Mapping and Validation of BrGOLDEN: A Dominant Gene Regulating Carotenoid Accumulation in Brassica rapa
Abstract
:1. Introduction
2. Results
2.1. Phenotypic Evaluation and Genetic Analysis
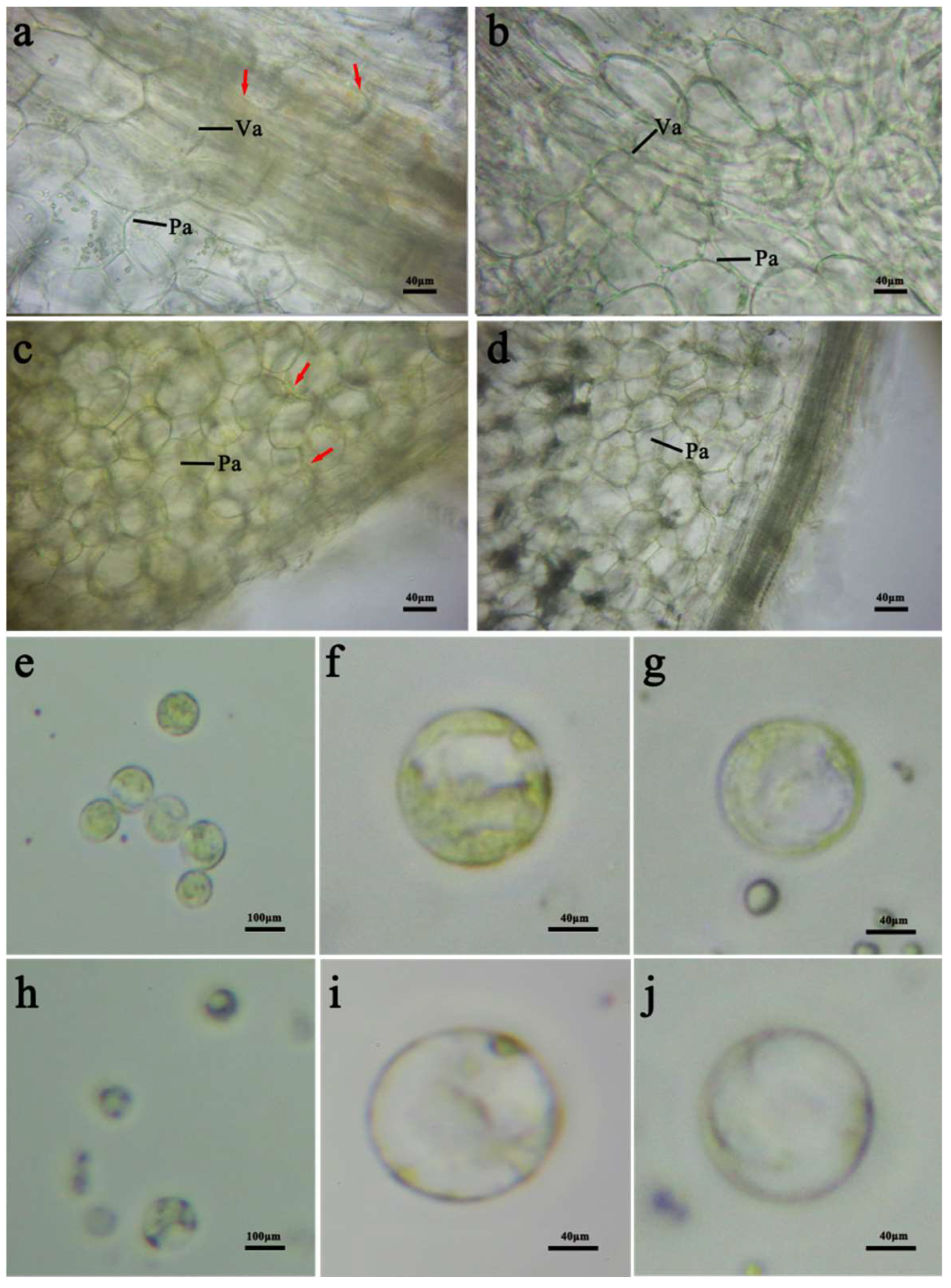
2.2. Comparison of Microstructure and the Carotenoid Component
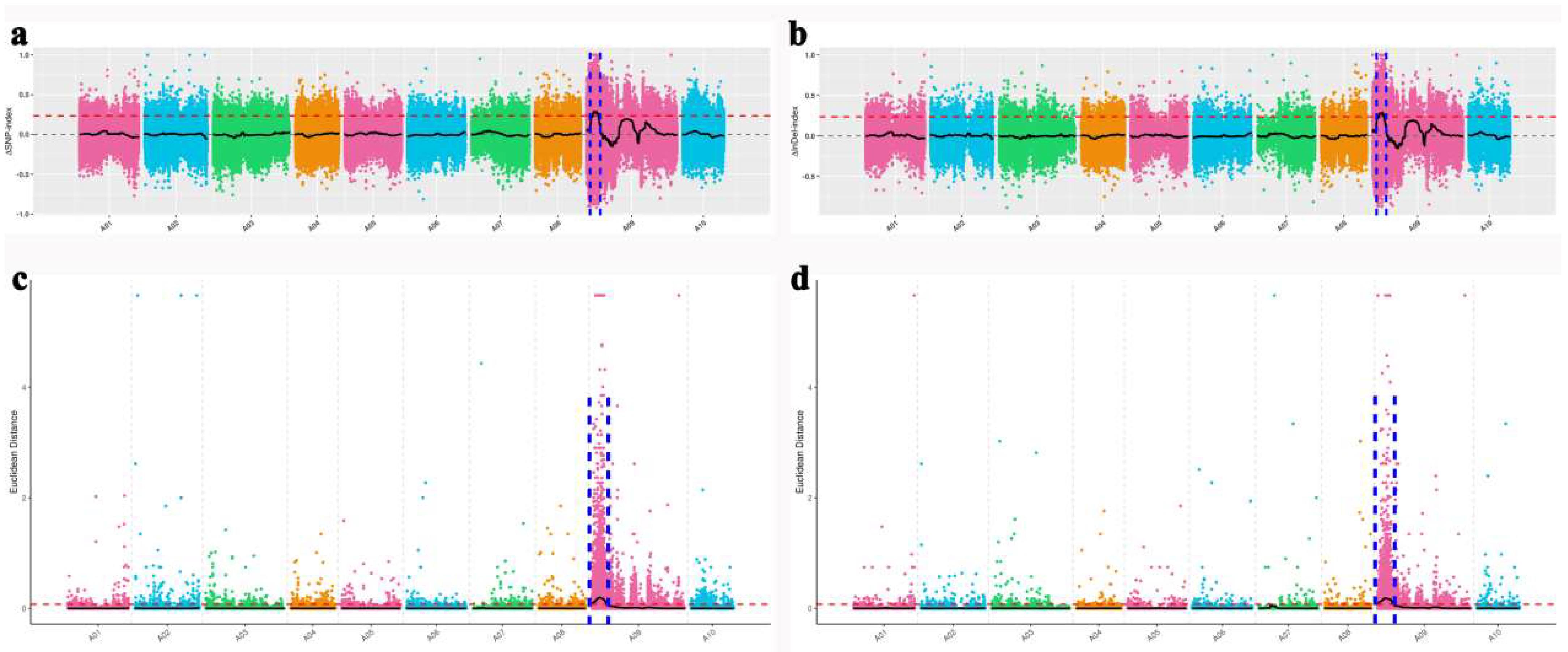
2.3. Primary Mapping for the Golden Inner Leaf Phenotype by BSA−seq
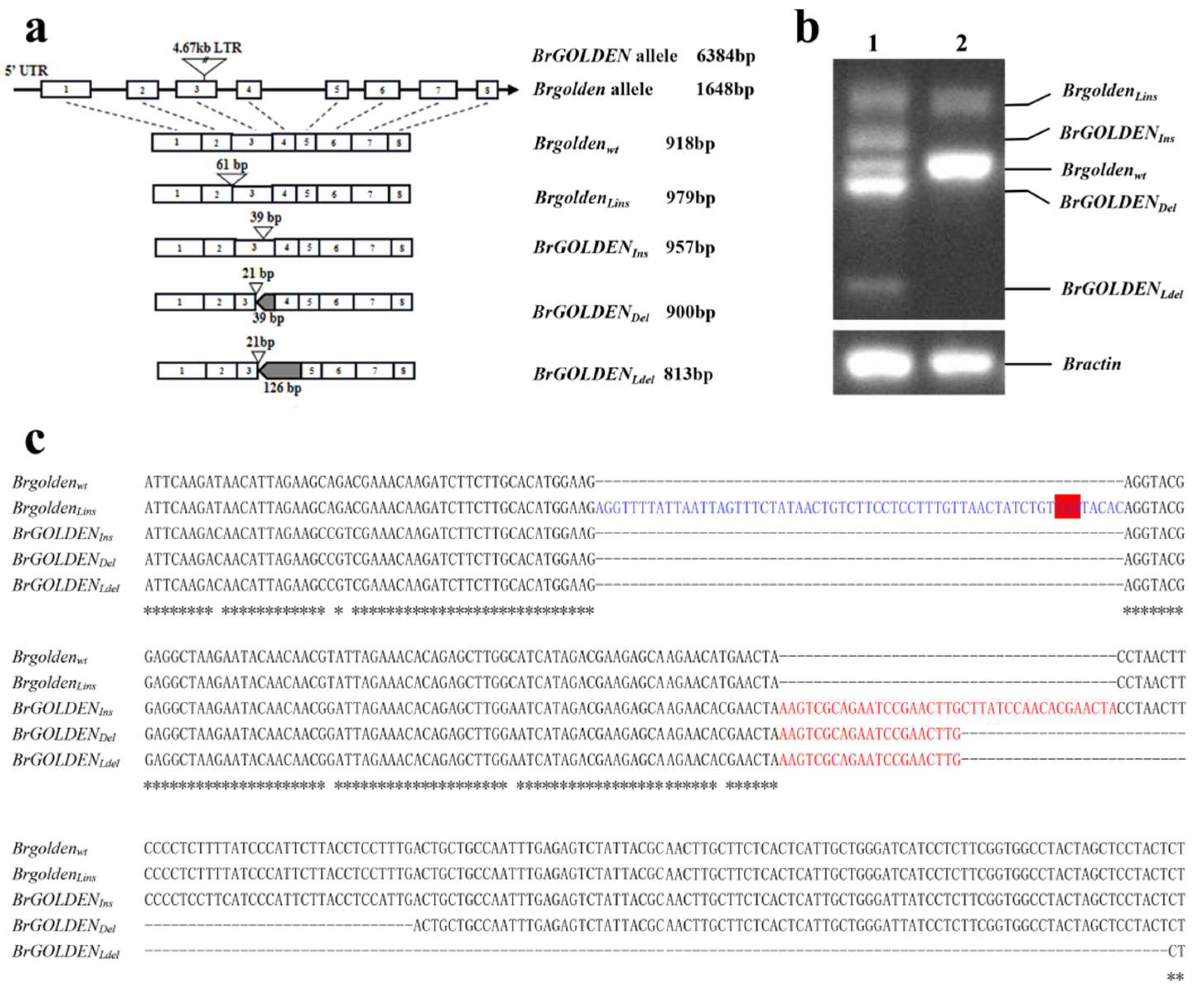
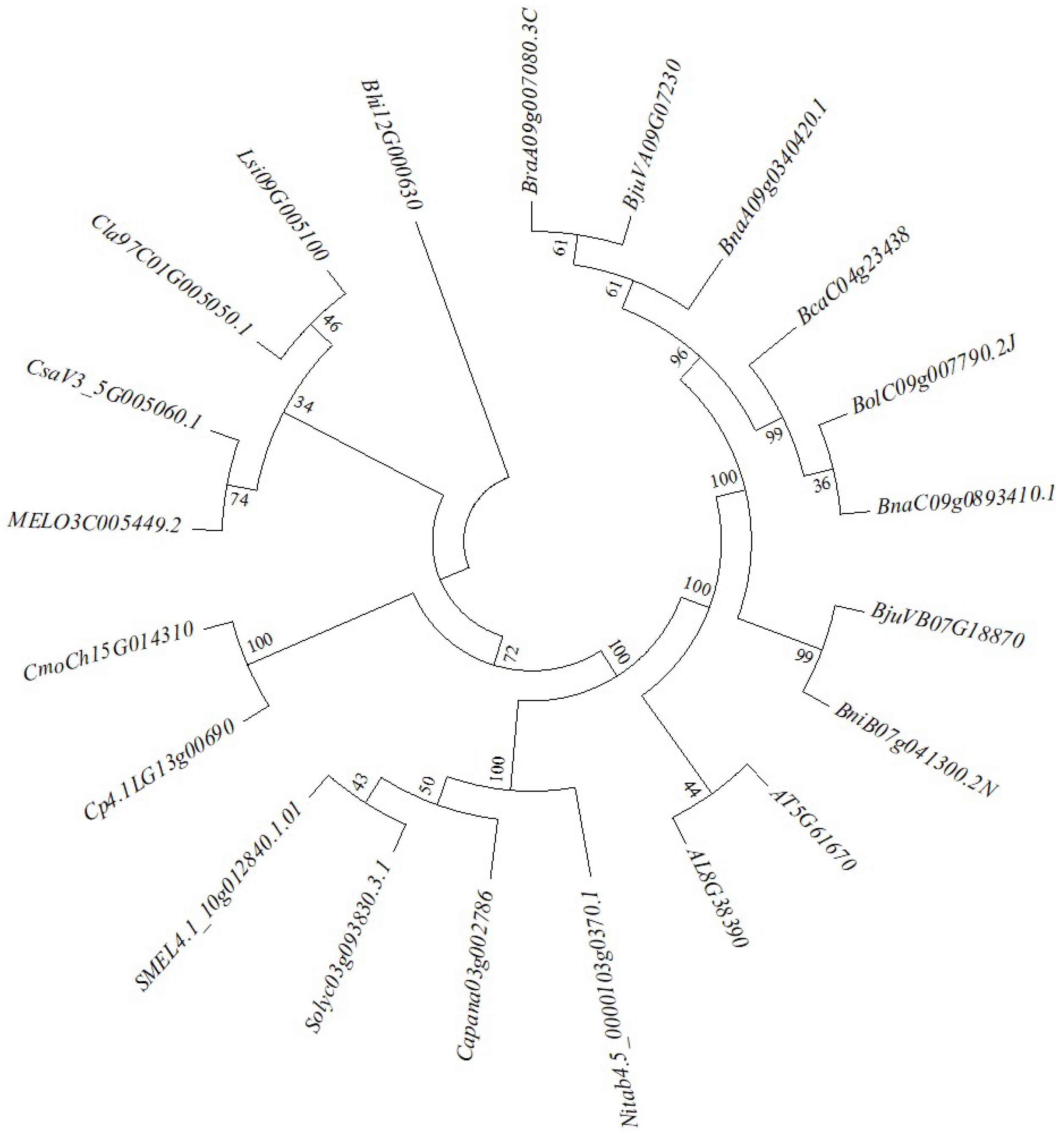
2.4. Verifying the BrGOLDEN Candidate Gene
2.5. Relative Expression Level Analysis of BrGOLDEN and Carotenoid Synthesis Pathway Genes
2.6. Brgolden Exhibits Distinct Expression Patterns
2.7. Ectopic Overexpression of BrGOLDEN Increases Carotenoid Accumulation in A. thaliana Calluses
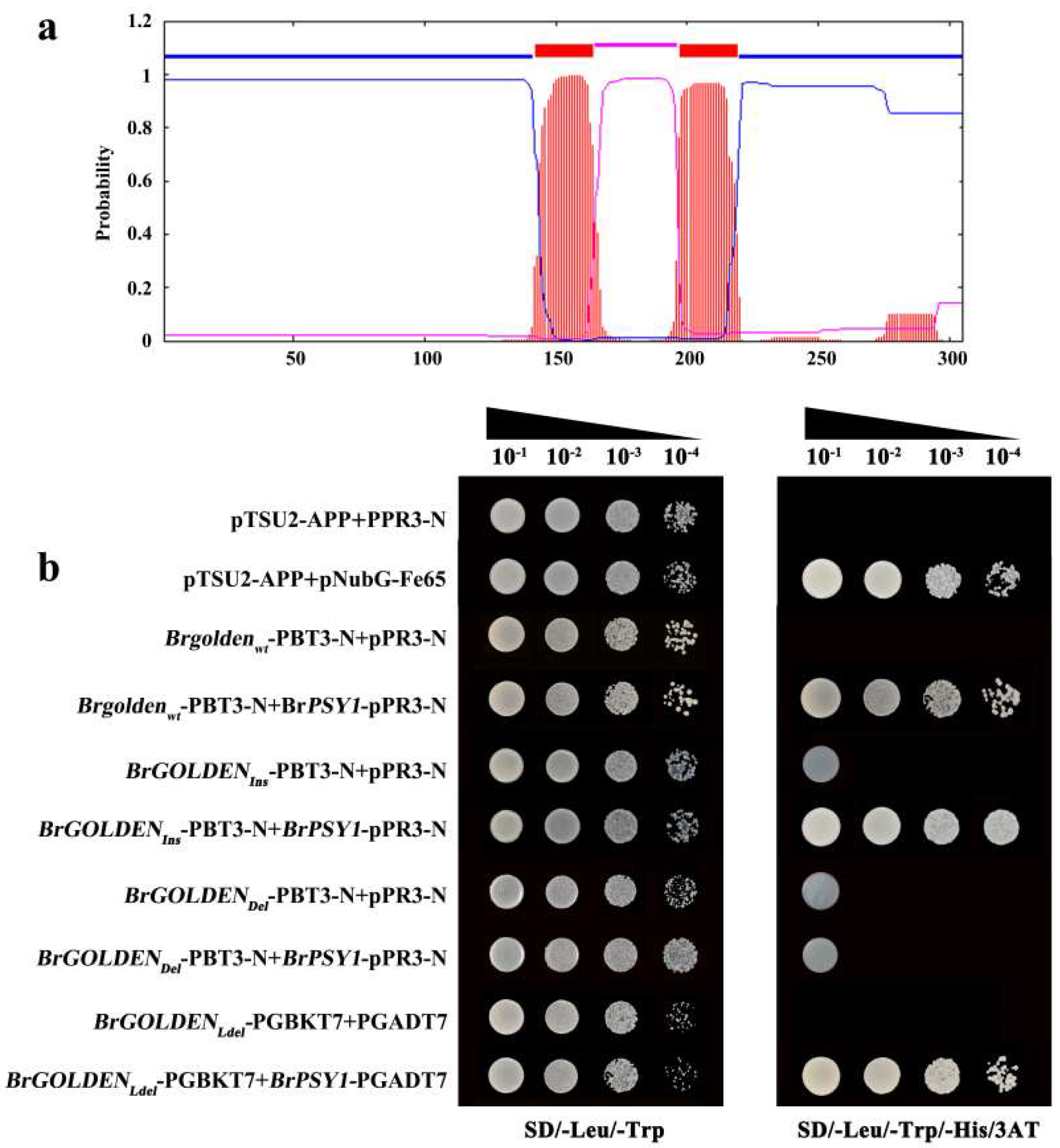
2.8. Identification of the Target Proteins Involved in BrGOLDEN Interactions
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Callus Induction
4.2. Genetic Analysis of the Golden Inner Leaf Phenotype
4.3. Protoplast Separation, Microstructure Observation, and Carotenoid Analysis
4.4. BSA−seq Mapping
4.5. Cloning and Sequencing of Candidate Genes
4.6. Quantitative Reverse−Transcription PCR
4.7. Construction of ProBrgolden:GUS Fusion Vector and Histochemical Staining of Transgenic Plants
4.8. Generation of Transgenic Plants Overexpressing BrGOLDEN
4.9. Y2H Assay
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bradshaw, J.E.; Pande, B.; Bryan, G.J.; Hackett, C.A.; McLean, K.; Stewart, H.E.; Waugh, R. Interval mapping of quantitative trait loci for resistance to late blight [Phytophthora infestans (Mont.) de Bary], height and maturity in a tetraploid population of potato (Solanum tuberosum subsp. tuberosum). Genetics 2004, 168, 983–995. [Google Scholar] [CrossRef] [Green Version]
- Walter, M.H.; Strack, D. Carotenoids and their cleavage products: Biosynthesis and functions. Nat. Prod. Rep. 2011, 28, 663–692. [Google Scholar] [CrossRef]
- Demmig-Adams, B.; Adams, W.W. Antioxidants in photosynthesis and human nutrition. Science 2002, 298, 2149–2153. [Google Scholar] [CrossRef]
- Milborrow, B.V. The pathway of biosynthesis of abscisic acid in vascular plants: A review of the present state of knowledge of ABA biosynthesis. J. Exp. Bot. 2001, 52, 1145–1164. [Google Scholar] [CrossRef]
- Bouvier, F.; Suire, C.; Mutterer, J.; Camara, B. Oxidative remodeling of chromoplast carotenoids: Identification of the carotenoid dioxygenase CsCCD and CsZCD genes involved in crocus secondary metabolite biogenesis. Plant Cell 2003, 15, 47–62. [Google Scholar] [CrossRef] [Green Version]
- Fester, T.; Hause, B.; Schmidt, D.; Halfmann, K.; Schmidt, J.; Wray, V.; Hanse, G.; Strack, D. Occurrence and localization of apocarotenoids in arbuscular mycorrhizal plant roots. Plant Cell Physiol. 2002, 43, 256–265. [Google Scholar] [CrossRef] [Green Version]
