Association of MARC1, ADCY5, and BCO1 Variants with the Lipid Profile, Suggests an Additive Effect for Hypertriglyceridemia in Mexican Adult Men
Abstract
:1. Introduction
2. Results
2.1. Baseline Clinical Characteristics of the Study Population
2.2. Association Analyses between the rs2642438 on MARC1 and rs56371916 on ADCY5 with the Lipid Profile
2.3. Conditional Analysis
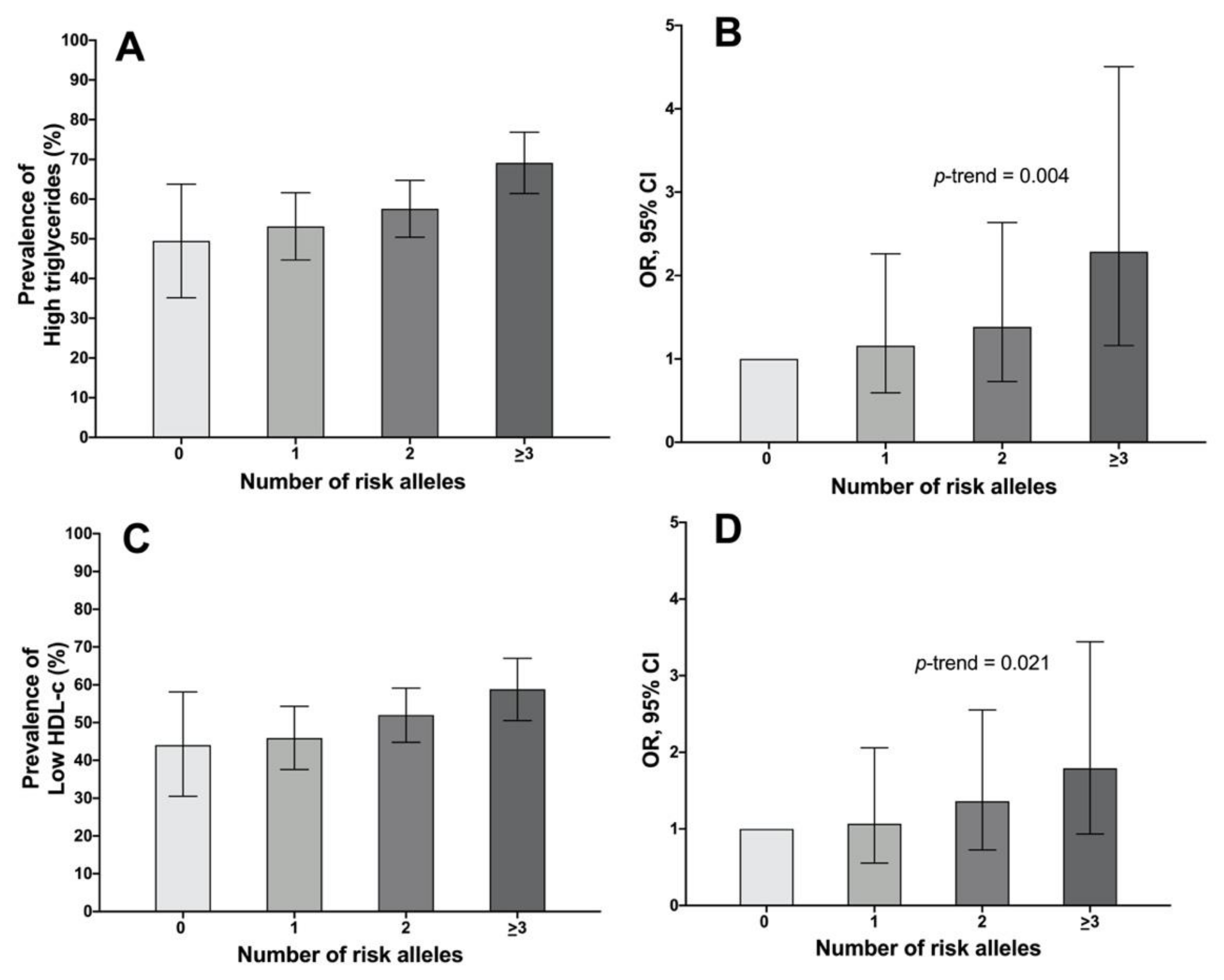
2.4. Association of the Genetic Risk Score with the Lipidic Profile
3. Discussion
4. Materials and Methods
4.1. Health Workers Cohort Study
4.2. Outcome
4.3. Genomic DNA Extraction and SNP Genotyping
4.4. Construction of the Genetic Risk Score
4.5. Covariates
4.6. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Kopin, L.; Lowenstein, C. Dyslipidemia. Ann. Intern. Med. 2017, 167, ITC81–ITC95. [Google Scholar] [CrossRef] [PubMed]
- Rivas-Gomez, B.; Almeda-Valdés, P.; Tusié-Luna, M.T.; Aguilar-Salinas, C.A. Dyslipidemia in mexico, a call for action. Rev. Investig. Clin. 2018, 70, 211–216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Escobedo-de la Peña, J.; de Jesús-Pérez, R.; Schargrodsky, H.; Champagne, B. Prevalence of Dyslipidemias in Mexico City and Its Relation to Other Cardiovascular Risk Factors. Results from the CARMELA Study. Gac. Med. Mex. 2014, 150, 128–136. [Google Scholar] [PubMed]
- Hernández-Alcaraz, C.; Aguilar-Salinas, C.A.; Mendoza-Herrera, K.; Pedroza-Tobías, A.; Villalpando, S.; Shamah-Levy, T.; Rivera-Dommarco, J.; Hernández-Ávila, M.; Barquera, S. Dyslipidemia Prevalence, Awareness, Treatment and Control in Mexico: Results of the Ensanut 2012. Salud Publica Mex. 2020, 62, 137–146. [Google Scholar] [CrossRef]
- Huerta-Chagoya, A.; Moreno-Macías, H.; Sevilla-González, M.; Rodríguez-Guillén, R.; Ordóñez-Sánchez, M.L.; Gómez-Velasco, D.; Muñóz-Hernández, L.; Segura-Kato, Y.; Arellano-Campos, O.; Cruz-Bautista, I.; et al. Contribution of Known Genetic Risk Variants to Dyslipidemias and Type 2 Diabetes in Mexico: A Population-Based Nationwide Study. Genes 2020, 11, 114. [Google Scholar] [CrossRef] [Green Version]
- Willer, C.J.; Mohlke, K.L. Finding Genes and Variants for Lipid Levels after Genome-Wide Association Analysis. Curr. Opin. Lipidol. 2012, 23, 98–103. [Google Scholar] [CrossRef] [Green Version]
- Sinnott-Armstrong, N.; Sousa, I.S.; Laber, S.; Rendina-Ruedy, E.; Nitter Dankel, S.E.; Ferreira, T.; Mellgren, G.; Karasik, D.; Rivas, M.; Pritchard, J.; et al. A Regulatory Variant at 3q21.1 Confers an Increased Pleiotropic Risk for Hyperglycemia and Altered Bone Mineral Density. Cell Metab. 2021, 33, 615-628.e13. [Google Scholar] [CrossRef]
- León-Reyes, G.; Rivera-Paredez, B.; Hidalgo-Bravo, A.; Flores, Y.N.; Salmerón, J.; Velázquez-Cruz, R. Common Variant Rs6564851 near the Beta-Carotene Oxygenase 1 Gene Is Associated with Plasma Triglycerides Levels in Middle-Aged Mexican Men Adults. Nutr. Res. 2022, 103, 30–39. [Google Scholar] [CrossRef]
- Emdin, C.A.; Haas, M.E.; Khera, A.v.; Aragam, K.; Chaffin, M.; Klarin, D.; Hindy, G.; Jiang, L.; Wei, W.Q.; Feng, Q.; et al. A Missense Variant in Mitochondrial Amidoxime Reducing Component 1 Gene and Protection against Liver Disease. PLoS Genet. 2020, 16, e1008629. [Google Scholar] [CrossRef] [Green Version]
