Trichostatin D as a Novel KLF2 Activator Attenuates TNFα-Induced Endothelial Inflammation
Abstract
1. Introduction
2. Results
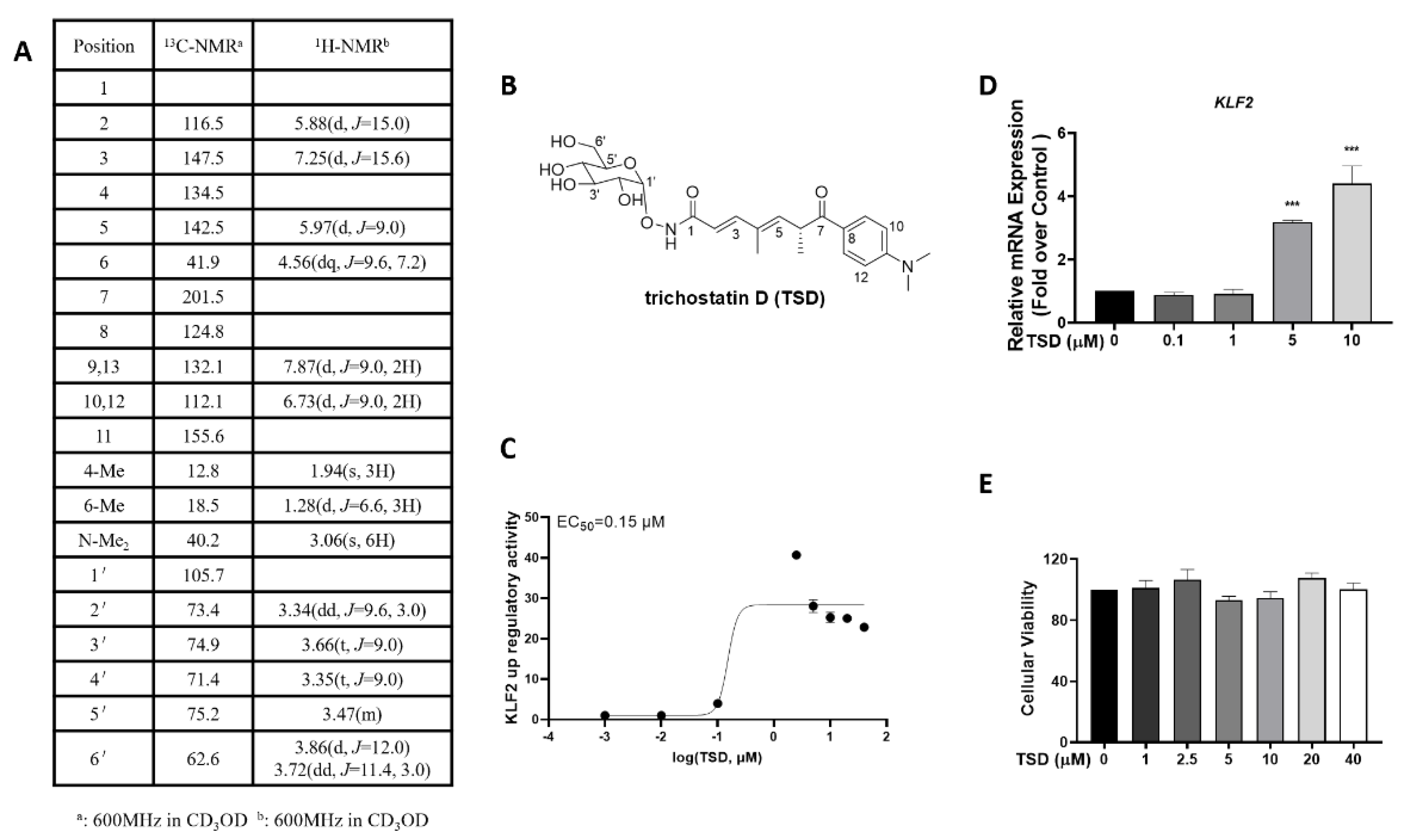
2.1. Isolation and Identification of TSD from Streptomyces sp. CPCC 203909
2.2. TSD Increases KLF2 Activity and mRNA Levels
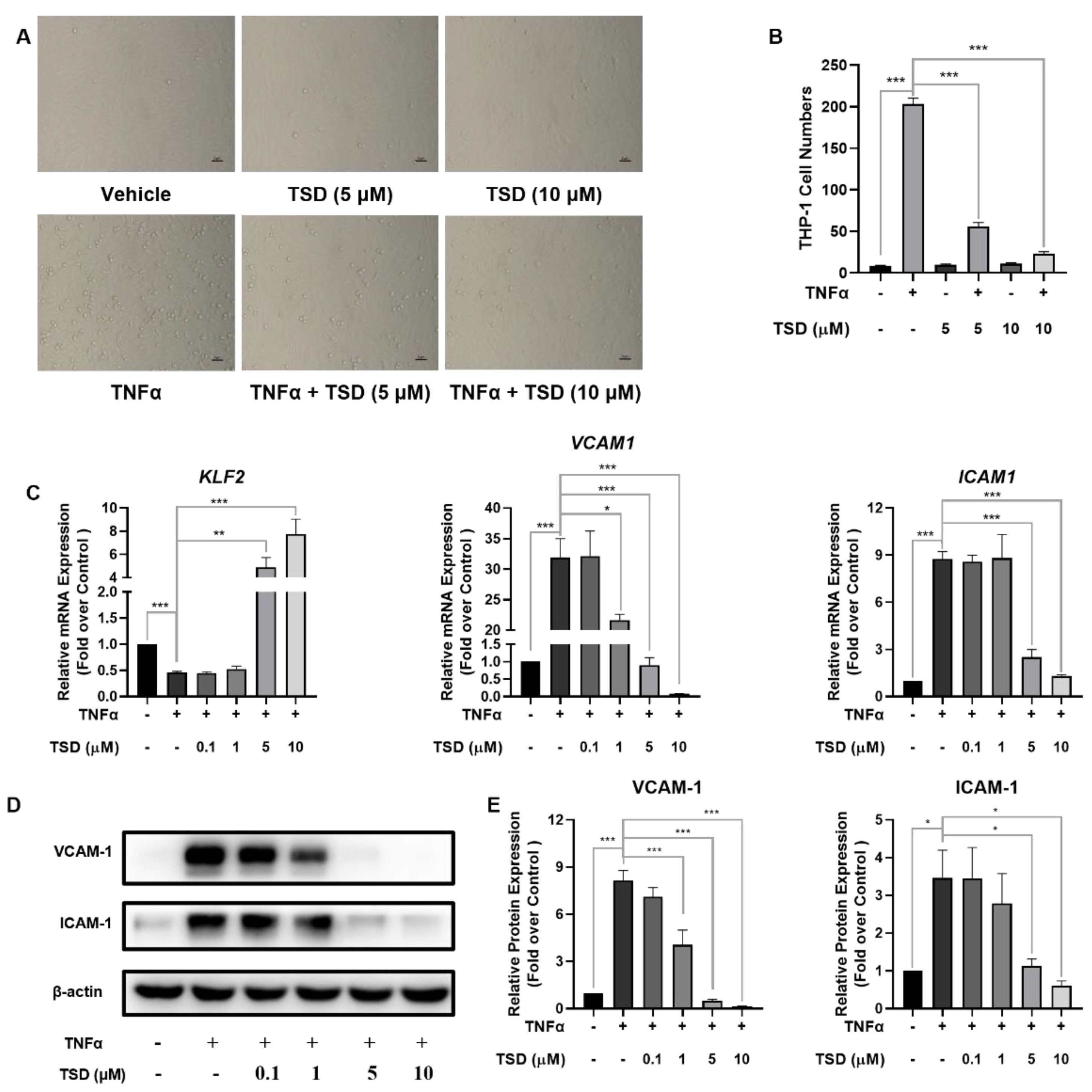
2.3. TSD Attenuated Monocyte Adhesion to HUVECs and Exhibited Anti-Inflammatory Effects
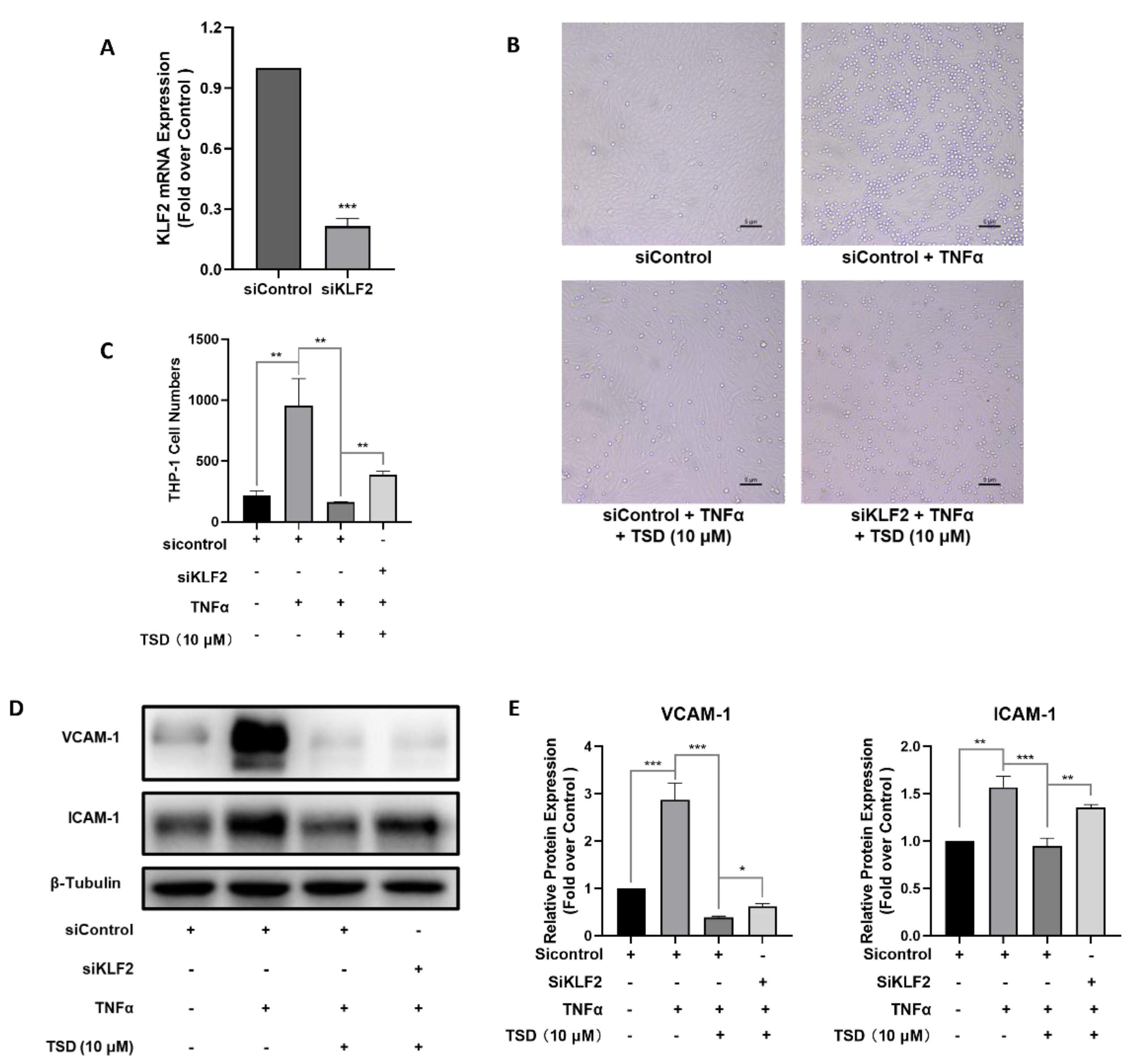
2.4. Anti-Inflammatory Effects of TSD in TNFα-Induced HUVECs Depend on KLF2
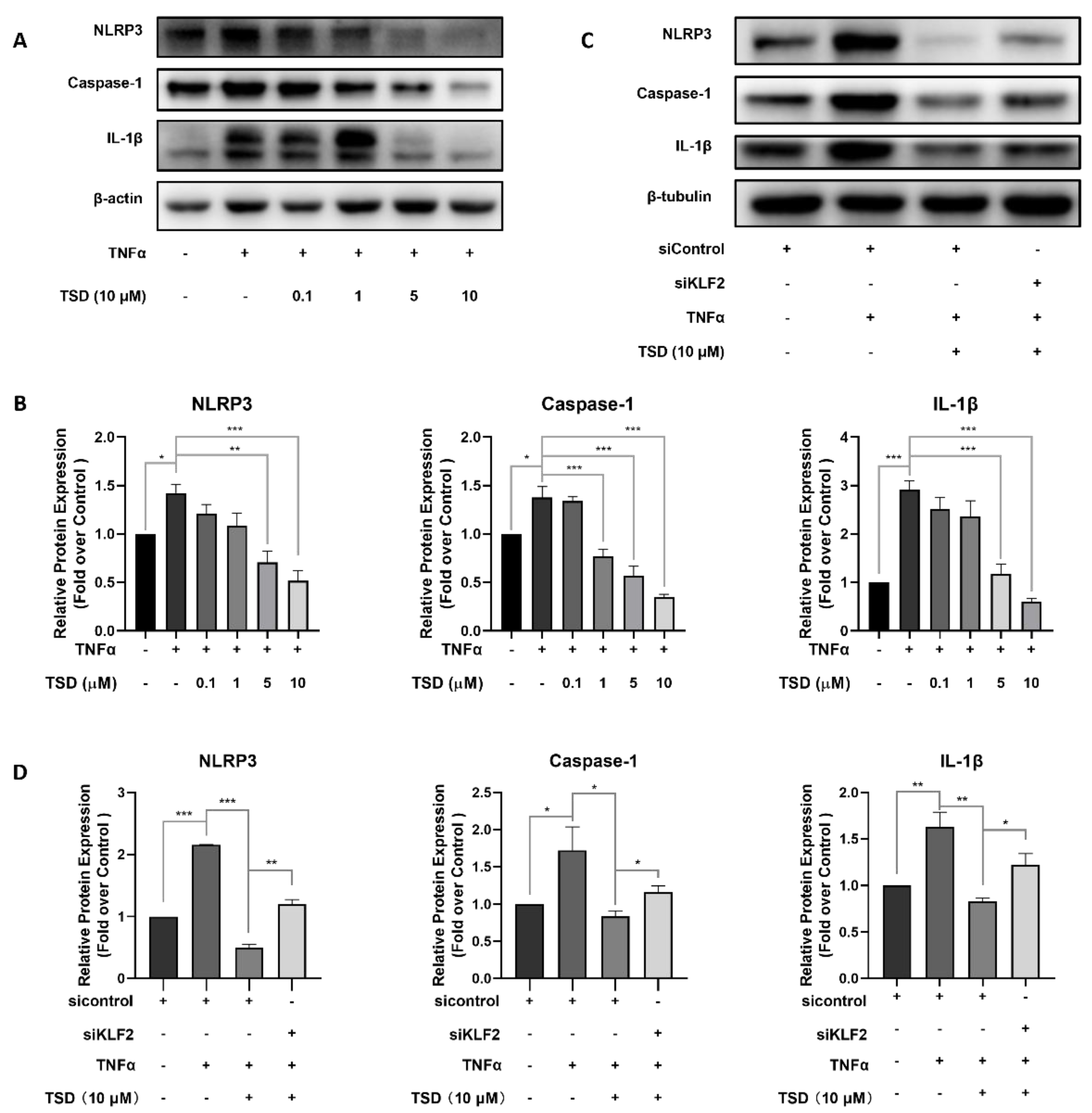
2.5. TSD Exerts Anti-Inflammatory Effects on Endothelial Cells through the KLF2/NLRP3/Caspase-1/IL-1β Signaling Pathway
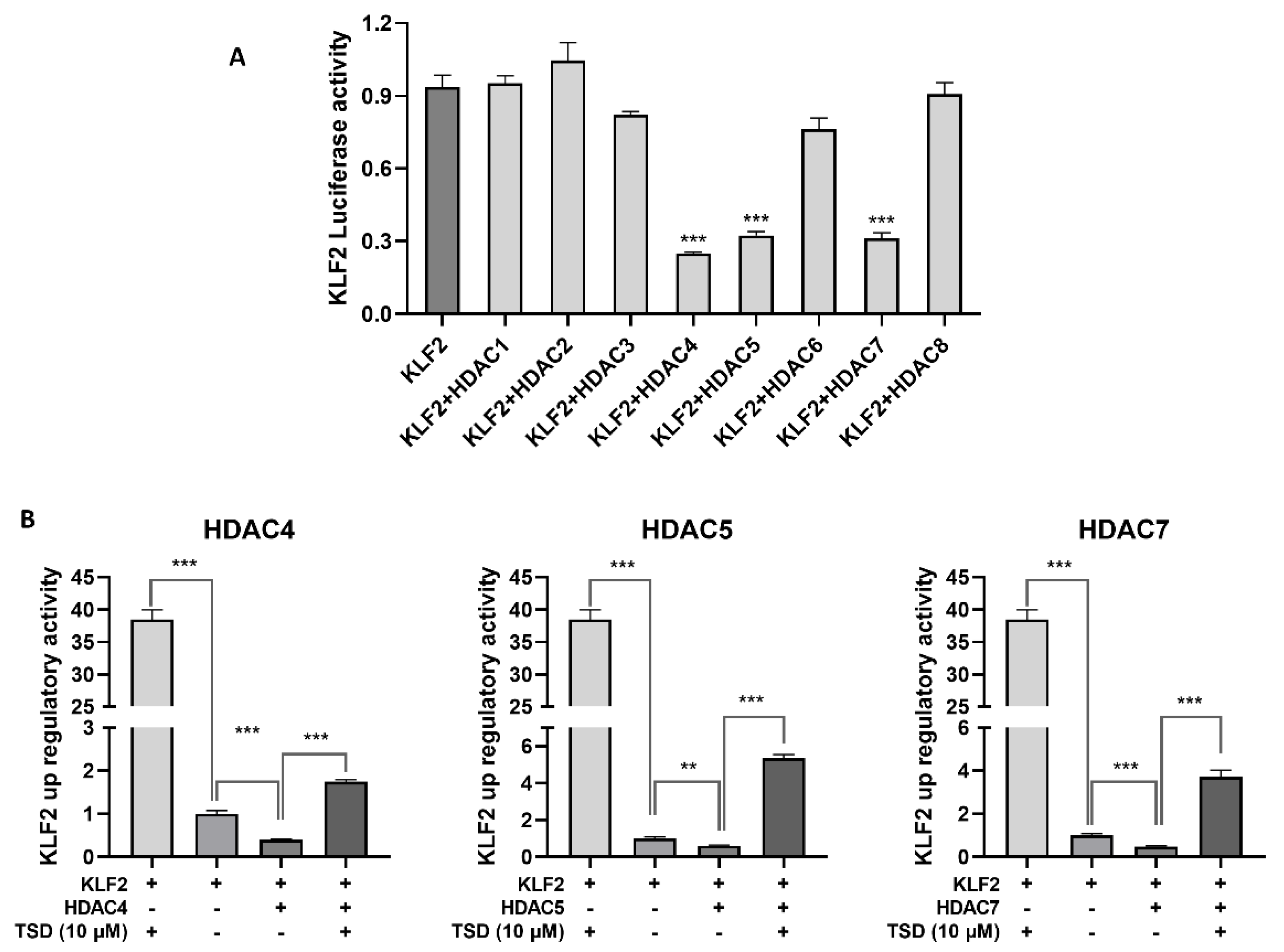
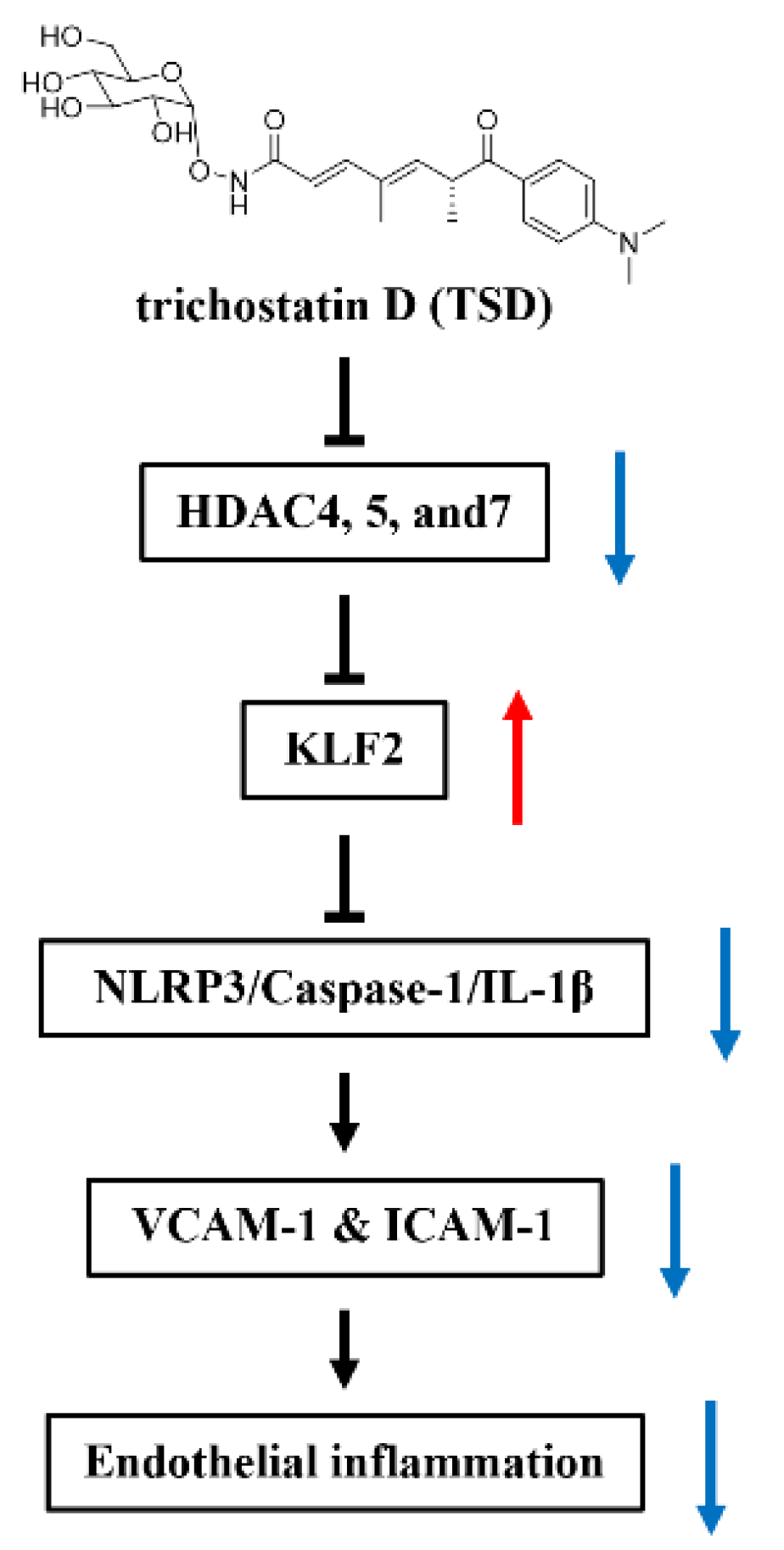
2.6. TSD Upregulates the Expression of KLF2 by Inhibiting HDAC4, HDAC5, and HDAC7
2.7. Interaction Modes between TSD and HDAC4, HDAC5, or HDAC7
3. Discussion
4. Materials and Methods
4.1. Fermentation of Streptomyces sp. CPCC 203909
4.2. Extraction, Isolation, Purification, and Structure Identification of Trichostatin D
4.3. Cell Culture
4.4. Cell Transfection and Luciferase Assay
4.5. Cell Viability Assay
4.6. RNA Isolation and RT-qPCR
4.7. Western Blot Analysis
4.8. Monocyte Adhesion Assay
4.9. Small Interfering RNA (SiRNA) Assay
4.10. Molecular Modeling Analysis
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kruk, M.E.; Gage, A.D.; Joseph, N.T.; Danaei, G.; García-Saisó, S.; Salomon, J.A. Mortality due to low-quality health systems in the universal health coverage era: A systematic analysis of amenable deaths in 137 countries. Lancet 2018, 392, 2203–2212. [Google Scholar] [CrossRef]