- Giuliano, G.; Al-Babili, S.; von Lintig, J. Carotenoid oxygenases: Cleave it or leave it. Trends Plant Sci. 2003, 8, 145–149. [Google Scholar] [CrossRef]
- Lewinsohn, E.; Sitrit, Y.; Bar, E.; Azulay, Y.; Ibdah, M.; Meir, A.; Yosef, E.; Zamir, D.; Tadmor, Y. Not just colors—carotenoid degradation as a link between pigmentation and aroma in tomato and watermelon fruit. Trends Food Sci. Technol. 2005, 16, 407–415. [Google Scholar] [CrossRef]
- Lu, S.; Li, L. Carotenoid metabolism: Biosynthesis, regulation, and beyond. J. Integr. Plant Biol. 2008, 50, 778–785. [Google Scholar] [CrossRef]
- Hirschberg, J. Carotenoid biosynthesis in flowering plants. Curr. Opin. Plant Biol. 2001, 4, 210–218. [Google Scholar] [CrossRef]
- Taylor, M.; Ramsay, G. Carotenoid biosynthesis in plant storage organs: Recent advances and prospects for improving plant food quality. Physiol. Plant. 2005, 124, 143–151. [Google Scholar] [CrossRef]
- DellaPenna, D.; Pogson, B.J. Vitamin synthesis in plants: Tocopherols and carotenoids. Annu. Rev. Plant Biol. 2006, 57, 711–738. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Yuan, H. Chromoplast biogenesis and carotenoid accumulation. Arch. Biochem. Biophys. 2013, 539, 102–109. [Google Scholar] [CrossRef]
- Cazzonelli, C.I.; Pogson, B.J. Source to sink: Regulation of carotenoid biosynthesis in plants. Trends Plant Sci. 2010, 15, 266–274. [Google Scholar] [CrossRef]
- Rodriguez-Concepcion, M. Supply of precursors for carotenoid biosynthesis in plants. Arch. Biochem. Biophys. 2010, 504, 118–122. [Google Scholar] [CrossRef]
- Brown, A.F.; Yousef, G.G.; Chebrolu, K.K.; Byrd, R.W.; Everhart, K.W.; Thomas, A.; Reid, R.W.; Parkin, I.A.P.; Sharpe, A.G.; Oliver, R.; et al. High-density single nucleotide polymorphism (SNP) array mapping in Brassica oleracea: Identification of QTL associated with carotenoid variation in broccoli florets. Theor. Appl. Genet. 2014, 127, 2051–2064. [Google Scholar] [CrossRef]
- Cunningham, F.X., Jr.; Chamovitz, D.; Misawa, N.; Gantt, E.; Hirschberg, J. Cloning and functional expression in Escherichia coli of a cyanobacterial gene for lycopene cyclase, the enzyme that catalyzes the biosynthesis of beta-carotene. FEBS Lett. 1993, 328, 130–138. [Google Scholar] [CrossRef] [Green Version]
- Cunningham, F.X., Jr.; Pogson, B.; Sun, Z.; McDonald, K.A.; DellaPenna, D.; Gantt, E. Functional analysis of the beta and epsilon lycopene cyclase enzymes of Arabidopsis reveals a mechanism for control of cyclic carotenoid formation. Plant Cell 1996, 8, 1613–1626. [Google Scholar] [CrossRef] [Green Version]
