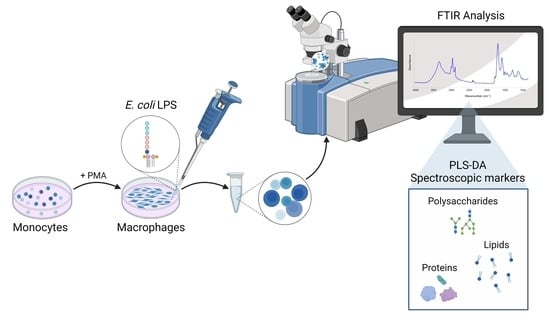
A Global Picture of Molecular Changes Associated to LPS Treatment in THP-1 Derived Human Macrophages by Fourier Transform Infrared Microspectroscopy
Abstract
1. Introduction
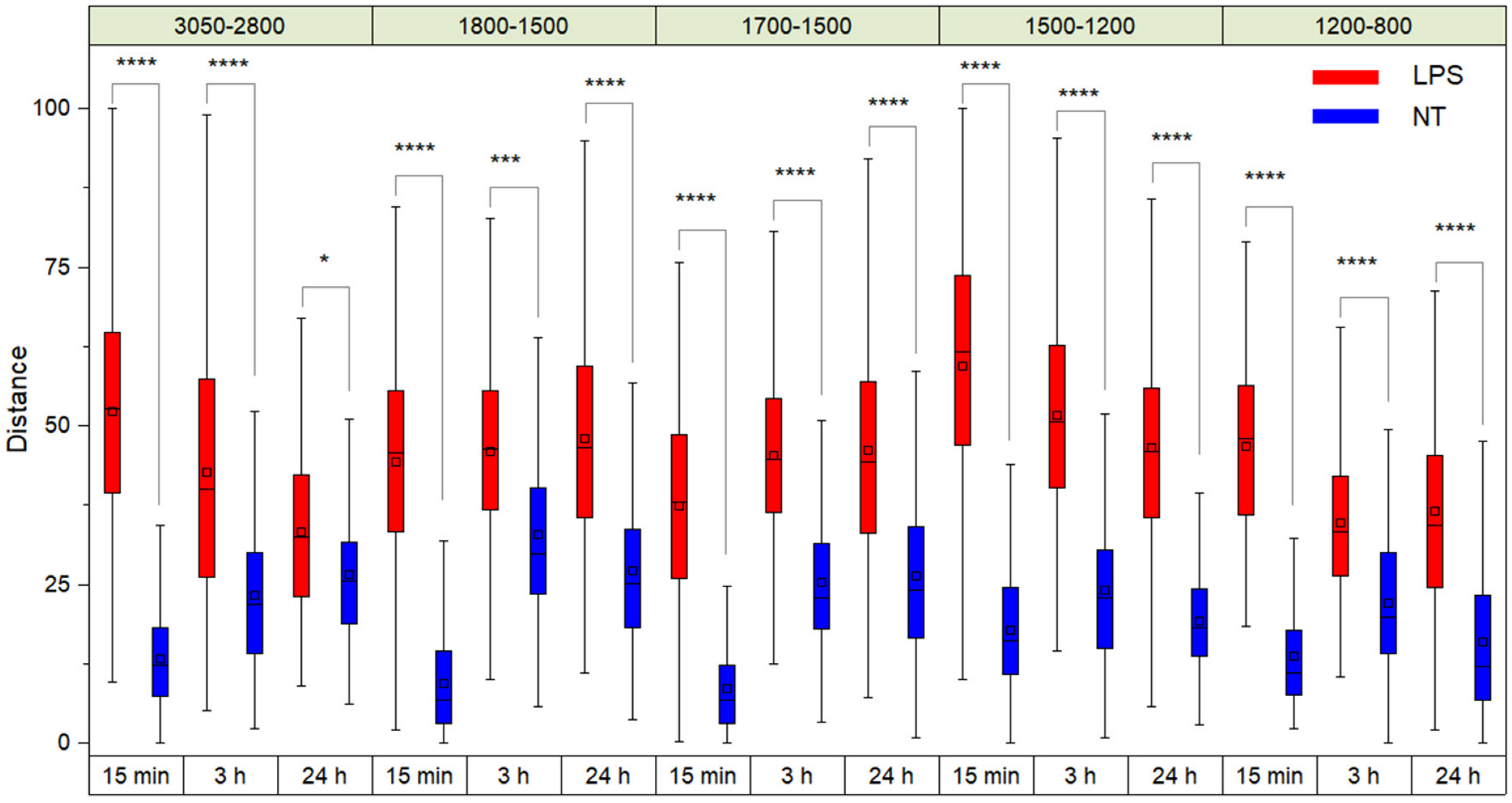
2. Results and Discussion
2.1. FTIR Analysis of Protein Secondary Structure Modifications in TDM Cells Exposed to LPS
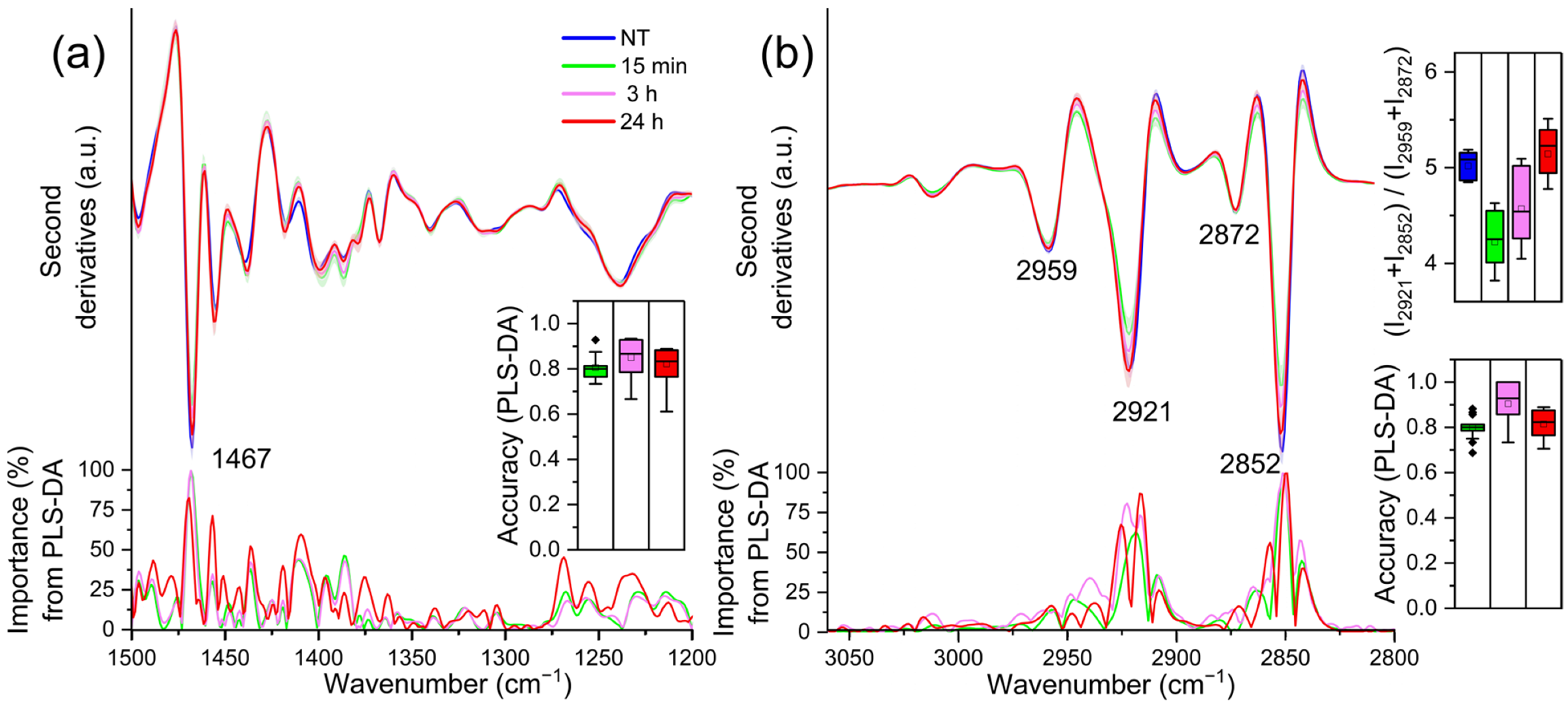
2.2. FTIR Analysis of Lipid Modifications in TDM Cells upon Exposure to LPS: Insights from the 1500–1200 cm−1 and 3050–2800 cm−1 Spectral Ranges
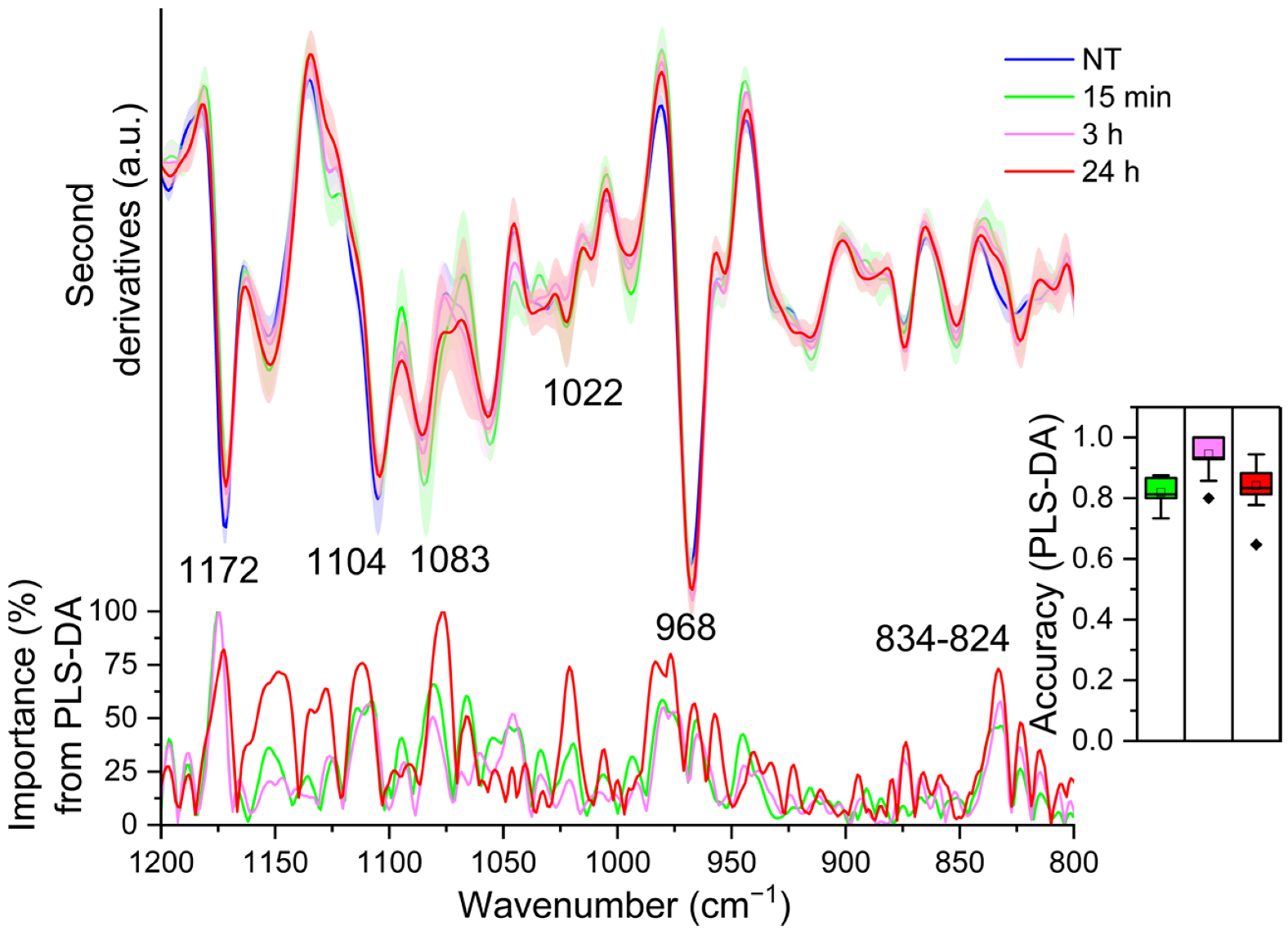
2.3. FTIR Analysis of TDM Exposed to LPS: Analysis of the Fingerprint Region
3. Materials and Methods
3.1. Cell Maintenance
3.2. Cell Differentiation
3.3. Cell Stimulation and Treatments
3.4. FTIR Microspectroscopy Analysis of Intact THP-1 Derived Macrophage-Like Cells
3.5. Multivariate Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Takeuchi, O.; Akira, S. Pattern Recognition Receptors and Inflammation. Cell 2010, 140, 805–820. [Google Scholar] [CrossRef] [PubMed]
- Viola, A.; Munari, F.; Sánchez-Rodríguez, R.; Scolaro, T.; Castegna, A. The Metabolic Signature of Macrophage Responses. Front. Immunol. 2019, 10, 1462. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Prados, J.-C.; Través, P.G.; Cuenca, J.; Rico, D.; Aragonés, J.; Martín-Sanz, P.; Cascante, M.; Boscá, L. Substrate Fate in Activated Macrophages: A Comparison between Innate, Classic, and Alternative Activation. J. Immunol. Baltim. Md 1950 2010, 185, 605–614. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, K.A.; Kagan, J.C. Toll-like Receptors and the Control of Immunity. Cell 2020, 180, 1044–1066. [Google Scholar] [CrossRef] [PubMed]
- Chanput, W.; Mes, J.J.; Wichers, H.J. THP-1 Cell Line: An in Vitro Cell Model for Immune Modulation Approach. Int. Immunopharmacol. 2014, 23, 37–45. [Google Scholar] [CrossRef]
- Sharif, O.; Bolshakov, V.N.; Raines, S.; Newham, P.; Perkins, N.D. Transcriptional Profiling of the LPS Induced NF-ΚB Response in Macrophages. BMC Immunol. 2007, 8, 1. [Google Scholar] [CrossRef]