- Sparacino-Watkins, C.E.; Tejero, J.; Sun, B.; Gauthier, M.C.; Thomas, J.; Ragireddy, V.; Merchan, B.A.; Wang, J.; Azarov, I.; Basu, P.; et al. Nitrite Reductase and Nitric-Oxide Synthase Activity of the Mitochondrial Molybdopterin Enzymes MARC1 and MARC2. J. Biol. Chem. 2014, 289, 10345–10358. [Google Scholar] [CrossRef]
- Innes, H.; Buch, S.; Hutchinson, S.; Guha, I.N.; Morling, J.R.; Barnes, E.; Irving, W.; Forrest, E.; Pedergnana, V.; Goldberg, D.; et al. Genome-Wide Association Study for Alcohol-Related Cirrhosis Identifies Risk Loci in MARC1 and HNRNPUL1. Gastroenterology 2020, 159, 1276-1289.e7. [Google Scholar] [CrossRef]
- Hudert, C.A.; Adams, L.A.; Alisi, A.; Anstee, Q.M.; Crudele, A.; Draijer, L.G.; Furse, S.; Hengstler, J.G.; Jenkins, B.; Karnebeek, K.; et al. Variants in Mitochondrial Amidoxime Reducing Component 1 and Hydroxysteroid 17-Beta Dehydrogenase 13 Reduce Severity of Nonalcoholic Fatty Liver Disease in Children and Suppress Fibrotic Pathways through Distinct Mechanisms. Hepatol. Commun. 2022, 6, 1934–1948. [Google Scholar] [CrossRef] [PubMed]
- Luukkonen, P.K.; Juuti, A.; Sammalkorpi, H.; Penttilä, A.K.; Orešič, M.; Hyötyläinen, T.; Arola, J.; Orho-Melander, M.; Yki-Järvinen, H. MARC1 Variant Rs2642438 Increases Hepatic Phosphatidylcholines and Decreases Severity of Non-Alcoholic Fatty Liver Disease in Humans. J. Hepatol. 2020, 73, 725–726. [Google Scholar] [CrossRef] [PubMed]
- Hanoune, J.; Defer, N. Regulation and Role of Adenylyl Cyclase Isoforms. Annu. Rev. Pharmacol. Toxicol. 2001, 41, 145–174. [Google Scholar] [CrossRef] [PubMed]
- Pieroni, J.P.; Miller, D.; Premont, R.T.; Lyengar, R. Type 5 Adenylyl Cyclase Distribution. Nature 1993, 363, 679. [Google Scholar] [CrossRef] [PubMed]
- Leech, C.A.; Castonguay, M.A.; Habener, J.F. Expression of Adenylyl Cyclase Subtypes in Pancreatic Beta-Cells. Biochem. Biophys. Res. Commun. 1999, 254, 703–706. [Google Scholar] [CrossRef] [PubMed]
- Eizirik, D.L.; Sammeth, M.; Bouckenooghe, T.; Bottu, G.; Sisino, G.; Igoillo-Esteve, M.; Ortis, F.; Santin, I.; Colli, M.L.; Barthson, J.; et al. The Human Pancreatic Islet Transcriptome: Expression of Candidate Genes for Type 1 Diabetes and the Impact of pro-Inflammatory Cytokines. PLoS Genet. 2012, 8, e1002552. [Google Scholar] [CrossRef]
- Defer, N.; Best-Belpomme, M.; Hanoune, J. Tissue Specificity and Physiological Relevance of Various Isoforms of Adenylyl Cyclase. Am. J. Physiol. Renal. Physiol. 2000, 279, F400–F416. [Google Scholar] [CrossRef] [Green Version]
- Halls, M.L.; Cooper, D.M.F. Adenylyl Cyclase Signalling Complexes—Pharmacological Challenges and Opportunities. Pharmacol. Ther. 2017, 172, 171–180. [Google Scholar] [CrossRef]
- Prentki, M.; Matschinsky, F.M. Ca2+, CAMP, and Phospholipid-Derived Messengers in Coupling Mechanisms of Insulin Secretion. Physiol. Rev. 1987, 67, 1185–1248. [Google Scholar] [CrossRef]