- Pecker, I.; Gabbay, R.; Cunningham, F.X., Jr.; Hirschberg, J. Cloning and characterization of the cDNA for lycopene beta-cyclase from tomato reveals decrease in its expression during fruit ripening. Plant Mol. Biol. 1996, 30, 807–819. [Google Scholar] [CrossRef]
- Ronen, G.; Cohen, M.; Zamir, D.; Hirschberg, J. Regulation of carotenoid biosynthesis during tomato fruit development: Expression of the gene for lycopene epsilon-cyclase is down-regulated during ripening and is elevated in the mutant Delta. Plant J. Cell Mol. Biol. 1999, 17, 341–351. [Google Scholar] [CrossRef]
- Nisar, N.; Li, L.; Lu, S.; Khin, N.C.; Pogson, B.J. Carotenoid Metabolism in Plants. Mol. Plant 2015, 8, 68–82. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez-Concepcion, M.; Boronat, A. Breaking new ground in the regulation of the early steps of plant isoprenoid biosynthesis. Curr. Opin. Plant Biol. 2015, 25, 17–22. [Google Scholar] [CrossRef]
- Hermanns, A.S.; Zhou, X.; Xu, Q.; Tadmor, Y.; Li, L. Carotenoid Pigment Accumulation in Horticultural Plants. Hortic. Plant J. 2020, 6, 343–360. [Google Scholar] [CrossRef]
- Ruiz-Sola, M.A.; Rodriguez-Concepcion, M. Carotenoid biosynthesis in Arabidopsis: A colorful pathway. Arab. Book 2012, 10, e0158. [Google Scholar] [CrossRef] [Green Version]
- Clotault, J.; Peltier, D.; Berruyer, R.; Thomas, M.; Briard, M.; Geoffriau, E. Expression of carotenoid biosynthesis genes during carrot root development. J. Exp. Bot. 2008, 59, 3563–3573. [Google Scholar] [CrossRef] [Green Version]
- Fujisawa, M.; Nakano, T.; Shima, Y.; Ito, Y. A Large-Scale Identification of Direct Targets of the Tomato MADS Box Transcription Factor RIPENING INHIBITOR Reveals the Regulation of Fruit Ripening. Plant Cell 2013, 25, 371–386. [Google Scholar] [CrossRef] [Green Version]
- Wu, M.B.; Xu, X.; Hu, X.W.; Liu, Y.D.; Cao, H.H.; Chan, H.E.; Gong, Z.H.; Yuan, Y.J.; Luo, Y.Q.; Feng, B.H.; et al. SlMYB72 Regulates the Metabolism of Chlorophylls, Carotenoids, and Flavonoids in Tomato Fruit(1). Plant Physiol. 2020, 183, 854–868. [Google Scholar] [CrossRef]
- Fraser, P.D.; Truesdale, M.R.; Bird, C.R.; Schuch, W.; Bramley, P.M. Carotenoid Biosynthesis during Tomato Fruit Development (Evidence for Tissue-Specific Gene Expression). Plant Physiol. 1994, 105, 405–413. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Huang, X.-Q.; Cao, T.-J.; Zhuang, Z.; Wang, R.; Lu, S. Heteromeric Geranylgeranyl Diphosphate Synthase Contributes to Carotenoid Biosynthesis in Ripening Fruits of Red Pepper (Capsicum annuum var. conoides). J. Agric. Food Chem. 2018, 66, 11691–11700. [Google Scholar] [CrossRef]
- Paul, P.; Dhandapani, V.; Li, X.; Choi, S.R.; Hur, Y.; Lim, Y.P. Identification of candidate genes involved in the biosynthesis of carotenoids in Brassica rapa. Hortic. Environ. Biotechnol. 2014, 55, 342–351. [Google Scholar] [CrossRef]