- Rossetti, C.; Peri, F. (Eds.) The Role of Toll-like Receptor 4 in Infectious and Non Infectious Inflammation; Progress in Inflammation Research; Springer International Publishing: Cham, Switerland, 2021; Volume 87, ISBN 978-3-030-56318-9. [Google Scholar]
- Swanson, K.V.; Deng, M.; Ting, J.P.-Y. The NLRP3 Inflammasome: Molecular Activation and Regulation to Therapeutics. Nat. Rev. Immunol. 2019, 19, 477–489. [Google Scholar] [CrossRef]
- Ma, J.; Wei, K.; Liu, J.; Tang, K.; Zhang, H.; Zhu, L.; Chen, J.; Li, F.; Xu, P.; Chen, J.; et al. Glycogen Metabolism Regulates Macrophage-Mediated Acute Inflammatory Responses. Nat. Commun. 2020, 11, 1769. [Google Scholar] [CrossRef]
- Regdon, Z.; Robaszkiewicz, A.; Kovács, K.; Rygielska, Ż.; Hegedűs, C.; Bodoor, K.; Szabó, É.; Virág, L. LPS Protects Macrophages from AIF-Independent Parthanatos by Downregulation of PARP1 Expression, Induction of SOD2 Expression, and a Metabolic Shift to Aerobic Glycolysis. Free Radic. Biol. Med. 2019, 131, 184–196. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, Y.; Wang, F.; Wang, Z.; Lu, Y.; Xu, Y.; Wang, K.; Shen, H.; Yang, P.; Li, S.; et al. Quantitative Profiling of Glycerophospholipids during Mouse and Human Macrophage Differentiation Using Targeted Mass Spectrometry. Sci. Rep. 2017, 7, 412. [Google Scholar] [CrossRef]
- Ami, D.; Natalello, A.; Doglia, S.M. Fourier Transform Infrared Microspectroscopy of Complex Biological Systems: From Intact Cells to Whole Organisms. Methods Mol. Biol. 2012, 895, 85–100. [Google Scholar] [CrossRef] [PubMed]
- Baker, M.J.; Trevisan, J.; Bassan, P.; Bhargava, R.; Butler, H.J.; Dorling, K.M.; Fielden, P.R.; Fogarty, S.W.; Fullwood, N.J.; Heys, K.A.; et al. Using Fourier Transform IR Spectroscopy to Analyze Biological Materials. Nat. Protoc. 2014, 9, 1771–1791. [Google Scholar] [CrossRef] [PubMed]
- Mantsch, H.H. The Evolution of Biomedical Vibrational Spectroscopy: A Personal Perspective. Biomed. Spectrosc. Imaging 2015, 4, 315–329. [Google Scholar] [CrossRef]
- Mantsch, H.H. Biomedical Vibrational Spectroscopy in the Era of Artificial Intelligence. Molecules 2021, 26, 1439. [Google Scholar] [CrossRef]
- Miller, L.M.; Dumas, P. From Structure to Cellular Mechanism with Infrared Microspectroscopy. Curr. Opin. Struct. Biol. 2010, 20, 649–656. [Google Scholar] [CrossRef]