- Herrington, W.; Lacey, B.; Sherliker, P.; Armitage, J.; Lewington, S. Epidemiology of Atherosclerosis and the Potential to Reduce the Global Burden of Atherothrombotic Disease. Circ. Res. 2016, 118, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Xian, X.; Wang, Z.; Bi, Y.; Chen, Q.; Han, X.; Tang, D.; Chen, R. Research Progress on the Relationship between Atherosclerosis and Inflammation. Biomolecules 2018, 8, 80. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, T.; Liu, J.; Chen, X.; Zhang, L.; Pi, J.; Sun, H.; Li, L.; Bauer, R.; Wang, H.; Yu, Z.; et al. Endothelial Foxp1 Suppresses Atherosclerosis via Modulation of Nlrp3 Inflammasome Activation. Circ. Res. 2019, 125, 590–605. [Google Scholar] [CrossRef] [PubMed]
- Gimbrone, M.A., Jr.; García-Cardeña, G. Endothelial Cell Dysfunction and the Pathobiology of Atherosclerosis. Circ. Res. 2016, 118, 620–636. [Google Scholar] [CrossRef]
- Chatterjee, S. Endothelial Mechanotransduction, Redox Signaling and the Regulation of Vascular Inflammatory Pathways. Front. Physiol. 2018, 9, 524. [Google Scholar] [CrossRef]
- Yau, J.W.; Teoh, H.; Verma, S. Endothelial cell control of thrombosis. BMC Cardiovasc. Disord. 2015, 15, 130. [Google Scholar] [CrossRef]
- Kasikara, C.; Doran, A.C.; Cai, B.; Tabas, I. The role of non-resolving inflammation in atherosclerosis. J. Clin. Investig. 2018, 128, 2713–2723. [Google Scholar] [CrossRef]
- Maguire, E.M.; Pearce, S.W.A.; Xiao, Q. Foam cell formation: A new target for fighting atherosclerosis and cardiovascular disease. Vasc. Pharmacol. 2019, 112, 54–71. [Google Scholar] [CrossRef]
- Jain, M.K.; Sangwung, P.; Hamik, A. Regulation of an inflammatory disease: Krüppel-like factors and atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2014, 34, 499–508. [Google Scholar] [CrossRef]