- Zhang, L.; Dai, Y.; Yue, L.X.; Chen, G.H.; Yuan, L.Y.; Zhang, S.F.; Li, F.; Zhang, H.; Li, G.L.; Zhu, S.D.; et al. Heat stress response in Chinese cabbage (Brassica rapa L.) revealed by transcriptome and physiological analysis. PeerJ 2022, 10, e13427. [Google Scholar] [CrossRef]
- Feng, H.; Li, Y.; Liu, Z.; Liu, J. Mapping of or, a gene conferring orange color on the inner leaf of the Chinese cabbage (Brassica rapa L. ssp pekinensis). Mol. Breed. 2012, 29, 235–244. [Google Scholar] [CrossRef]
- Matsumoto, E.; Yasui, C.; Ohi, M.; Tsukada, M. Linkage analysis of RFLP markers for clubroot resistance and pigmentation in Chinese cabbage (Brassica rapa ssp. pekinensis). Euphytica 1998, 104, 79–86. [Google Scholar] [CrossRef]
- Zhang, F.; Wang, G.; Wang, M.; Liu, X.; Zhao, X.; Yu, Y.; Zhang, D.; Yu, S. Identification of SCAR markers linked to or, a gene inducing beta-carotene accumulation in Chinese cabbage. Euphytica 2008, 164, 463–471. [Google Scholar] [CrossRef]
- Zhang, J.; Li, H.; Zhang, M.; Hui, M.; Wang, Q.; Li, L.; Zhang, L. Fine mapping and identification of candidate Br-or gene controlling orange head of Chinese cabbage (Brassica rapa L. ssp. pekinensis). Mol. Breed. 2013, 32, 799–805. [Google Scholar] [CrossRef]
- Lee, S.; Lee, S.C.; Byun, D.H.; Lee, D.Y.; Park, J.Y.; Lee, J.H.; Lee, H.O.; Sung, S.H.; Yang, T.J. Association of molecular markers derived from the BrCRTISO1 gene with prolycopene-enriched orange-colored leaves in Brassica rapa [corrected]. Theor. Appl. Genet. 2014, 127, 179–191. [Google Scholar] [CrossRef]
- Li, P.; Zhang, S.; Zhang, S.; Li, F.; Zhang, H.; Liu, X.; Wu, J.; Wang, X.; Sun, R. Carotenoid identification and molecular analysis of carotenoid isomerase-encoding BrCRTISO, the candidate gene for inner leaf orange coloration in Chinese cabbage. Mol. Breed. 2015, 35, 72. [Google Scholar] [CrossRef]
- Su, T.; Yu, S.; Zhang, J.W.F.; Yu, Y.; Zhang, D.; Zhao, X.; Wang, W. Loss of Function of the Carotenoid Isomerase Gene BrCRTISO Confers Orange Color to the Inner Leaves of Chinese Cabbage (Brassica rapa L. ssp pekinensis). Plant Mol. Biol. Report. 2015, 33, 648–659. [Google Scholar] [CrossRef]
- Zou, C.L.; Zheng, Y.; Wang, P.; Zhang, X.; Wang, Y.H.; Liu, Z.Y.; Feng, H. Fine mapping and characterization of the or gene in Chinese cabbage (Brassica rapa L. ssp pekinensis). Genet. Mol. Res. 2016, 15. [Google Scholar] [CrossRef]
- Yuan, H.; Zhang, J.; Nageswaran, D.; Li, L. Carotenoid metabolism and regulation in horticultural crops. Hortic. Res. 2015, 2, 15036. [Google Scholar] [CrossRef]