- Finlayson, D.; Rinaldi, C.; Baker, M.J. Is Infrared Spectroscopy Ready for the Clinic? Anal. Chem. 2019, 91, 12117–12128. [Google Scholar] [CrossRef]
- Morais, C.L.M.; Lima, K.M.G.; Singh, M.; Martin, F.L. Tutorial: Multivariate Classification for Vibrational Spectroscopy in Biological Samples. Nat. Protoc. 2020, 15, 2143–2162. [Google Scholar] [CrossRef]
- Ami, D.; Mereghetti, P.; Natalello, A. Contribution of Infrared Spectroscopy to the Understanding of Amyloid Protein Aggregation in Complex Systems. Front. Mol. Biosci. 2022, 9, 822852. [Google Scholar] [CrossRef]
- Ami, D.; Doglia, S.M.; Mereghetti, P. Multivariate Analysis for Fourier Transform Infrared Spectra of Complex Biological Systems and Processes; IntechOpen: London, UK, 2013. [Google Scholar]
- Barth, A. Infrared Spectroscopy of Proteins. Biochim. Biophys. Acta-Bioenerg. 2007, 1767, 1073–1101. [Google Scholar] [CrossRef]
- Tamm, L.K.; Tatulian, S.A. Infrared Spectroscopy of Proteins and Peptides in Lipid Bilayers. Q. Rev. Biophys. 1997, 30, 365–429. [Google Scholar] [CrossRef]
- Seshadri, S.; Khurana, R.; Fink, A.L. Fourier Transform Infrared Spectroscopy in Analysis of Protein Deposits. Methods Enzymol. 1999, 309, 559–576. [Google Scholar] [CrossRef] [PubMed]
- Natalello, A.; Doglia, S.M.; Carey, J.; Grandori, R. Role of Flavin Mononucleotide in the Thermostability and Oligomerization of Escherichia Coli Stress-Defense Protein WrbA. Biochemistry 2007, 46, 543–553. [Google Scholar] [CrossRef] [PubMed]
- Meijer, K.; Weening, D.; de Vries, M.P.; Priebe, M.G.; Vonk, R.J.; Roelofsen, H. Quantitative Proteomics Analyses of Activation States of Human THP-1 Macrophages. J. Proteom. 2015, 128, 164–172. [Google Scholar] [CrossRef] [PubMed]
- Kolseth, I.B.M.; Reine, T.M.; Vuong, T.T.; Meen, A.J.; Fan, Q.; Jenssen, T.G.; Grønning-Wang, L.M.; Kolset, S.O. Serglycin Is Part of the Secretory Repertoire of LPS-Activated Monocytes. Immun. Inflamm. Dis. 2015, 3, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Dhungana, S.; Merrick, B.A.; Tomer, K.B.; Fessler, M.B. Quantitative Proteomics Analysis of Macrophage Rafts Reveals Compartmentalized Activation of the Proteasome and of Proteasome-Mediated ERK Activation in Response to Lipopolysaccharide. Mol. Cell. Proteom. 2009, 8, 201–213. [Google Scholar] [CrossRef] [PubMed]
