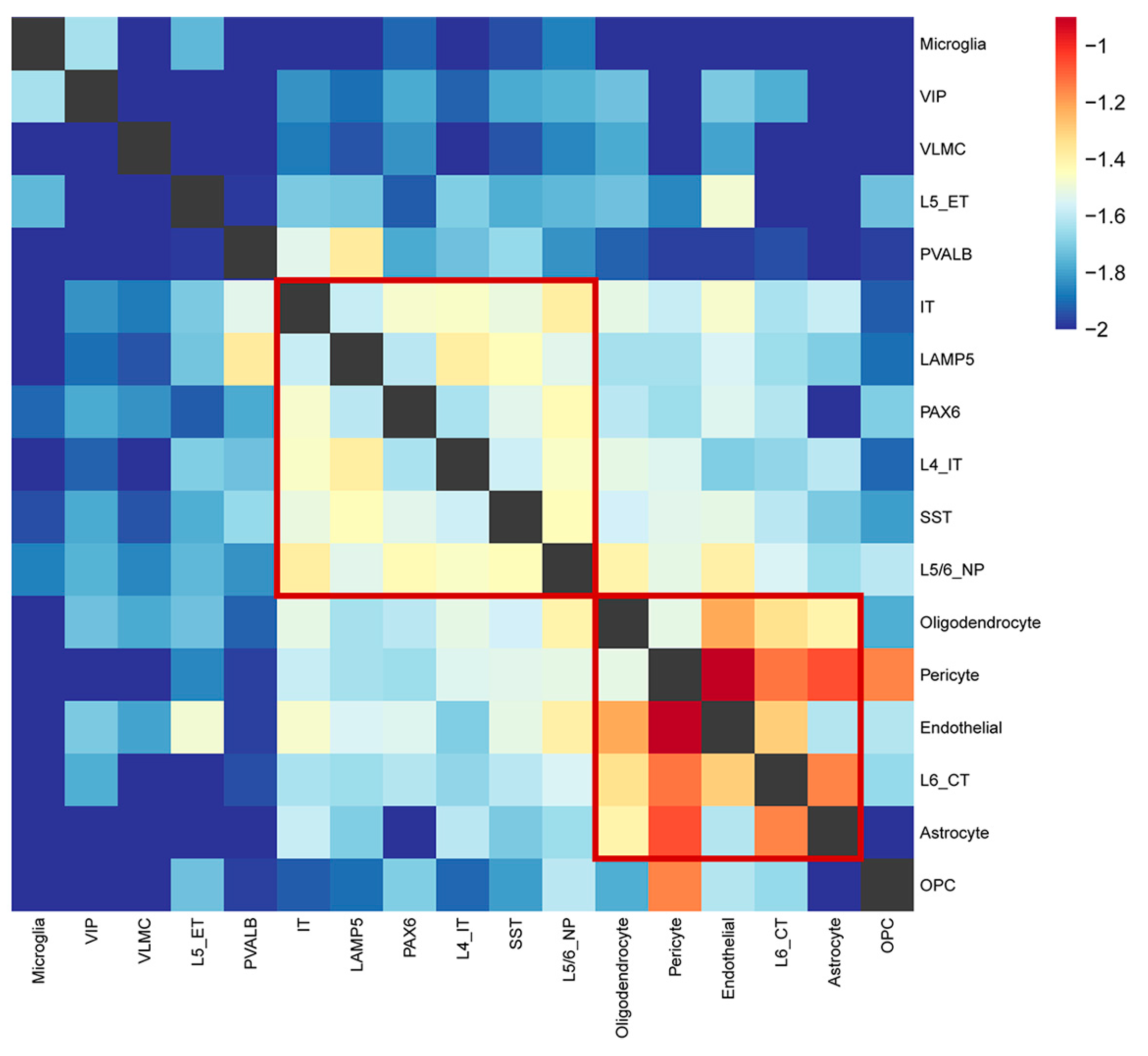
2.6. The Relationships between Each Cell Type Identifying Two Major Modules Underlying SCZ
3. Discussion
4. Materials and Methods
4.1. Data Collection
4.2. Impute Cell Fractions with CIBERSORTx
4.3. Bulk RNA-Seq Differentially Expressed Gene Analysis
4.4. Impute Cell-Type-Specific Gene Expression with CIBERSORTx
4.5. Gene Ontology (GO) Analysis
4.6. Jaccard Similarity between Each Cell Types
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kuperberg, G.; Heckers, S. Schizophrenia and cognitive function. Curr. Opin. Neurobiol. 2000, 10, 205–210. [Google Scholar] [CrossRef]
- Zhang, T.; Zhu, L.; Ni, T.; Liu, D.; Chen, G.; Yan, Z.; Lin, H.; Guan, F.; Rice, J.P. Voltage-gated calcium channel activity and complex related genes and schizophrenia: A systematic investigation based on Han Chinese population. J. Psychiatr. Res. 2018, 106, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Rapoport, J.L.; Addington, A.; Frangou, S.; Psych, M.R.C. The neurodevelopmental model of schizophrenia: Update 2005. Mol. Psychiatry 2005, 10, 434–449. [Google Scholar] [CrossRef] [PubMed]
- Potkin, S.G.; Turner, J.; Brown, G.G.; McCarthy, G.; Greve, D.N.; Glover, G.H.; Manoach, D.S.; Belger, A.; Diaz, M.; Wible, C.G.; et al. Working memory and DLPFC inefficiency in schizophrenia: The FBIRN study. Schizophr. Bull. 2009, 35, 19–31. [Google Scholar] [CrossRef]
- Finn, E.S.; Huber, L.; Jangraw, D.C.; Molfese, P.J.; Bandettini, P.A. Layer-dependent activity in human prefrontal cortex during working memory. Nat. Neurosci. 2019, 22, 1687–1695. [Google Scholar] [CrossRef]
- Sullivan, C.R.; Koene, R.H.; Hasselfeld, K.; O’Donovan, S.; Ramsey, A.; McCullumsmith, R.E. Neuron-specific deficits of bioenergetic processes in the dorsolateral prefrontal cortex in schizophrenia. Mol. Psychiatry 2019, 24, 1319–1328. [Google Scholar] [CrossRef]
- Smucny, J.; Dienel, S.J.; Lewis, D.A.; Carter, C.S. Mechanisms underlying dorsolateral prefrontal cortex contributions to cognitive dysfunction in schizophrenia. Neuropsychopharmacology 2021, 47, 292–308. [Google Scholar] [CrossRef]
- Koboldt, D.C.; Mosher, T.M.; Kelly, B.J.; Sites, E.; Bartholomew, D.; Hickey, S.E.; McBride, K.; Wilson, R.K.; White, P. A de novo nonsense mutation in ASXL3 shared by siblings with Bainbridge–Ropers syndrome. Mol. Case Study 2018, 4, a002410. [Google Scholar] [CrossRef]
- Braff, D.L.; Heaton, R.; Kuck, J.; Cullum, M.; Moranville, J.; Grant, I.; Zisook, S. The Generalized Pattern of Neuropsychological Deficits in Outpatients with Chronic Schizophrenia with Heterogeneous Wisconsin Card Sorting Test Results. Arch. Gen. Psychiatry 1991, 48, 891–898. [Google Scholar] [CrossRef]
- Blackwood, D.H.R. Auditory P300 and Eye Tracking Dysfunction in Schizophrenic Pedigrees. Arch. Gen. Psychiatry 1991, 48, 899–909. [Google Scholar] [CrossRef]