- Michelmore, R.W.; Paran, I.; Kesseli, R.V. Identification of markers linked to disease-resistance genes by bulked segregant analysis: A rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. USA 1991, 88, 9828–9832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hill, J.T.; Demarest, B.L.; Bisgrove, B.W.; Gorsi, B.; Su, Y.C.; Yost, H.J. MMAPPR: Mutation Mapping Analysis Pipeline for Pooled RNA-seq. Genome Res. 2013, 23, 687–697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fekih, R.; Takagi, H.; Tamiru, M.; Abe, A.; Natsume, S.; Yaegashi, H.; Sharma, S.; Sharma, S.; Kanzaki, H.; Matsumura, H. MutMap plus: Genetic Mapping and Mutant Identification without Crossing in Rice. PLoS ONE 2013, 8, e68529. [Google Scholar] [CrossRef]
- Lu, S.; Van Eck, J.; Zhou, X.; Lopez, A.B.; O’Halloran, D.M.; Cosman, K.M.; Conlin, B.J.; Paolillo, D.J.; Garvin, D.F.; Vrebalov, J.; et al. The cauliflower or gene encodes a DnaJ cysteine-rich domain-containing protein that mediates high levels of beta-carotene accumulation. Plant Cell 2006, 18, 3594–3605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, T.; Zhou, F.; Huang, X.-Q.; Chen, W.-C.; Kong, M.-J.; Zhou, C.-F.; Zhuang, Z.; Li, L.; Lu, S. ORANGE Represses Chloroplast Biogenesis in Etiolated Arabidopsis Cotyledons via Interaction with TCP14. Plant Cell 2019, 31, 2996–3014. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Welsch, R.; Yang, Y.; Alvarez, D.; Riediger, M.; Yuan, H.; Fish, T.; Liu, J.; Thannhauser, T.W.; Li, L. Arabidopsis OR proteins are the major posttranscriptional regulators of phytoene synthase in controlling carotenoid biosynthesis. Proc. Natl. Acad. Sci. USA 2015, 112, 3558–3563. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Paolillo, D.J.; Parthasarathy, M.V.; DiMuzio, E.M.; Garvin, D.F. A novel gene mutation that confers abnormal patterns of beta-carotene accumulation in cauliflower (Brassica oleracea var. botrytis). Plant J. 2001, 26, 59–67. [Google Scholar] [CrossRef]
- Yuan, H.; Owsiany, K.; Sheeja, T.E.; Zhou, X.; Rodriguez, C.; Li, Y.; Welsch, R.; Chayut, N.; Yang, Y.; Thannhauser, T.W.; et al. A Single Amino Acid Substitution in an ORANGE Protein Promotes Carotenoid Overaccumulation in Arabidopsis. Plant Physiol. 2015, 169, 421–431. [Google Scholar] [CrossRef] [Green Version]
- Domonkos, I.; Kis, M.; Gombos, Z.; Ughy, B. Carotenoids, versatile components of oxygenic photosynthesis. Prog. Lipid Res. 2013, 52, 539–561. [Google Scholar] [CrossRef]
- Maass, D.; Arango, J.; Wuest, F.; Beyer, P.; Welsch, R. Carotenoid Crystal Formation in Arabidopsis and Carrot Roots Caused by Increased Phytoene Synthase Protein Levels. PLoS ONE 2009, 4, e6373. [Google Scholar] [CrossRef]
- Bai, C.; Rivera, S.M.; Medina, V.; Alves, R.; Vilaprinyo, E.; Sorribas, A.; Canela, R.; Capell, T.; Sandmann, G.; Christou, P.; et al. An in vitro system for the rapid functional characterization of genes involved in carotenoid biosynthesis and accumulation. Plant J. 2014, 77, 464–475. [Google Scholar] [CrossRef] [PubMed]