- Balka, K.R.; De Nardo, D. Understanding Early TLR Signaling through the Myddosome. J. Leukoc. Biol. 2019, 105, 339–351. [Google Scholar] [CrossRef]
- Kieser, K.J.; Kagan, J.C. Multi-Receptor Detection of Individual Bacterial Products by the Innate Immune System. Nat. Rev. Immunol. 2017, 17, 376–390. [Google Scholar] [CrossRef]
- Casal, H.L.; Mantsch, H.H. Polymorphic Phase Behaviour of Phospholipid Membranes Studied by Infrared Spectroscopy. BBA-Rev. Biomembr. 1984, 779, 381–401. [Google Scholar] [CrossRef]
- Lewis, R.N.A.H.; McElhaney, R.N. Fourier Transform Infrared Spectroscopy in the Study of Lipid Phase Transitions in Model and Biological Membranes. Practical Considerations. In Methods in Molecular Biology; Dopico, A.M., Ed.; Humana Press: Totowa, NJ, USA, 2007; pp. 207–226. ISBN 978-1-59745-519-0. [Google Scholar]
- Banyay, M.; Sarkar, M.; Gräslund, A. A Library of IR Bands of Nucleic Acids in Solution. Biophys. Chem. 2003, 104, 477–488. [Google Scholar] [CrossRef]
- Dučić, T.; Stamenković, S.; Lai, B.; Andjus, P.; Lučić, V. Multimodal Synchrotron Radiation Microscopy of Intact Astrocytes from the HSOD1 G93A Rat Model of Amyotrophic Lateral Sclerosis. Anal. Chem. 2019, 91, 1460–1471. [Google Scholar] [CrossRef]
- Ami, D.; Duse, A.; Mereghetti, P.; Cozza, F.; Ambrosio, F.; Ponzini, E.; Grandori, R.; Lunetta, C.; Tavazzi, S.; Pezzoli, F.; et al. Tear-Based Vibrational Spectroscopy Applied to Amyotrophic Lateral Sclerosis. Anal. Chem. 2021, 93, 16995–17002. [Google Scholar] [CrossRef] [PubMed]
- Batista-Gonzalez, A.; Vidal, R.; Criollo, A.; Carreño, L.J. New Insights on the Role of Lipid Metabolism in the Metabolic Reprogramming of Macrophages. Front. Immunol. 2020, 10, 2993. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.W.; Mok, H.J.; Lee, D.Y.; Park, S.C.; Kim, G.-S.; Lee, S.-E.; Lee, Y.-S.; Kim, K.P.; Kim, H.D. UPLC-QqQ/MS-Based Lipidomics Approach To Characterize Lipid Alterations in Inflammatory Macrophages. J. Proteome Res. 2017, 16, 1460–1469. [Google Scholar] [CrossRef] [PubMed]