- Kim, R.; Healey, K.L.; Sepulveda-Orengo, M.T.; Reissner, K.J. Astroglial correlates of neuropsychiatric disease: From astrocytopathy to astrogliosis. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2018, 87, 126–146. [Google Scholar] [CrossRef] [PubMed]
- Samartzis, L.; Dima, D.; Fusar-Poli, P.; Kyriakopoulos, M. White Matter Alterations in Early Stages of Schizophrenia: A Systematic Review of Diffusion Tensor Imaging Studies. J. Neuroimaging 2014, 24, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Skene, N.G.; Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium; Bryois, J.; Bakken, T.E.; Breen, G.; Crowley, J.J.; Gaspar, H.A.; Giusti-Rodriguez, P.; Hodge, R.D.; Miller, J.A.; et al. Genetic identification of brain cell types underlying schizophrenia. Nat. Genet. 2018, 50, 825–833. [Google Scholar] [CrossRef] [PubMed]
- Lam, M.; Schizophrenia Working Group of the Psychiatric Genomics Consortium; Chen, C.-Y.; Li, Z.; Martin, A.R.; Bryois, J.; Ma, X.; Gaspar, H.; Ikeda, M.; Benyamin, B.; et al. Comparative genetic architectures of schizophrenia in East Asian and European populations. Nat. Genet. 2019, 51, 1670–1678. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.M.; Steen, C.B.; Liu, C.L.; Gentles, A.J.; Chaudhuri, A.A.; Scherer, F.; Khodadoust, M.S.; Esfahani, M.S.; Luca, B.A.; Steiner, D.; et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 2019, 37, 773–782. [Google Scholar] [CrossRef] [PubMed]
- Tasic, B.; Yao, Z.; Graybuck, L.T.; Smith, K.A.; Nguyen, T.N.; Bertagnolli, D.; Goldy, J.; Garren, E.; Economo, M.N.; Viswanathan, S.; et al. Shared and distinct transcriptomic cell types across neocortical areas. Nature 2018, 563, 72–78. [Google Scholar] [CrossRef]
- Hodge, R.D.; Bakken, T.E.; Miller, J.A.; Smith, K.A.; Barkan, E.R.; Graybuck, L.T.; Close, J.L.; Long, B.; Johansen, N.; Penn, O.; et al. Conserved cell types with divergent features in human versus mouse cortex. Nature 2019, 573, 61–68. [Google Scholar] [CrossRef]
- Collado-Torres, L.; Burke, E.E.; Peterson, A.; Shin, J.; Straub, R.E.; Rajpurohit, A.; Semick, S.A.; Ulrich, W.S.; BrainSeq Consortium; Price, A.J.; et al. Regional Heterogeneity in Gene Expression, Regulation, and Coherence in the Frontal Cortex and Hippocampus across Development and Schizophrenia. Neuron 2018, 103, 203–216.e8. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Smyth, G.K. limma: Linear Models for Microarray Data. In Bioinformatics and Computational Biology Solutions Using R and Bioconductor; Springer: New York, NY, USA, 2005; pp. 397–420. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Steen, C.B.; Liu, C.L.; Alizadeh, A.A.; Newman, A.M. Profiling Cell Type Abundance and Expression in Bulk Tissues with CIBERSORTx. Methods Mol. Biol. 2020, 2117, 135–157. [Google Scholar] [CrossRef]