- McConnell, B.B.; Yang, V.W. Mammalian Krüppel-like factors in health and diseases. Physiol. Rev. 2010, 90, 1337–1381. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Lu, H.; Liang, W.; Hu, W.; Zhang, J.; Chen, Y.E. Krüppel-like factors and vascular wall homeostasis. J. Mol. Cell Biol. 2017, 9, 352–363. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Shen, Q. IRF2BP2 prevents ox-LDL-induced inflammation and EMT in endothelial cells via regulation of KLF2. Exp. Ther. Med. 2021, 21, 481. [Google Scholar] [CrossRef] [PubMed]
- Chiu, Y.L.; Tsai, W.C.; Wu, C.H.; Wu, C.H.; Cheng, C.C.; Lin, W.S.; Tsai, T.N.; Wu, L.S. Ginkgo biloba Induces Thrombomodulin Expression and Tissue-Type Plasminogen Activator Secretion via the Activation of Krüppel-Like Factor 2 within Endothelial Cells. Am. J. Chin. Med. 2020, 48, 357–372. [Google Scholar] [CrossRef]
- Wu, C.H.; Chiu, Y.L.; Hsieh, C.Y.; Tsung, G.S.; Wu, L.S.; Cheng, C.C.; Tsai, T.N. Cilostazol Induces eNOS and TM Expression via Activation with Sirtuin 1/Krüppel-like Factor 2 Pathway in Endothelial Cells. Int. J. Mol. Sci. 2021, 22, 10287. [Google Scholar] [CrossRef]
- Novodvorsky, P.; Chico, T.J. The role of the transcription factor KLF2 in vascular development and disease. Prog. Mol. Biol. Transl. Sci. 2014, 124, 155–188. [Google Scholar]
- Tang, Y.; Wa, Q.; Peng, L.; Zheng, Y.; Chen, J.; Chen, X.; Zou, X.; Shen, H.; Huang, S. Salvianolic Acid B Suppresses ER Stress-Induced NLRP3 Inflammasome and Pyroptosis via the AMPK/FoxO4 and Syndecan-4/Rac1 Signaling Pathways in Human Endothelial Progenitor Cells. Oxidative Med. Cell. Longev. 2022, 2022, 8332825. [Google Scholar] [CrossRef]
- Jin, H.; Zhu, Y.; Wang, X.D.; Luo, E.F.; Li, Y.P.; Wang, B.L.; Chen, Y.F. BDNF corrects NLRP3 inflammasome-induced pyroptosis and glucose metabolism reprogramming through KLF2/HK1 pathway in vascular endothelial cells. Cell. Signal. 2021, 78, 109843. [Google Scholar] [CrossRef]