- Hodson, D.J.; Mitchell, R.K.; Marselli, L.; Pullen, T.J.; Brias, S.G.; Semplici, F.; Everett, K.L.; Cooper, D.M.F.; Bugliani, M.; Marchetti, P.; et al. ADCY5 Couples Glucose to Insulin Secretion in Human Islets. Diabetes 2014, 63, 3009–3021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.Z.; Matsushita, M.M.; Robertson, P.; Rieder, M.; Girirajan, S.; Antonacci, F.; Lipe, H.; Eichler, E.E.; Nickerson, D.A.; Bird, T.D.; et al. Autosomal Dominant Familial Dyskinesia and Facial Myokymia: Single Exome Sequencing Identifies a Mutation in Adenylyl Cyclase 5. Arch. Neurol. 2012, 69, 630–635. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.O.; Rauch, A.; Mazzaferro, E.; Preuss, M.; Carobbio, S.; Bayrak, C.S.; Chami, N.; Wang, Z.; Schick, U.M.; Yang, N.; et al. Genome-Wide Discovery of Genetic Loci That Uncouple Excess Adiposity from Its Comorbidities. Nat. Metab. 2021, 3, 228–243. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Lian, F.; Kong, Y.; Huang, L.; Xu, L.; Wu, Y.; Ma, H.; Yang, L. Carotenoid Metabolic (BCO1) Polymorphisms and Personal Behaviors Modify the Risk of Coronary Atherosclerosis: A Nested Case-Control Study in Han Chinese with Dyslipidaemia (2013–2016). Asia Pac. J. Clin. Nutr. 2019, 28, 192–202. [Google Scholar] [CrossRef] [PubMed]
- Amengual, J.; Coronel, J.; Marques, C.; Aradillas-García, C.; Morales, J.M.V.; Andrade, F.C.D.; Erdman, J.W.; Teran-Garcia, M. β-Carotene Oxygenase 1 Activity Modulates Circulating Cholesterol Concentrations in Mice and Humans. J. Nutr. 2020, 150, 2023–2030. [Google Scholar] [CrossRef]
- Below, J.E.; Parra, E.J.; Gamazon, E.R.; Torres, J.; Krithika, S.; Candille, S.; Lu, Y.; Manichakul, A.; Peralta-Romero, J.; Duan, Q.; et al. Meta-Analysis of Lipid-Traits in Hispanics Identifies Novel Loci, Population-Specific Effects, and Tissue-Specific Enrichment of EQTLs. Sci. Rep. 2016, 6, 19429. [Google Scholar] [CrossRef] [Green Version]
- Schneider, C.v.; Schneider, K.M.; Conlon, D.M.; Park, J.; Vujkovic, M.; Zandvakili, I.; Ko, Y.A.; Trautwein, C.; Carr, R.M.; Strnad, P.; et al. A Genome-First Approach to Mortality and Metabolic Phenotypes in MTARC1 p.Ala165Thr (Rs2642438) Heterozygotes and Homozygotes. Med 2021, 2, 851-863.e3. [Google Scholar] [CrossRef]
- Janik, M.K.; Smyk, W.; Kruk, B.; Szczepankiewicz, B.; Górnicka, B.; Lebiedzińska-Arciszewska, M.; Potes, Y.; Simões, I.C.M.; Weber, S.N.; Lammert, F.; et al. MARC1 p.A165T Variant Is Associated with Decreased Markers of Liver Injury and Enhanced Antioxidant Capacity in Autoimmune Hepatitis. Sci. Rep. 2021, 11, 24407. [Google Scholar] [CrossRef]
- Bernal-Reyes, R.; Castro-Narro, G.; Malé-Velázquez, R.; Carmona-Sánchez, R.; González-Huezo, M.S.; García-Juárez, I.; Chávez-Tapia, N.; Aguilar-Salinas, C.; Aiza-Haddad, I.; Ballesteros-Amozurrutia, M.A.; et al. The Mexican Consensus on Nonalcoholic Fatty Liver Disease. Rev. Gastroenterol. Mex. 2019, 84, 69–99. [Google Scholar] [CrossRef]
- Ott, G.; Havemeyer, A.; Clement, B. The Mammalian Molybdenum Enzymes of MARC. J. Biol. Inorg. Chem. 2015, 20, 265–275. [Google Scholar] [CrossRef]
- Abdel-Maksoud, M.F.; Hokanson, J.E. The Complex Role of Triglycerides in Cardiovascular Disease. Semin. Vasc. Med. 2002, 2, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Cyr, A.R.; Huckaby, L.v.; Shiva, S.S.; Zuckerbraun, B.S. Nitric Oxide and Endothelial Dysfunction. Crit. Care Clin. 2020, 36, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.Y.; Liu, L. Remnant-like Lipoprotein Particles Impair Endothelial Function: Direct and Indirect Effects on Nitric Oxide Synthase. J. Lipid Res. 2007, 48, 1673–1680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kajikawa, M.; Higashi, Y. Triglycerides and Endothelial Function: Molecular Biology to Clinical Perspective. Curr. Opin. Lipidol. 