- Olona, A.; Hateley, C.; Muralidharan, S.; Wenk, M.R.; Torta, F.; Behmoaras, J. Sphingolipid Metabolism during Toll-like Receptor 4 (TLR4)-mediated Macrophage Activation. Br. J. Pharmacol. 2021, 178, 4575–4587. [Google Scholar] [CrossRef] [PubMed]
- Conde, T.A.; Mendes, L.; Gaspar, V.M.; Mano, J.F.; Melo, T.; Domingues, M.R.; Duarte, I.F. Differential Modulation of the Phospholipidome of Proinflammatory Human Macrophages by the Flavonoids Quercetin, Naringin and Naringenin. Molecules 2020, 25, 3460. [Google Scholar] [CrossRef]
- Castoldi, A.; Monteiro, L.B.; van Teijlingen Bakker, N.; Sanin, D.E.; Rana, N.; Corrado, M.; Cameron, A.M.; Hässler, F.; Matsushita, M.; Caputa, G.; et al. Triacylglycerol Synthesis Enhances Macrophage Inflammatory Function. Nat. Commun. 2020, 11, 4107. [Google Scholar] [CrossRef]
- Everts, B.; Amiel, E.; Huang, S.C.-C.; Smith, A.M.; Chang, C.-H.; Lam, W.Y.; Redmann, V.; Freitas, T.C.; Blagih, J.; van der Windt, G.J.W.; et al. TLR-Driven Early Glycolytic Reprogramming via the Kinases TBK1-IKKɛ Supports the Anabolic Demands of Dendritic Cell Activation. Nat. Immunol. 2014, 15, 323–332. [Google Scholar] [CrossRef]
- Moon, J.-S.; Lee, S.; Park, M.-A.; Siempos, I.I.; Haslip, M.; Lee, P.J.; Yun, M.; Kim, C.K.; Howrylak, J.; Ryter, S.W.; et al. UCP2-Induced Fatty Acid Synthase Promotes NLRP3 Inflammasome Activation during Sepsis. J. Clin. Investig. 2015, 125, 665–680. [Google Scholar] [CrossRef]
- Gazi, E.; Dwyer, J.; Lockyer, N.P.; Gardner, P.; Shanks, J.H.; Roulson, J.; Hart, C.A.; Clarke, N.W.; Brown, M.D. Biomolecular Profiling of Metastatic Prostate Cancer Cells in Bone Marrow Tissue Using FTIR Microspectroscopy: A Pilot Study. Anal. Bioanal. Chem. 2007, 387, 1621–1631. [Google Scholar] [CrossRef]
- Kacuráková, M. FT-IR Study of Plant Cell Wall Model Compounds: Pectic Polysaccharides and Hemicelluloses. Carbohydr. Polym. 2000, 43, 195–203. [Google Scholar] [CrossRef]
- Brézillon, S.; Untereiner, V.; Lovergne, L.; Tadeo, I.; Noguera, R.; Maquart, F.X.; Wegrowski, Y.; Sockalingum, G.D. Glycosaminoglycan Profiling in Different Cell Types Using Infrared Spectroscopy and Imaging. Anal. Bioanal. Chem. 2014, 406, 5795–5803. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, H.T.; Untereiner, V.; Cinque, G.; Ibrahim, S.A.; Götte, M.; Nguyen, N.Q.; Rivet, R.; Sockalingum, G.D.; Brézillon, S. Infrared Microspectroscopy and Imaging Analysis of Inflammatory and Non-Inflammatory Breast Cancer Cells and Their GAG Secretome. Molecules 2020, 25, 4300. [Google Scholar] [CrossRef] [PubMed]