- Luo, J.; Wang, X.; Jiang, X.; Liu, C.; Li, Y.; Han, X.; Zuo, X.; Li, Y.; Li, N.; Xu, Y.; et al. Rutaecarpine derivative R3 attenuates atherosclerosis via inhibiting NLRP3 inflammasome-related inflammation and modulating cholesterol transport. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2020, 34, 1398–1411. [Google Scholar] [CrossRef]
- Sharma, A.; Tate, M.; Mathew, G.; Vince, J.E.; Ritchie, R.H.; de Haan, J.B. Oxidative Stress and NLRP3-Inflammasome Activity as Significant Drivers of Diabetic Cardiovascular Complications: Therapeutic Implications. Front. Physiol. 2018, 9, 114. [Google Scholar] [CrossRef]
- Xu, Y.; Xu, S.; Liu, P.; Koroleva, M.; Zhang, S.; Si, S.; Jin, Z.G. Suberanilohydroxamic Acid as a Pharmacological Kruppel-Like Factor 2 Activator That Represses Vascular Inflammation and Atherosclerosis. J. Am. Heart Assoc. 2017, 6, e007134. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Liu, P.; Xu, S.; Koroleva, M.; Zhang, S.; Si, S.; Jin, Z.G. Tannic acid as a plant-derived polyphenol exerts vasoprotection via enhancing KLF2 expression in endothelial cells. Sci. Rep. 2017, 7, 6686. [Google Scholar] [CrossRef] [PubMed]
- Hayakawa, Y.; Nakai, M.; Furihata, K.; Shin-ya, K.; Seto, H. Trichostatin D, a new inducer of phenotypic reversion in transformed cells. J. Antibiot. 2000, 53, 179–183. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dabravolski, S.A.; Sukhorukov, V.N.; Kalmykov, V.A.; Grechko, A.V.; Shakhpazyan, N.K.; Orekhov, A.N. The Role of KLF2 in the Regulation of Atherosclerosis Development and Potential Use of KLF2-Targeted Therapy. Biomedicines 2022, 10, 254. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.H.; Tan, Y.; Yu, S.; Lu, L.; Deng, Y. Pinitol Protects Against Ox-Low-Density Lipoprotein-Induced Endothelial Inflammation and Monocytes Attachment. J. Cardiovasc. Pharmacol. 2022, 79, 368–374. [Google Scholar] [CrossRef]