2019, 30, 364–369. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, T.J.; Theusch, E.; Haldar, T.; Ranatunga, D.K.; Jorgenson, E.; Medina, M.W.; Kvale, M.N.; Kwok, P.Y.; Schaefer, C.; Krauss, R.M.; et al. A Large Electronic-Health-Record-Based Genome-Wide Study of Serum Lipids. Nat. Genet. 2018, 50, 401–413. [Google Scholar] [CrossRef] [PubMed]
- Haidar, B.; Denis, M.; Marcil, M.; Krimbou, L.; Genest, J. Apolipoprotein A-I Activates Cellular CAMP Signaling through the ABCA1 Transporter. J. Biol. Chem. 2004, 279, 9963–9969. [Google Scholar] [CrossRef] [Green Version]
- Haidar, B.; Denis, M.; Krimbou, L.; Marcil, M.; Genest, J. CAMP Induces ABCA1 Phosphorylation Activity and Promotes Cholesterol Efflux from Fibroblasts. J. Lipid Res. 2002, 43, 2087–2094. [Google Scholar] [CrossRef] [Green Version]
- Srivastava, N.; Cefalu, A.B.; Averna, M.; Srivastava, R.A.K. Rapid Degradation of ABCA1 Protein Following CAMP Withdrawal and Treatment with PKA Inhibitor Suggests ABCA1 Is a Short-Lived Protein Primarily Regulated at the Transcriptional Level. J. Diabetes Metab. Disord. 2020, 19, 363–371. [Google Scholar] [CrossRef]
- Keller, H.; Dreyer, C.; Medin, J.; Mahfoudi, A.; Ozato, K.; Wahli, W. Fatty Acids and Retinoids Control Lipid Metabolism through Activation of Peroxisome Proliferator-Activated Receptor-Retinoid X Receptor Heterodimers. Proc. Natl. Acad. Sci. USA 1993, 90, 2160–2164. [Google Scholar] [CrossRef] [Green Version]
- Grimaldi, P.A. Peroxisome Proliferator-Activated Receptors as Sensors of Fatty Acids and Derivatives. Cell Mol. Life Sci. 2007, 64, 2459–2464. [Google Scholar] [CrossRef]
- Villacorta, L.; Schopfer, F.J.; Zhang, J.; Freeman, B.A.; Chen, Y.E. PPARgamma and Its Ligands: Therapeutic Implications in Cardiovascular Disease. Clin. Sci. 2009, 116, 205–218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Airanthi, M.; Widjaja-Adhi, K.; Lobo, G.P.; Golczak, M.; von Lintig, J. A Genetic Dissection of Intestinal Fat-Soluble Vitamin and Carotenoid Absorption. Hum. Mol. Genet. 2015, 24, 3206–3219. [Google Scholar] [CrossRef] [Green Version]
- Rigotti, A.; Miettinen, H.E.; Krieger, M. The Role of the High-Density Lipoprotein Receptor SR-BI in the Lipid Metabolism of Endocrine and Other Tissues. Endocr. Rev. 2003, 24, 357–387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trigatti, B.L.; Krieger, M.; Rigotti, A. Influence of the HDL Receptor SR-BI on Lipoprotein Metabolism and Atherosclerosis. Arter. Thromb. Vasc. Biol. 2003, 23, 1732–1738. [Google Scholar] [CrossRef] [PubMed]
- Leiva, A.; Verdejo, H.; Benítez, M.L.; Martínez, A.; Busso, D.; Rigotti, A. Mechanisms Regulating Hepatic SR-BI Expression and Their Impact on HDL Metabolism. Atherosclerosis 2011, 217, 299–307. [Google Scholar] [CrossRef]
- Bietrix, F.; Yan, D.; Nauze, M.; Rolland, C.; Bertrand-Michel, J.; Coméra, C.; Schaak, S.; Barbaras, R.; Groen, A.K.; Perret, B.; et al. Accelerated Lipid Absorption in Mice Overexpressing Intestinal SR-BI. J. Biol. Chem. 2006, 281, 7214–7219. [Google Scholar] [CrossRef] [Green Version]