- Makatsori, E.; Karamanos, N.K.; Papadogiannakis, N.; Hjerpe, A.; Anastassiou, E.D.; Tsegenidis, T. Synthesis and Distribution of Glycosaminoglycans in Human Leukemic B- and T-Cells and Monocytes Studied Using Specific Enzymic Treatments and High-Performance Liquid Chromatography. Biomed. Chromatogr. 2001, 15, 413–417. [Google Scholar] [CrossRef] [PubMed]
- Taylor, K.R.; Gallo, R.L. Glycosaminoglycans and Their Proteoglycans: Host-associated Molecular Patterns for Initiation and Modulation of Inflammation. FASEB J. 2006, 20, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Derenne, A.; Derfoufi, K.-M.; Cowper, B.; Delporte, C.; Goormaghtigh, E. FTIR Spectroscopy as an Analytical Tool to Compare Glycosylation in Therapeutic Monoclonal Antibodies. Anal. Chim. Acta 2020, 1112, 62–71. [Google Scholar] [CrossRef]
- Kirschbaum, C.; Greis, K.; Mucha, E.; Kain, L.; Deng, S.; Zappe, A.; Gewinner, S.; Schöllkopf, W.; von Helden, G.; Meijer, G.; et al. Unravelling the Structural Complexity of Glycolipids with Cryogenic Infrared Spectroscopy. Nat. Commun. 2021, 12, 1201. [Google Scholar] [CrossRef]
- Kačuráková, M.; Mathlouthi, M. FTIR and Laser-Raman Spectra of Oligosaccharides in Water: Characterization of the Glycosidic Bond. Carbohydr. Res. 1996, 284, 145–157. [Google Scholar] [CrossRef]
- Pan, N.C.; Pereira, H.C.B.; da Silva, M.d.L.C.; Vasconcelos, A.F.D.; Celligoi, M.A.P.C. Improvement Production of Hyaluronic Acid by Streptococcus Zooepidemicus in Sugarcane Molasses. Appl. Biochem. Biotechnol. 2017, 182, 276–293. [Google Scholar] [CrossRef]
- Synytsya, A. Fourier Transform Raman and Infrared Spectroscopy of Pectins. Carbohydr. Polym. 2003, 54, 97–106. [Google Scholar] [CrossRef]
- Foster, A.B.; Martlew, E.F.; Stacey, M.; Taylor, P.J.M.; Webber, J.M. 236. Amino-Sugars and Related Compounds. Part VIII. Some Properties of 2-Deoxy-2-Sulphoamino-D-Glucose, Heparin, and Related Substances. J. Chem. Soc. Resumed 1961, 1204–1208. [Google Scholar] [CrossRef]