- Bayly-Jones, C.; Pang, S.S.; Spicer, B.A.; Whisstock, J.C.; Dunstone, M.A. Ancient but Not Forgotten: New Insights into MPEG1, a Macrophage Perforin-Like Immune Effector. Front. Immunol. 2020, 11, 2690. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Kong, W.; Wang, S. Peripheral Blood Biomarkers CXCL12 and TNFRSF13C Associate with Cerebrospinal Fluid Biomarkers and Infiltrating Immune Cells in Alzheimer Disease. J. Mol. Neurosci. 2021, 71, 1485–1494. [Google Scholar] [CrossRef] [PubMed]
- De La Cruz, J.M.; Bamford, R.N.; Burdine, R.D.; Roessler, E.; Barkovich, J.A.; Donnai, D.; Schier, A.F.; Muenke, M. A loss-of-function mutation in the CFC domain of TDGF1 is associated with human forebrain defects. Qual. Life Res. 2002, 110, 422–428. [Google Scholar] [CrossRef] [PubMed]
- Crist, A.M.; Hinkle, K.M.; Wang, X.; Moloney, C.M.; Matchett, B.J.; Labuzan, S.A.; Frankenhauser, I.; Azu, N.O.; Liesinger, A.M.; Lesser, E.R.; et al. Transcriptomic analysis to identify genes associated with selective hippocampal vulnerability in Alzheimer’s disease. Nat. Commun. 2021, 12, 2311. [Google Scholar] [CrossRef]
- Liu, H.; Irwanto, A.; Tian, H.; Fu, X.; Yu, Y.; Yu, G.; Low, H.; Chu, T.; Li, Y.; Shi, B.; et al. Identification of IL18RAP/IL18R1 and IL12B as Leprosy Risk Genes Demonstrates Shared Pathogenesis between Inflammation and Infectious Diseases. Am. J. Hum. Genet. 2012, 91, 935–941. [Google Scholar] [CrossRef]
- Tunca, Z.; Akdede, B.K.; Özerdem, A.; Alkın, T.; Polat, S.; Ceylan, D.; Bayın, M.; Kocuk, N.C.; Şimşek, S.; Resmi, H.; et al. Diverse Glial Cell Line-Derived Neurotrophic Factor (GDNF) Support Between Mania and Schizophrenia: A Comparative Study in Four Major Psychiatric Disorders. Eur. Psychiatry 2015, 30, 198–204. [Google Scholar] [CrossRef]
- Griffin, P.; Sheehan, P.W.; Dimitry, J.M.; Guo, C.; Kanan, M.F.; Lee, J.; Zhang, J.; Musiek, E.S. REV-ERBα mediates complement expression and diurnal regulation of microglial synaptic phagocytosis. eLife 2020, 9, e58765. [Google Scholar] [CrossRef]
- Notter, T. Astrocytes in schizophrenia. Brain Neurosci. Adv. 2021, 5, 23982128211009148. [Google Scholar] [CrossRef]
- Schnieder, T.P.; Dwork, A.J. Searching for Neuropathology: Gliosis in Schizophrenia. Biol. Psychiatry 2011, 69, 134–139. [Google Scholar] [CrossRef]
- Steiner, J.; Bernstein, H.-G.; Bielau, H.; Farkas, N.; Winter, J.; Dobrowolny, H.; Brisch, R.; Gos, T.; Mawrin, C.; Myint, A.M.; et al. S100B-immunopositive glia is elevated in paranoid as compared to residual schizophrenia: A morphometric study. J. Psychiatr. Res. 2008, 42, 868–876. [Google Scholar] [CrossRef] [PubMed]
- Carrier, M.; Guilbert, J.; Lévesque, J.P.; Tremblay, M.È.; Desjardins, M. Structural and Functional Features of Developing Brain Capillaries, and Their Alteration in Schizophrenia. Front. Cell. Neurosci. 2021, 14, 456. [Google Scholar] [CrossRef] [PubMed]