- Hoekstra, M.; Ouweneel, A.B.; Price, J.; van der Geest, R.; van der Sluis, R.J.; Geerling, J.J.; Nahon, J.E.; van Eck, M. SR-BI Deficiency Disassociates Obesity from Hepatic Steatosis and Glucose Intolerance Development in High Fat Diet-Fed Mice. J. Nutr. Biochem. 2021, 89, 108564. [Google Scholar] [CrossRef]
- Jakobs, H.H.; Mikula, M.; Havemeyer, A.; Strzalkowska, A.; Borowa-Chmielak, M.; Dzwonek, A.; Gajewska, M.; Hennig, E.E.; Ostrowski, J.; Clement, B. The N-Reductive System Composed of Mitochondrial Amidoxime Reducing Component (MARC), Cytochrome B5 (CYB5B) and Cytochrome B5 Reductase (CYB5R) Is Regulated by Fasting and High Fat Diet in Mice. PLoS ONE 2014, 9, e105371. [Google Scholar] [CrossRef]
- Dommel, S.; Hoffmann, A.; Berger, C.; Kern, M.; Klöting, N.; Kannt, A.; Blüher, M. Effects of Whole-Body Adenylyl Cyclase 5 ( Adcy5) Deficiency on Systemic Insulin Sensitivity and Adipose Tissue. Int. J. Mol. Sci. 2021, 22, 4353. [Google Scholar] [CrossRef]
- Tang, W.; Ma, W.; Ding, H.; Lin, M.; Xiang, L.; Lin, G.; Zhang, Z. Adenylyl Cyclase 1 as a Major Isoform to Generate CAMP Signaling for ApoA-1-Mediated Cholesterol Efflux Pathway. J. Lipid Res. 2018, 59, 635–645. [Google Scholar] [CrossRef]
- Salazar, M.R.; Carbajal, H.A.; Espeche, W.G.; Leiva Sisnieguez, C.E.; Balbín, E.; Dulbecco, C.A.; Aizpurúa, M.; Marillet, A.G.; Reaven, G.M. Relation among the Plasma Triglyceride/High-Density Lipoprotein Cholesterol Concentration Ratio, Insulin Resistance, and Associated Cardio-Metabolic Risk Factors in Men and Women. Am. J. Cardiol. 2012, 109, 1749–1753. [Google Scholar] [CrossRef] [PubMed]
- Salazar, M.R.; Carbajal, H.A.; Espeche, W.G.; Aizpurúa, M.; Marillet, A.G.; Leiva Sisnieguez, C.E.; Leiva Sisnieguez, B.C.; Stavile, R.N.; March, C.E.; Reaven, G.M. Use of the Triglyceride/High-Density Lipoprotein Cholesterol Ratio to Identify Cardiometabolic Risk: Impact of Obesity? J. Investig. Med. 2017, 65, 323–327. [Google Scholar] [CrossRef] [PubMed]
- Rangel-Baltazar, E.; Cuevas-Nasu, L.; Shamah-Levy, T.; Rodríguez-Ramírez, S.; Méndez-Gómez-Humarn, I.; Rivera, J.A. Association between High Waist-to-Height Ratio and Cardiovascular Risk among Adults Sampled by the 2016 Half-Way National Health and Nutrition Survey in Mexico (ENSANUT MC 2016). Nutrients 2019, 11, 1402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klop, B.; do Rego, A.T.; Cabezas, M.C. Alcohol and Plasma Triglycerides. Curr. Opin. Lipidol. 2013, 24, 321–326. [Google Scholar] [CrossRef]
- Barquera, S.; Hernandez-Barrera, L.; Tolentino, M.L.; Espinosa, J.; Shu, W.N.; Rivera, J.A.; Popkin, B.M. Energy Intake from Beverages Is Increasing among Mexican Adolescents and Adults. J. Nutr. 2008, 138, 2454–2461. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Pimienta, T.G.; Batis, C.; Lutter, C.K.; Rivera, J.A. Sugar-Sweetened Beverages Are the Main Sources of Added Sugar Intake in the Mexican Population. J. Nutr. 2016, 146, 1888S–1896S. [Google Scholar] [CrossRef] [Green Version]
- Pérez-López, F.R.; Larrad-Mur, L.; Kallen, A.; Chedraui, P.; Taylor, H.S. Gender Differences in Cardiovascular Disease: Hormonal and Biochemical Influences. Reprod. Sci. 2010, 17, 511–531. [Google Scholar] [CrossRef] [Green Version]