- Chen, C.; Chen, J.; Tao, X.; Fu, M.; Cheng, B.; Chen, X. Activation of GPR30 with G1 inhibits oscillatory shear stress-induced adhesion of THP-1 monocytes to HAECs by increasing KLF2. Aging 2021, 13, 11942–11953. [Google Scholar] [CrossRef]
- Wang, X.; Huo, R.; Liang, Z.; Xu, C.; Chen, T.; Lin, J.; Li, L.; Lin, W.; Pan, B.; Fu, X.; et al. Simvastatin Inhibits NLRP3 Inflammasome Activation and Ameliorates Lung Injury in Hyperoxia-Induced Bronchopulmonary Dysplasia via the KLF2-Mediated Mechanism. Oxidative Med. Cell. Longev. 2022, 2022, 8336070. [Google Scholar] [CrossRef]
- Hu, R.; Wang, M.Q.; Ni, S.H.; Wang, M.; Liu, L.Y.; You, H.Y.; Wu, X.H.; Wang, Y.J.; Lu, L.; Wei, L.B. Salidroside ameliorates endothelial inflammation and oxidative stress by regulating the AMPK/NF-κB/NLRP3 signaling pathway in AGEs-induced HUVECs. Eur. J. Pharmacol. 2020, 867, 172797. [Google Scholar] [CrossRef]
- Wan, Z.; Fan, Y.; Liu, X.; Xue, J.; Han, Z.; Zhu, C.; Wang, X. NLRP3 inflammasome promotes diabetes-induced endothelial inflammation and atherosclerosis. Diabetes Metab. Syndr. Obes. Targets Ther. 2019, 12, 1931–1942. [Google Scholar] [CrossRef]
- Chen, M.L.; Zhu, X.H.; Ran, L.; Lang, H.D.; Yi, L.; Mi, M.T. Trimethylamine-N-Oxide Induces Vascular Inflammation by Activating the NLRP3 Inflammasome through the SIRT3-SOD2-mtROS Signaling Pathway. J. Am. Heart Assoc. 2017, 6, e006347. [Google Scholar] [CrossRef]
- Lee, D.Y.; Chiu, J.J. Atherosclerosis and flow: Roles of epigenetic modulation in vascular endothelium. J. Biomed. Sci. 2019, 26, 56. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Wei, A.; Chen, F.; Chen, X.; Ding, W.; Ding, Z.; Wu, Z.; Du, R.; Cao, W. Enhancing PPARγ by HDAC inhibition reduces foam cell formation and atherosclerosis in ApoE deficient mice. Pharmacol. Res. 2020, 160, 105059. [Google Scholar] [CrossRef] [PubMed]