- Quiñones, G.M.; Mayeli, A.; Yushmanov, V.E.; Hetherington, H.P.; Ferrarelli, F. Reduced GABA/glutamate in the thalamus of individuals at clinical high risk for psychosis. Neuropsychopharmacology 2020, 46, 1133–1139. [Google Scholar] [CrossRef]
- Benson, C.A.; Powell, H.R.; Liput, M.; Dinham, S.; Freedman, D.A.; Ignatowski, T.A.; Stachowiak, E.K.; Stachowiak, M.K. Immune Factor, TNFα, Disrupts Human Brain Organoid Development Similar to Schizophrenia—Schizophrenia Increases Developmental Vulnerability to TNFα. Front. Cell. Neurosci. 2020, 14, 233. [Google Scholar] [CrossRef] [PubMed]
- Nahar, L.; Delacroix, B.M.; Nam, H.W. The Role of Parvalbumin Interneurons in Neurotransmitter Balance and Neurological Disease. Front. Psychiatry 2021, 12, 679960. [Google Scholar] [CrossRef] [PubMed]
- Marín, O. Interneuron dysfunction in psychiatric disorders. Nat. Rev. Neurosci. 2012, 13, 107–120. [Google Scholar] [CrossRef] [PubMed]
- Duchatel, R.J.; Harms, L.R.; Meehan, C.L.; Michie, P.T.; Bigland, M.J.; Smith, D.W.; Jobling, P.; Hodgson, D.M.; Tooney, P.A. Reduced cortical somatostatin gene expression in a rat model of maternal immune activation. Psychiatry Res. 2019, 282, 112621. [Google Scholar] [CrossRef]
- Harrington, E.P.; Bergles, D.E.; Calabresi, P.A. Immune cell modulation of oligodendrocyte lineage cells. Neurosci. Lett. 2020, 715, 134601. [Google Scholar] [CrossRef]
- Madsen, P.; Desu, H.L.; Vaccari, J.P.D.R.; Florimon, Y.; Ellman, D.G.; Keane, R.W.; Clausen, B.H.; Lambertsen, K.L.; Brambilla, R. Oligodendrocytes modulate the immune-inflammatory response in EAE via TNFR2 signaling. Brain Behav. Immun. 2020, 84, 132–146. [Google Scholar] [CrossRef]
- Almonte, A.G.; Sweatt, J.D. Serine proteases, serine protease inhibitors, and protease-activated receptors: Roles in synaptic function and behavior. Brain Res. 2011, 1407, 107–122. [Google Scholar] [CrossRef]
- DeMichele-Sweet, M.A.A.; Weamer, E.A.; Klei, L.; Vrana, D.T.; Hollingshead, D.J.; Seltman, H.J.; Sims, R.; Foroud, T.; Hernandez, I.; Moreno-Grau, S.; et al. Genetic risk for schizophrenia and psychosis in Alzheimer disease. Mol. Psychiatry 2017, 23, 963–972. [Google Scholar] [CrossRef] [PubMed]
- Shukla, D.K.; Wijtenburg, S.A.; Chen, H.; Chiappelli, J.J.; Kochunov, P.; Hong, L.E.; Rowland, L.M. Anterior Cingulate Glutamate and GABA Associations on Functional Connectivity in Schizophrenia. Schizophr. Bull. 2018, 45, 647–658. [Google Scholar] [CrossRef]
- Nutma, E.; van Gent, D.; Amor, S.; Peferoen, L.A.N. Astrocyte and Oligodendrocyte Cross-Talk in the Central Nervous System. Cells 2020, 9, 600. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Liu, Z.-W.; Wen, L.; Qiao, H.-F.; Zhou, W.-X.; Zhang, Y.-X. Interleukin-1β enhances NMDA receptor-mediated current but inhibits excitatory synaptic transmission. Brain Res. 2005, 1034, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Ren, K.; Dubner, R. Neuron–glia crosstalk gets serious: Role in pain hypersensitivity. Curr. Opin. Anaesthesiol. 2008, 21, 570–579. [Google Scholar] [CrossRef] [PubMed]