- Gallagher, D.; Heymsfield, S.B.; Heo, M.; Jebb, S.A.; Murgatroyd, P.R.; Sakamoto, Y. Healthy Percentage Body Fat Ranges: An Approach for Developing Guidelines Based on Body Mass Index. Am. J. Clin. Nutr. 2000, 72, 694–701. [Google Scholar] [CrossRef] [Green Version]
- Liao, R.S.; Ma, S.; Miao, L.; Li, R.; Yin, Y.; Raj, G.v. Androgen Receptor-Mediated Non-Genomic Regulation of Prostate Cancer Cell Proliferation. Transl. Androl. Urol. 2013, 2, 187–196. [Google Scholar] [CrossRef]
- Badeau, R.M.; Metso, J.; Wähälä, K.; Tikkanen, M.J.; Jauhiainen, M. Human Macrophage Cholesterol Efflux Potential Is Enhanced by HDL-Associated 17beta-Estradiol Fatty Acyl Esters. J. Steroid Biochem. Mol. Biol. 2009, 116, 44–49. [Google Scholar] [CrossRef]
- Denova-Gutiérrez, E.; Flores, Y.N.; Gallegos-Carrillo, K.; Ramírez-Palacios, P.; Rivera-Paredez, B.; Muñoz-Aguirre, P.; Velázquez-Cruz, R.; Torres-Ibarra, L.; Meneses-León, J.; Méndez-Hernández, P.; et al. Health Workers Cohort Study: Methods and Study Design. Salud Publica Mex. 2016, 58, 708–716. [Google Scholar] [CrossRef] [PubMed]
- Tate, J.R.; Berg, K.; Couderc, R.; Dati, F.; Kostner, G.M.; Marcovina, S.M.; Rifai, N.; Sakurabayashi, I.; Steinmetz, A. International Federation of Clinical Chemistry and Laboratory Medicine (IFCC) Standardization Project for the Measurement of Lipoprotein(a). Phase 2: Selection and Properties of a Proposed Secondary Reference Material for Lipoprotein(a). Clin. Chem. Lab. Med. 1999, 37, 949–958. [Google Scholar] [CrossRef] [PubMed]
- Saklayen, M.G. The Global Epidemic of the Metabolic Syndrome. Curr. Hypertens. Rep. 2018, 20, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernández-Avila, M.; Romieu, I.; Parra, S.; Hernández-Avila, J.; Madrigal, H.; Willett, W. Validity and Reproducibility of a Food Frequency Questionnaire to Assess Dietary Intake of Women Living in Mexico City. Salud Publica Mex. 1998, 40, 133–140. [Google Scholar] [CrossRef] [PubMed]
| Men = 579 | Women = 1321 | p | |
|---|---|---|---|
| Age a, (years) | 46.3 (14.6) | 52.3 (14.9) | <0.001 |
| BMI a, (kg/m2) | 26.6 (24.3–29.2) | 26.9 (24.1–30.3) | 0.115 |
| Overweight, % | 48.7 | 40.4 | 0.0008 |
| Obesity, % | 19.9 | 26.3 | 0.0028 |
| Leisure time physical activity a (hour/week) | 1.7 (0.4–5) | 1.1 (0.2–3.5) | <0.001 |
| Active (>150 min/week), % | 42.3 | 31.1 | <0.001 |
| Smoking status, % | |||
| Current, % | 20.9 | 8.9 | <0.001 |
| Past, % | 39.2 | 22.5 | <0.001 |
| ALT a, (U/L) | 25 (19–35) | 20 (15–29) | <0.001 |
| AST a, (U/L) | 25 (21–31) | 23 (20–30) | 0.0001 |
| Serum total cholesterol a, (mg/dL) | 192 (168–222) | 199 (172–226) | 0.0003 |
| High total cholesterol b, % | 40.6 | 48.8 | 0.0008 |
| Serum HDL-c a, (mg/dL) | 39 (34–46) | 46 (39–54) | <0.001 |
| Low HDL-c c, % | 51.8 | 64.0 | <0.001 |
| Serum LDL-c a (mg/dL) | 115 (96–144) | 121 (99–146) | 0.007 |
| High LDL-c d, % | 70.5 | 74.2 | 0.094 |
| Serum triglycerides a, (mg/dL) | 168 (119–245) | 150 (109–201) | <0.0001 |
| High triglycerides e, % | 58.4 | 50.4 | 0.001 |
| Lipid-lowering treatment, % | 11.5 | 13.9 | 0.154 |
| Diet | |||
| Energy intake a (kcal/day) | 1936 (1457–2549) | 1687 (1242–2221) | <0.001 |
| Carbohydrate a (% energy) | 64.6 (58.1–70.5) | 66.5 (60.6–71.8) | <0.001 |
| Protein a (% energy) | 12.3 (10.6–14.1) | 12.5 (11.0–14.3) | 0.061 |