- Lv, X.; Qiu, J.; Hao, T.; Zhang, H.; Jiang, H.; Tan, Y. HDAC inhibitor Trichostatin A suppresses adipogenesis in 3T3-L1 preadipocytes. Aging 2021, 13, 17489–17498. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Liu, M.; Lee, L.; Davies, M.; Wang, Z.; Kim, H.; Feeley, B.T. Trichostatin A regulates fibro/adipogenic progenitor adipogenesis epigenetically and reduces rotator cuff muscle fatty infiltration. J. Orthop. Res. Off. Publ. Orthop. Res. Soc. 2021, 39, 1452–1462. [Google Scholar] [CrossRef]
- Das, M.; Laha, D.; Kanji, S.; Joseph, M.; Aggarwal, R.; Iwenofu, O.H.; Pompili, V.J.; Jain, M.K.; Das, H. Induction of Krüppel-like factor 2 reduces K/BxN serum-induced arthritis. J. Cell. Mol. Med. 2019, 23, 1386–1395. [Google Scholar] [CrossRef]
- Lee, D.Y.; Lin, T.E.; Lee, C.I.; Zhou, J.; Huang, Y.H.; Lee, P.L.; Shih, Y.T.; Chien, S.; Chiu, J.J. MicroRNA-10a is crucial for endothelial response to different flow patterns via interaction of retinoid acid receptors and histone deacetylases. Proc. Natl. Acad. Sci. USA 2017, 114, 2072–2077. [Google Scholar] [CrossRef]
- Libby, P. Interleukin-1 Beta as a Target for Atherosclerosis Therapy: Biological Basis of CANTOS and Beyond. J. Am. Coll. Cardiol. 2017, 70, 2278–2289. [Google Scholar] [CrossRef]
- Chistiakov, D.A.; Melnichenko, A.A.; Grechko, A.V.; Myasoedova, V.A.; Orekhov, A.N. Potential of anti-inflammatory agents for treatment of atherosclerosis. Exp. Mol. Pathol. 2018, 104, 114–124. [Google Scholar] [CrossRef]
- Mo, W.; Chen, Z.; Zhang, X.; Dai, G.; Ma, D.; Pan, J.; Zhang, X.; Wu, G.; Fan, W. N6-Methyladenosine Demethylase FTO (Fat Mass and Obesity-Associated Protein) as a Novel Mediator of Statin Effects in Human Endothelial Cells. Arterioscler. Thromb. Vasc. Biol. 2022, 42, 644–658. [Google Scholar]