- Chayut, N.; Yuan, H.; Ohali, S.; Meir, A.; Sa’ar, U.; Tzuri, G.; Zheng, Y.; Mazourek, M.; Gepstein, S.; Zhou, X.; et al. Distinct Mechanisms of the ORANGE Protein in Controlling Carotenoid Flux. Plant Physiol. 2017, 173, 376–389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Welsch, R.; Zhou, X.; Yuan, H.; Alvarez, D.; Sun, T.; Schlossarek, D.; Yang, Y.; Shen, G.; Zhang, H.; Rodriguez-Concepcion, M.; et al. Clp Protease and OR Directly Control the Proteostasis of Phytoene Synthase, the Crucial Enzyme for Carotenoid Biosynthesis in Arabidopsis. Mol. Plant 2018, 11, 149–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shevtsov, M.B.; Chen, Y.L.; Gollnick, P.; Antson, A.A. Crystal structure of Bacillus subtilis anti-TRAP protein, an antagonist of TRAP/RNA interaction. Proc. Natl. Acad. Sci. USA 2005, 102, 17600–17605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, M.; Deng, L.; Guo, S.; Yuan, G.; Li, C.; Li, C. Alternative transcription and feedback regulation suggest that SlIDI1 is involved in tomato carotenoid synthesis in a complex way. Hortic. Res. 2022, 9, uhab045. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Sola, M.A.; Coman, D.; Beck, G.; Barja, M.V.; Colinas, M.; Graf, A.; Welsch, R.; Ruetimann, P.; Buehlmann, P.; Bigler, L.; et al. Arabidopsis GERANYLGERANYL DIPHOSPHATE SYNTHASE 11 is a hub isozyme required for the production of most photosynthesis-related isoprenoids. New Phytol. 2016, 209, 252–264. [Google Scholar] [CrossRef]
- Camagna, M.; Grundmann, A.; Bar, C.; Koschmieder, J.; Beyer, P.; Welsch, R. Enzyme Fusion Removes Competition for Geranylgeranyl Diphosphate in Carotenogenesis. Plant Physiol. 2019, 179, 1013–1027. [Google Scholar] [CrossRef] [Green Version]
- Kaul, S.; Koo, H.L.; Jenkins, J.; Rizzo, M.; Rooney, T.; Tallon, L.J.; Feldblyum, T.; Nierman, W.; Benito, M.I.; Lin, X.Y. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000, 408, 796–815. [Google Scholar]
- Wang, X.; Wang, H.; Wang, J.; Sun, R.; Wu, J.; Liu, S.; Bai, Y.; Mun, J.-H.; Bancroft, I.; Cheng, F. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 2011, 43, 1035–1039. [Google Scholar] [CrossRef] [Green Version]
- Matsumoto, T.; Wu, J.Z.; Kanamori, H.; Katayose, Y.; Fujisawa, M.; Namiki, N.; Mizuno, H.; Yamamoto, K.; Antonio, B.A.; Baba, T.; et al. The map-based sequence of the rice genome. Nature 2005, 436, 793–800. [Google Scholar]