| MUFAs a (% energy) | 8.4 (6.8–10.5) | 8.6 (7.0–10.4) | 0.210 |
| PUFAs a (% energy) | 1.8 (1.5–2.2) | 1.9 (1.6–2.2) | 0.199 |
| Alcohol a (g/day) | 2.8 (0.6–7.5) | 0.6 (0–1.8) | <0.001 |
| rs2642438 MARC1 | rs56371916 ADCY5 | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Model | High Total Cholesterol a OR (95% CI) | Low HDL-c b OR (95% CI) | High TG c OR (95% CI) | High LDL-c d OR (95% CI) | Model | High Total Cholesterol a OR (95% CI) | Low HDL-c b OR (95% CI) | High TG c OR (95% CI) | High LDL-c d OR (95% CI) |
| Additive | |||||||||
| 0.86 (0.62–1.21) | 1.26 (0.90–1.75) | 1.57 (1.10–2.24) | 1.37 (0.94–1.98) | 0.93 (0.73–1.19) | 1.27 (0.99–1.63) | 1.03 (0.80–1.33) | 0.94 (0.72–1.23) | ||
| p | 0.390 | 0.181 | 0.013 | 0.101 | p | 0.569 | 0.060 | 0.830 | 0.647 |
| Codominant | |||||||||
| GG * | TT * | ||||||||
| GA | 0.83 (0.57–1.23) | 1.24 (0.85–1.83) | 1.44 (0.96–2.14) | 1.41 (0.92–2.15) | TC | 0.85 (0.59–1.23) | 1.26 (0.88–1.81) | 1.27 (0.87–1.86) | 0.74 (0.50–1.10) |
| p | 0.357 | 0.263 | 0.075 | 0.114 | p | 0.396 | 0.211 | 0.209 | 0.140 |
| AA | 0.89 (0.28–2.84) | 1.65 (0.51–5.31) | 4.58 (0.95–22.03) | 1.57 (0.42–5.89) | CC | 0.93 (0.54–1.59) | 1.62 (0.94–2.79) | 0.90 (0.52–1.55) | 1.07 (0.58–1.95) |
| p | 0.838 | 0.263 | 0.057 | 0.503 | p | 0.784 | 0.082 | 0.694 | 0.834 |
| Recessive | |||||||||
| GG + GA * | TT + TC * | ||||||||
| AA | 0.93 (0.29–2.96) | 1.55 (0.48–4.98) | 4.16 (0.87–19.9) | 1.44 (0.38–5.36) | CC | 1.00 (0.60–1.67) | 1.45 (0.87–2.41) | 0.80 (0.47–1.34) | 1.24 (0.70–2.18) |
| p | 0.903 | 0.461 | 0.075 | 0.590 | p | 0.993 | 0.159 | 0.387 | 0.457 |
| Dominant | |||||||||
| GG * | TT * | ||||||||
| GA + AA | 0.84 (0.58–1.22) | 1.27 (0.88–1.85) | 1.54 (1.04–2.28) | 1.42 (0.94–2.14) | TC + CC | 0.87 (0.62–1.23) | 1.33 (0.95–1.88) | 1.17 (0.82–1.67) | 0.81 (0.55–1.17) |
| p | 0.355 | 0.207 | 0.030 | 0.096 | p | 0.427 | 0.097 | 0.377 | 0.255 |
| Number of Risk Alleles | Low HDL-c | High Triglycerides | High LDL-c | High Total Cholesterol | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Model SNP/Gene | n (%) | OR (IC 95%) | p | OR (IC 95%) | p | OR (IC 95%) | p | OR (IC 95%) | p | |
| rs2642438 MARC1 rs6564851 BCO1 | 0 * | 119(20.5) | ||||||||
| 1 | 262(45.1) | 1.34 (0.85–2.12) | 0.205 | 1.03 (0.65–1.65) | 0.890 | 0.86 (0.53–1.40) | 0.543 | 0.71 (0.45–1.12) | 0.143 | |
| 2 | 161(27.7) | 1.83 (1.11–3.02) | 0.018 | 1.69 (1.00–2.85) | 0.048 | 1.15 (0.67–1.97) | 0.616 | 0.85 (0.52–1.39) | 0.520 | |
| ≥3 | 39(6.7) | 0.71 (0.32–1.58) | 0.408 | 3.83 (1.55–10.10) | 0.005 | 1.49 (0.60–3.68) | 0.385 | 0.71 (0.32–1.58) | 0.398 | |
| rs2642438 MARC1 rs56371916 ADCY5 | 0 * | 552(29.1) | ||||||||
| 1 | 812(42.7) | 1.40 (0.93–2.12) | 0.105 | 1.35 (0.89–2.05) | 0.162 | 0.77 (0.50–1.20) | 0.249 | 0.87 (0.58–1.31) | 0.504 | |
| 2 | 446(23.5) | 1.42 (0.87–2.31) | 0.162 | 1.39 (0.84–2.30) | 0.195 | 1.28 (0.74–2.22) | 0.382 | 0.78 (0.48–1.27) | 0.321 | |
| ≥3 | 90(4.7) | 3.46 (1.24–9.64) | 0.018 | 1.83 (0.68–4.88) | 0.229 | 1.38 (0.48–4.04) | 0.551 | 0.92 (0.36–2.33) | 0.865 | |
| rs56371916 ADCY5 rs6564851 BCO1 | 0 * | 80(13.8) | ||||||||
| 1 | 195(33.5) | 0.91 (0.52–1.58) | 0.733 | 1.11 (0.63–1.95) | 0.723 | 0.83 (0.45–1.52) | 0.546 | 0.98 (0.56–1.70) | 0.941 | |