- Wang, X.; Wu, Z.; He, Y.; Zhang, H.; Tian, L.; Zheng, C.; Shang, T.; Zhu, Q.; Li, D.; He, Y. Humanin prevents high glucose-induced monocyte adhesion to endothelial cells by targeting KLF2. Mol. Immunol. 2018, 101, 245–250. [Google Scholar] [CrossRef]
- Deng, Y.; Lei, T.; Li, H.; Mo, X.; Wang, Z.; Ou, H. ERK5/KLF2 activation is involved in the reducing effects of puerarin on monocyte adhesion to endothelial cells and atherosclerotic lesion in apolipoprotein E-deficient mice. Biochim. et Biophys. Acta Mol. Basis Dis. 2018, 1864, 2590–2599. [Google Scholar] [CrossRef] [PubMed]
- Cernotta, N.; Clocchiatti, A.; Florean, C.; Brancolini, C. Ubiquitin-dependent degradation of HDAC4, a new regulator of random cell motility. Mol. Biol. Cell 2011, 22, 278–289. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.; Lee, S.M.; Lee, J.W.; Kim, I.; Pack, C.G.; Ha, C.H. Yeast beta-glucan mediates histone deacetylase 5-induced angiogenesis in vascular endothelial cells. Int. J. Biol. Macromol. 2022, 211, 556–567. [Google Scholar] [CrossRef] [PubMed]
- Tian, R.; Li, R.; Liu, Y.; Liu, J.; Pan, T.; Zhang, R.; Liu, B.; Chen, E.; Tang, Y.; Qu, H. Metformin ameliorates endotoxemia-induced endothelial pro-inflammatory responses via AMPK-dependent mediation of HDAC5 and KLF2. Biochim. et Biophys. Acta Mol. Basis Dis. 2019, 1865, 1701–1712. [Google Scholar] [CrossRef]
- Palomo, M.; Vera, M.; Martin, S.; Torramadé-Moix, S.; Martinez-Sanchez, J.; Belen Moreno, A.; Carreras, E.; Escolar, G.; Cases, A.; Díaz-Ricart, M. Up-regulation of HDACs, a harbinger of uraemic endothelial dysfunction, is prevented by defibrotide. J. Cell. Mol. Med. 2020, 24, 1713–1723. [Google Scholar] [CrossRef]