- Picelli, S.; Faridani, O.R.; Bjorklund, A.K.; Winberg, G.; Sagasser, S.; Sandberg, R. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014, 9, 171–181. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]


| Board Class | Subtype | Top Marker Genes |
|---|---|---|
| GABAergic (inhibitory) | LAMP5 | LAMP5, GGT8P, NDNF, DUSP4, CA13, SFTA3, C1QL2, ANKRD20A11P |
| SST | SST, MTHFD2P6, MAFB, ISOC1, KLHL14, AHR, NPY | |
| VIP | VIP, TOX2, ZNF322P1, CBLN1, CXCL14, PPAPDC1A, ADARB2, ADAM33, CHRNA2, KCNJ2, SSTR1, PRSS8 | |
| PAX6 | PAX6, GRIP2, CA4, SCGN, NABP1 | |
| PVALB | PVALB, FAM150B, CNTNAP3P2, WFDC2, STON2, LHX6, GLP1R, SCUBE3, TAC1, MFI2, C8ORF4 | |
| Glutamatergic (excitatory) | L4_ IT | RORB, GRIK1, RPS3P6, HLHE22, ACNG5, CDC168, AIM2, ASCL1 |
| L5_ET | FEZF2, SCN7A, ONECUT1, DCN, MORN2 | |
| L5/6_NP | FEZF2, MYBPHL, CYP26B1, DYRK2, CABP7, RSAD2 | |
| L5/6_IT_CAR3 | THEMIS, GPR21, C6ORF48, THTPA, IL7R | |
| L6_CT | FEZF2, FAM95C, ANKRD20A1, CPZ, ETV4, VWA2 | |
| L6b | FEZF2, KRT17, TBC1D26, SLITRK6, P4HA3, TBCC | |
| IT | LINC00507, RPL9P17, RORB, RPL31P31, LCN15, THEMIS, LINC00343, SNHG7, SEMA6D, PRSS12, LINC01474, LINC01202 | |
| Non-neuron | Astrocyte | FGFR3, ETNPPL, MT1G, FOS |
| Endothelial | CLDN5 | |
| Microglia | C1QC | |
| Oligodendrocyte | OPALIN, MOBP, COL18A1 | |
| OPC | MYT1 | |
| Pericyte | MUSTN1 | |
| VLMC | CYP1B1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, W.; Song, W.; Liu, Z.; Maharjan, D.T.; Liang, J.; Lin, G.N. An Integrative Analysis of Identified Schizophrenia-Associated Brain Cell Types and Gene Expression Changes. Int. J. Mol. Sci. 2022, 23, 11581. https://doi.org/10.3390/ijms231911581
Cai W, Song W, Liu Z, Maharjan DT, Liang J, Lin GN. An Integrative Analysis of Identified Schizophrenia-Associated Brain Cell Types and Gene Expression Changes. International Journal of Molecular Sciences. 2022; 23(19):11581. https://doi.org/10.3390/ijms231911581
Chicago/Turabian StyleCai, Wenxiang, Weichen Song, Zhe Liu, Dhruba Tara Maharjan, Jisheng Liang, and Guan Ning Lin. 2022. "An Integrative Analysis of Identified Schizophrenia-Associated Brain Cell Types and Gene Expression Changes" International Journal of Molecular Sciences 23, no. 19: 11581. https://doi.org/10.3390/ijms231911581
APA StyleCai, W., Song, W., Liu, Z., Maharjan, D. T., Liang, J., & Lin, G. N. (2022). An Integrative Analysis of Identified Schizophrenia-Associated Brain Cell Types and Gene Expression Changes. International Journal of Molecular Sciences, 23(19), 11581. https://doi.org/10.3390/ijms231911581