- Macia, A.; Blanco-Jimenez, E.; Garcia-Perez, J.L. Retrotransposons in pluripotent cells: Impact and new roles in cellular plasticity. Biochim. Biophys. Acta-Gene Regul. Mech. 2015, 1849, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Paolillo, D.J.; Garvin, D.F.; Parthasarathy, M.V. The chromoplasts of Or mutants of cauliflower (Brassica oleracea L. var. botrytis). Protoplasma 2004, 224, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Yazdani, M.; Sun, Z.X.; Yuan, H.; Zeng, S.H.; Thannhauser, T.W.; Vrebalov, J.; Ma, Q.Y.; Xu, Y.M.; Fei, Z.J.; Van Eck, J.; et al. Ectopic expression of ORANGE promotes carotenoid accumulation and fruit development in tomato. Plant Biotechnol. J. 2019, 17, 33–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tzuri, G.; Zhou, X.J.; Chayut, N.; Yuan, H.; Portnoy, V.; Meir, A.; Sa’ar, U.; Baumkoler, F.; Mazourek, M.; Lewinsohn, E.; et al. A ‘golden’ SNP in CmOr governs the fruit flesh color of melon (Cucumis melo). Plant J. 2015, 82, 267–279. [Google Scholar] [CrossRef]
- Mathur, J.; Koncz, C. Establishment and maintenance of cell suspension cultures. Methods Mol. Biol. 1998, 82, 27–30. [Google Scholar]
- Fantini, E.; Falcone, G.; Frusciante, S.; Giliberto, L.; Giuliano, G. Dissection of Tomato Lycopene Biosynthesis through Virus-Induced Gene Silencing. Plant Physiol. 2013, 163, 986–998. [Google Scholar] [CrossRef]
- Meynert, A.M.; Ansari, M.; FitzPatrick, D.R.; Taylor, M.S. Variant detection sensitivity and biases in whole genome and exome sequencing. BMC Bioinform. 2014, 15, 247. [Google Scholar] [CrossRef] [Green Version]
- Islam, M.S.; Zeng, L.; Thyssen, G.N.; Delhom, C.D.; Kim, H.J.; Li, P.; Fang, D.D. Mapping by sequencing in cotton (Gossypium hirsutum) line MD52ne identified candidate genes for fiber strength and its related quality attributes. Theor. Appl. Genet. 2016, 129, 1071–1086. [Google Scholar] [CrossRef]
- Wang, N.; Liu, Z.; Zhang, Y.; Li, C.; Feng, H. Identification and fine mapping of a stay-green gene (Brnye1) in pakchoi (Brassica campestris L. ssp chinensis). Theor. Appl. Genet. 2018, 131, 673–684. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Hong, B.; Tong, Z.; Ma, N.; Kasuga, M.; Yamaguchi-Shinozaki, K.; Gao, J.-P. Expression of the Arabidopsis DREB1A gene in transgenic chrysanthemum enhances tolerance to low temperature. J. Hortic. Sci. Biotechnol. 2006, 81, 1002–1008. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, Z.; Liang, J.; Wu, J.; Cheng, F.; Wang, X. Three genes encoding AOP2, a protein involved in aliphatic glucosinolate biosynthesis, are differentially expressed in Brassica rapa. J. Exp. Bot. 2015, 66, 6205–6218. [Google Scholar] [CrossRef] [PubMed]





| Populations | Generations | Total Plants | Non−Golden Inner Leaves | Golden Inner Leaves | Expected Ratio | χ2 Test (Chi−Squared Test) |
|---|---|---|---|---|---|---|
| Tri−crossed hybrid lines | P1 (1900262, aa genotype) | 20 | 20 | |||
| P2 (1900264, Aa genotype) | 20 | 20 | ||||
| P2 × P1 | 151 | 80 | 71 | 1:1 | 0.54 |
| Component Name | Core Sequence | Numbers | Predictive Function |
|---|---|---|---|