- Li, M.; van Esch, B.; Henricks, P.A.J.; Folkerts, G.; Garssen, J. The Anti-inflammatory Effects of Short Chain Fatty Acids on Lipopolysaccharide- or Tumor Necrosis Factor α-Stimulated Endothelial Cells via Activation of GPR41/43 and Inhibition of HDACs. Front. Pharmacol. 2018, 9, 533. [Google Scholar] [CrossRef]
- Inoue, K.; Kobayashi, M.; Yano, K.; Miura, M.; Izumi, A.; Mataki, C.; Doi, T.; Hamakubo, T.; Reid, P.C.; Hume, D.A.; et al. Histone deacetylase inhibitor reduces monocyte adhesion to endothelium through the suppression of vascular cell adhesion molecule-1 expression. Arterioscler. Thromb. Vasc. Biol. 2006, 26, 2652–2659. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lei, L.; Chen, M.; Wang, C.; Jiang, X.; Li, Y.; Wang, W.; Li, S.; Zhao, L.; Sheng, R.; Han, J.; et al. Trichostatin D as a Novel KLF2 Activator Attenuates TNFα-Induced Endothelial Inflammation. Int. J. Mol. Sci. 2022, 23, 13477. https://doi.org/10.3390/ijms232113477
Lei L, Chen M, Wang C, Jiang X, Li Y, Wang W, Li S, Zhao L, Sheng R, Han J, et al. Trichostatin D as a Novel KLF2 Activator Attenuates TNFα-Induced Endothelial Inflammation. International Journal of Molecular Sciences. 2022; 23(21):13477. https://doi.org/10.3390/ijms232113477
Chicago/Turabian StyleLei, Lijuan, Minghua Chen, Chenyin Wang, Xinhai Jiang, Yinghong Li, Weizhi Wang, Shunwang Li, Liping Zhao, Ren Sheng, Jiangxue Han, and et al. 2022. "Trichostatin D as a Novel KLF2 Activator Attenuates TNFα-Induced Endothelial Inflammation" International Journal of Molecular Sciences 23, no. 21: 13477. https://doi.org/10.3390/ijms232113477
APA StyleLei, L., Chen, M., Wang, C., Jiang, X., Li, Y., Wang, W., Li, S., Zhao, L., Sheng, R., Han, J., Zhang, Y., Chen, Y., Yan, B., Wu, Y., Yu, L., Si, S., & Xu, Y. (2022). Trichostatin D as a Novel KLF2 Activator Attenuates TNFα-Induced Endothelial Inflammation. International Journal of Molecular Sciences, 23(21), 13477. https://doi.org/10.3390/ijms232113477