- Parker, F.S. Applications of Infrared Spectroscopy in Biochemistry, Biology, and Medicine; Springer US: Boston, MA, USA, 1971; ISBN 978-1-4684-1874-3. [Google Scholar]
- Zhao, Y.; Mahajan, G.; Kothapalli, C.R.; Sun, X.-L. Sialylation Status and Mechanical Properties of THP-1 Macrophages upon LPS Stimulation. Biochem. Biophys. Res. Commun. 2019, 518, 573–578. [Google Scholar] [CrossRef] [PubMed]
- Su, L.; Athamna, M.; Wang, Y.; Wang, J.; Freudenberg, M.; Yue, T.; Wang, J.; Moresco, E.M.Y.; He, H.; Zor, T.; et al. Sulfatides Are Endogenous Ligands for the TLR4-MD-2 Complex. Proc. Natl. Acad. Sci. USA 2021, 118, e2105316118. [Google Scholar] [CrossRef] [PubMed]
- Artusa, V.; Ciaramelli, C.; D’Aloia, A.; Facchini, F.A.; Gotri, N.; Bruno, A.; Costa, B.; Palmioli, A.; Airoldi, C.; Peri, F. Green and Roasted Coffee Extracts Inhibit Interferon-β Release in LPS-Stimulated Human Macrophages. Front. Pharmacol. 2022, 13, 806010. [Google Scholar] [CrossRef] [PubMed]
- Facchini, F.A.; Minotti, A.; Luraghi, A.; Romerio, A.; Gotri, N.; Matamoros-Recio, A.; Iannucci, A.; Palmer, C.; Wang, G.; Ingram, R.; et al. Synthetic Glycolipids as Molecular Vaccine Adjuvants: Mechanism of Action in Human Cells and In Vivo Activity. J. Med. Chem. 2021, 64, 12261–12272. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yan, Y.-B. Probing Conformational Changes of Proteins by Quantitative Second-Derivative Infrared Spectroscopy. Anal. Biochem. 2005, 340, 89–98. [Google Scholar] [CrossRef]
- Song, C.L.; Kazarian, S.G. Effect of Controlled Humidity and Tissue Hydration on Colon Cancer Diagnostic via FTIR Spectroscopic Imaging. Anal. Chem. 2020, 92, 9691–9698. [Google Scholar] [CrossRef]
- Pérez-Enciso, M.; Tenenhaus, M. Prediction of Clinical Outcome with Microarray Data: A Partial Least Squares Discriminant Analysis (PLS-DA) Approach. Hum. Genet. 2003, 112, 581–592. [Google Scholar] [CrossRef]
- Breiman, L.; Friedman, J.; Stone, C.J.; Olshen, R.A. Classification and Regression Trees; The Wadsworth and Brooks-Cole Statistics-Probability Series; Taylor & Francis: Abingdon-on-Thames, UK, 1984; ISBN 978-0-412-04841-8. [Google Scholar]