| 2 | 213(36.6) | 1.15 (0.67–1.98) | 0.617 | 1.39 (0.79–2.42) | 0.252 | 0.83 (0.46–1.51) | 0.544 | 0.87 (0.50–1.49) | 0.603 | |
| ≥3 | 94(16.2) | 1.61 (0.85–3.06) | 0.147 | 1.80 (0.93–3.50) | 0.082 | 0.94 (0.47–1.89) | 0.870 | 0.91 (0.48–1.71) | 0.760 | |
| rs2642438 MARC1 rs56371916 ADCY5 rs6564851 BCO1 | 0 * | 53(9.2) | ||||||||
| 1 | 160(27.6) | 1.08 (0.56–2.08) | 0.823 | 1.16 (0.59–2.28) | 0.662 | 0.59 (0.28–1.24) | 0.164 | 0.73 (0.38–1.40) | 0.347 | |
| 2 | 207(35.8) | 1.37 (0.73–2.58) | 0.326 | 1.32 (0.69–2.54) | 0.397 | 0.70 (0.34–1.44) | 0.335 | 0.68 (0.36–1.32) | 0.223 | |
| ≥3 | 159(27.5) | 1.80 (0.93–3.46) | 0.079 | 2.23 (1.13–4.42) | 0.022 | 0.93 (0.44–1.98) | 0.858 | 0.69 (0.36–1.32) | 0.258 | |
| Number of Risk Alleles (rs2642438-A MARC1, rs56371916-C ADCY5, rs6564851-A BCO1) | |||||
|---|---|---|---|---|---|
| Characteristic | 0 * | 1 | 2 | ≥3 | p |
| n = 53 (9.2%) | n = 160 (27.5%) | n = 207 (35.8%) | n = 159 (27.5%) | (0 vs. ≥ 3) | |
| Triglycerides, mg/dL a | 153 (115–234) | 156 (114–241) | 171 (120–249) | 171 (129–253) | 0.086 |
| High triglycerides, % b | 52.8 | 53.1 | 57.5 | 66.7 | 0.069 |
| HDL-c, mg/dL a | 41 (35.4–49.1) | 41 (35–47) | 39 (34–45.4) | 37.7 (33.6–44) | 0.004 |
| Low HDL-c, % c | 45.3 | 45.6 | 52.7 | 59.1 | 0.079 |
| Total cholesterol, mg/dL a | 199 (177–229) | 192 (167–224) | 190 (166–221) | 192 (168–217) | 0.073 |
| High total cholesterol, % d | 49.1 | 41.3 | 38.7 | 39.6 | 0.225 |
| LDL-c, mg/dL a | 112 (100–146) | 114 (95–147) | 115 (96–140) | 116 (98–142) | 0.406 |
| High LDL-c, % e | 76.5 | 67.5 | 69 | 73.6 | 0.675 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rivera-Paredez, B.; Aparicio-Bautista, D.I.; Argoty-Pantoja, A.D.; Patiño, N.; Flores Morales, J.; Salmerón, J.; León-Reyes, G.; Velázquez-Cruz, R. Association of MARC1, ADCY5, and BCO1 Variants with the Lipid Profile, Suggests an Additive Effect for Hypertriglyceridemia in Mexican Adult Men. Int. J. Mol. Sci. 2022, 23, 11815. https://doi.org/10.3390/ijms231911815
Rivera-Paredez B, Aparicio-Bautista DI, Argoty-Pantoja AD, Patiño N, Flores Morales J, Salmerón J, León-Reyes G, Velázquez-Cruz R. Association of MARC1, ADCY5, and BCO1 Variants with the Lipid Profile, Suggests an Additive Effect for Hypertriglyceridemia in Mexican Adult Men. International Journal of Molecular Sciences. 2022; 23(19):11815. https://doi.org/10.3390/ijms231911815
Chicago/Turabian StyleRivera-Paredez, Berenice, Diana I. Aparicio-Bautista, Anna D. Argoty-Pantoja, Nelly Patiño, Jeny Flores Morales, Jorge Salmerón, Guadalupe León-Reyes, and Rafael Velázquez-Cruz. 2022. "Association of MARC1, ADCY5, and BCO1 Variants with the Lipid Profile, Suggests an Additive Effect for Hypertriglyceridemia in Mexican Adult Men" International Journal of Molecular Sciences 23, no. 19: 11815. https://doi.org/10.3390/ijms231911815
APA StyleRivera-Paredez, B., Aparicio-Bautista, D. I., Argoty-Pantoja, A. D., Patiño, N., Flores Morales, J., Salmerón, J., León-Reyes, G., & Velázquez-Cruz, R. (2022). Association of MARC1, ADCY5, and BCO1 Variants with the Lipid Profile, Suggests an Additive Effect for Hypertriglyceridemia in Mexican Adult Men. International Journal of Molecular Sciences, 23(19), 11815. https://doi.org/10.3390/ijms231911815