| MBS | CAACTG | 2 | MYB binding site involved in drought-inducibility |
| RY-element | CATGCATG | 1 | Cis-acting regulatory element involved in seed-specific regulation |
| GATA-motif | GATAGGA | 1 | Part of a light-responsive element |
| TCA-element | CCATCTTTTT | 1 | Cis-acting element involved in salicylic acid responsiveness |
| Motif I | gGTACGTGGCG | 1 | Cis-acting regulatory element root specific |
| Box 4 | ATTAAT | 1 | Part of a conserved DNA module involved in light responsiveness |
| ARE | AAACCA | 4 | Cis-acting regulatory element essential for the anaerobic induction |
| ABRE | ACGTG | 3 | Cis-acting element involved in the abscisic acid responsiveness |
| TATA-box | TATATA | 19 | Core promoter element around −30 of transcription start |
| TATA-box | TATA | 15 | |
| TATA-box | TATAAAA | 5 | |
| TATA-box | TATTTAAA | 1 | |
| TATA-box | TATAA | 3 | |
| TATA-box | ccTATAAAaa | 1 | |
| TCT-motif | TCTTAC | 2 | Part of a light-responsive element |
| CAAT-box | CCAAT | 9 | Common cis-acting element in promoter and enhancer regions |
| CAAT-box | CAAT | 10 | |
| I-box | GTATAAGGCC | 1 | Part of a light-responsive element |
| G-box | CACGTG | 2 | Cis-acting regulatory element involved in light responsiveness |
| G-box | TACGTG | 1 | |
| motif_sequence | 10 | Short-function | |
| W box | TTGACC | 1 | Cis-acting regulatory element essential for the pathogen induction |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Zhang, S.; Dai, Y.; Wang, S.; Wang, C.; Li, F.; Zhang, H.; Chen, G.; Yuan, L.; Hou, J.; et al. Mapping and Validation of BrGOLDEN: A Dominant Gene Regulating Carotenoid Accumulation in Brassica rapa. Int. J. Mol. Sci. 2022, 23, 12442. https://doi.org/10.3390/ijms232012442
Zhang L, Zhang S, Dai Y, Wang S, Wang C, Li F, Zhang H, Chen G, Yuan L, Hou J, et al. Mapping and Validation of BrGOLDEN: A Dominant Gene Regulating Carotenoid Accumulation in Brassica rapa. International Journal of Molecular Sciences. 2022; 23(20):12442. https://doi.org/10.3390/ijms232012442
Chicago/Turabian StyleZhang, Lei, Shifan Zhang, Yun Dai, Shaoxing Wang, Chenggang Wang, Fei Li, Hui Zhang, Guohu Chen, Lingyun Yuan, Jinfeng Hou, and et al. 2022. "Mapping and Validation of BrGOLDEN: A Dominant Gene Regulating Carotenoid Accumulation in Brassica rapa" International Journal of Molecular Sciences 23, no. 20: 12442. https://doi.org/10.3390/ijms232012442
APA StyleZhang, L., Zhang, S., Dai, Y., Wang, S., Wang, C., Li, F., Zhang, H., Chen, G., Yuan, L., Hou, J., Tang, X., Zhu, S., Sun, R., Li, G., & Zhang, S. (2022). Mapping and Validation of BrGOLDEN: A Dominant Gene Regulating Carotenoid Accumulation in Brassica rapa. International Journal of Molecular Sciences, 23(20), 12442. https://doi.org/10.3390/ijms232012442