- Dunkhunthod, B.; Talabnin, C.; Murphy, M.; Thumanu, K.; Sittisart, P.; Eumkeb, G. Gymnema inodorum (Lour.) Decne. Extract Alleviates Oxidative Stress and Inflammatory Mediators Produced by RAW264.7 Macrophages. Oxidative Med. Cell. Longev. 2021, 2021, 8658314. [Google Scholar] [CrossRef]
- Paemanee, A.; Rattanabunyong, S.; Ketngamkum, Y.; Siriwaseree, J.; Pongpamorn, P.; Romyanon, K.; Tangphatsornruang, S.; Kuaprasert, B.; Choowongkomon, K. Mass Spectrometry and Synchrotron-FTIR Microspectroscopy Reveal the Anti-inflammatory Activity of Bua Bok Extracts. Phytochem. Anal. 2022, 33, 1086–1098. [Google Scholar] [CrossRef]
- Hatano, S.; Watanabe, H. Regulation of Macrophage and Dendritic Cell Function by Chondroitin Sulfate in Innate to Antigen-Specific Adaptive Immunity. Front. Immunol. 2020, 11, 232. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ami, D.; Franco, A.R.; Artusa, V.; Mereghetti, P.; Peri, F.; Natalello, A. A Global Picture of Molecular Changes Associated to LPS Treatment in THP-1 Derived Human Macrophages by Fourier Transform Infrared Microspectroscopy. Int. J. Mol. Sci. 2022, 23, 13447. https://doi.org/10.3390/ijms232113447
Ami D, Franco AR, Artusa V, Mereghetti P, Peri F, Natalello A. A Global Picture of Molecular Changes Associated to LPS Treatment in THP-1 Derived Human Macrophages by Fourier Transform Infrared Microspectroscopy. International Journal of Molecular Sciences. 2022; 23(21):13447. https://doi.org/10.3390/ijms232113447
Chicago/Turabian StyleAmi, Diletta, Ana Rita Franco, Valentina Artusa, Paolo Mereghetti, Francesco Peri, and Antonino Natalello. 2022. "A Global Picture of Molecular Changes Associated to LPS Treatment in THP-1 Derived Human Macrophages by Fourier Transform Infrared Microspectroscopy" International Journal of Molecular Sciences 23, no. 21: 13447. https://doi.org/10.3390/ijms232113447
APA StyleAmi, D., Franco, A. R., Artusa, V., Mereghetti, P., Peri, F., & Natalello, A. (2022). A Global Picture of Molecular Changes Associated to LPS Treatment in THP-1 Derived Human Macrophages by Fourier Transform Infrared Microspectroscopy. International Journal of Molecular Sciences, 23(21), 13447. https://doi.org/10.3390/ijms232113447