Arabidopsis thaliana SHOOT MERISTEMLESS Substitutes for Medicago truncatula SINGLE LEAFLET1 to Form Complex Leaves and Petals
Abstract
:1. Introduction
2. Results
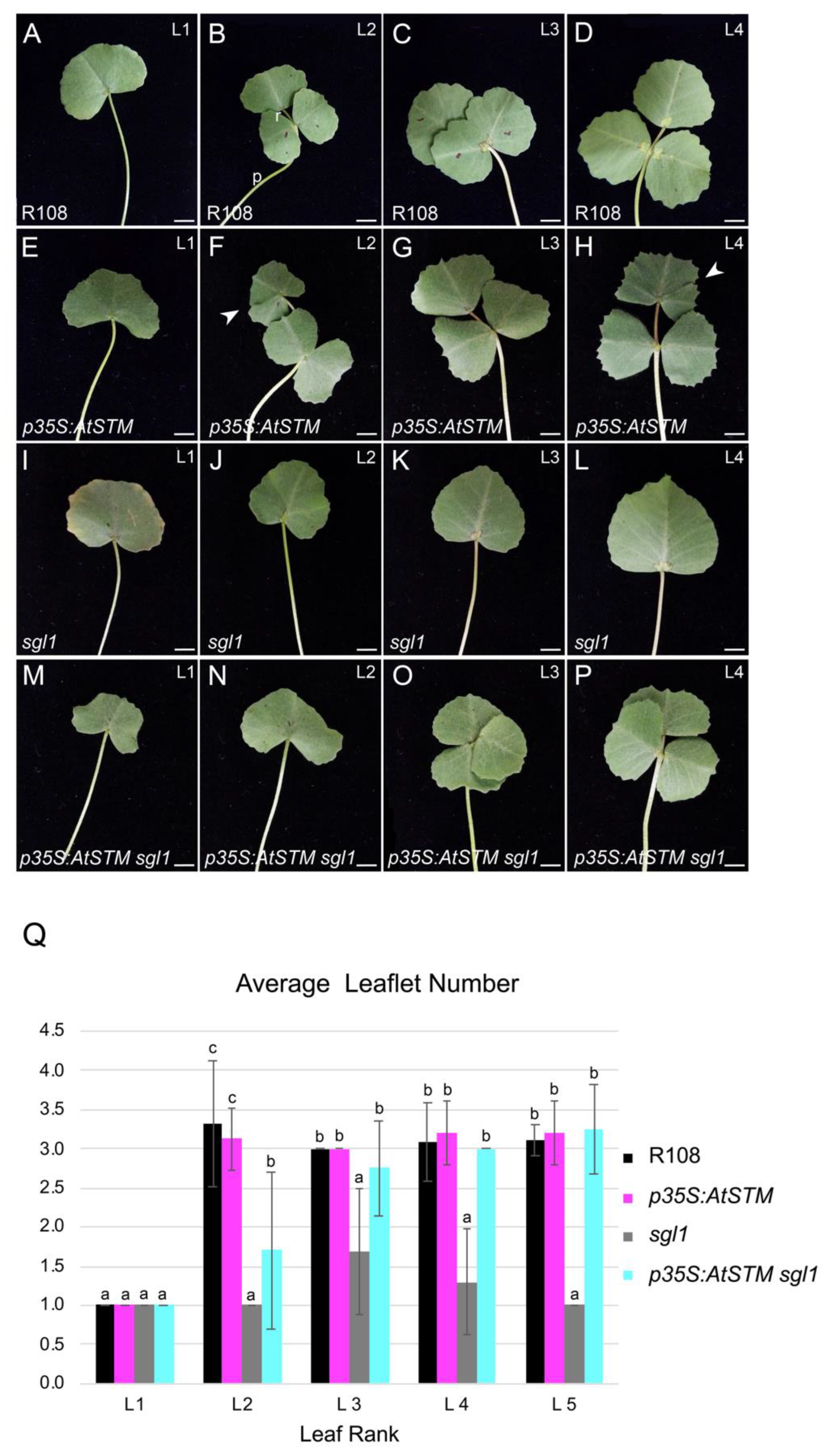
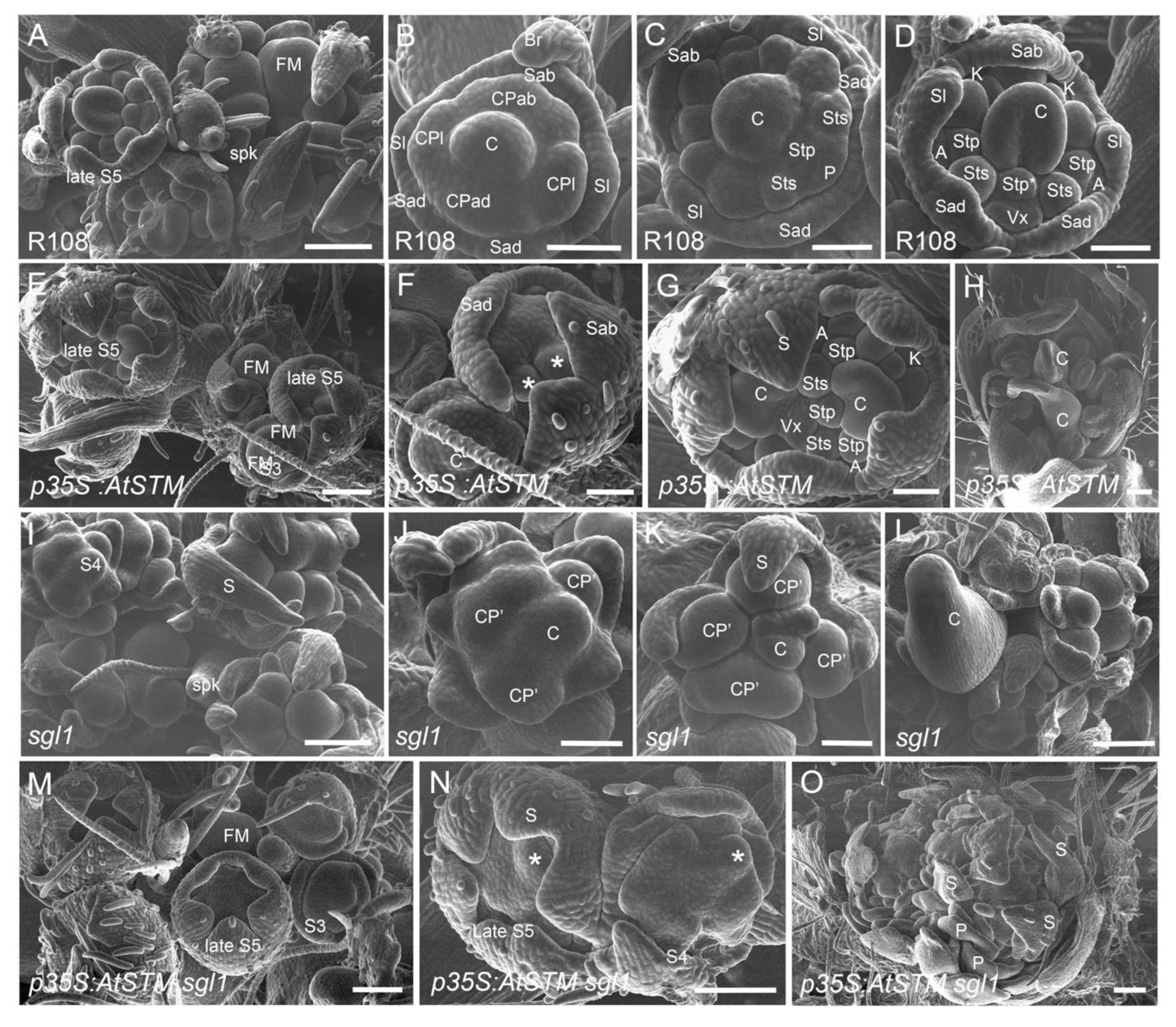
2.1. AtSTM Substitutes for SGL1 in M. truncatula to Form Compound Leaves
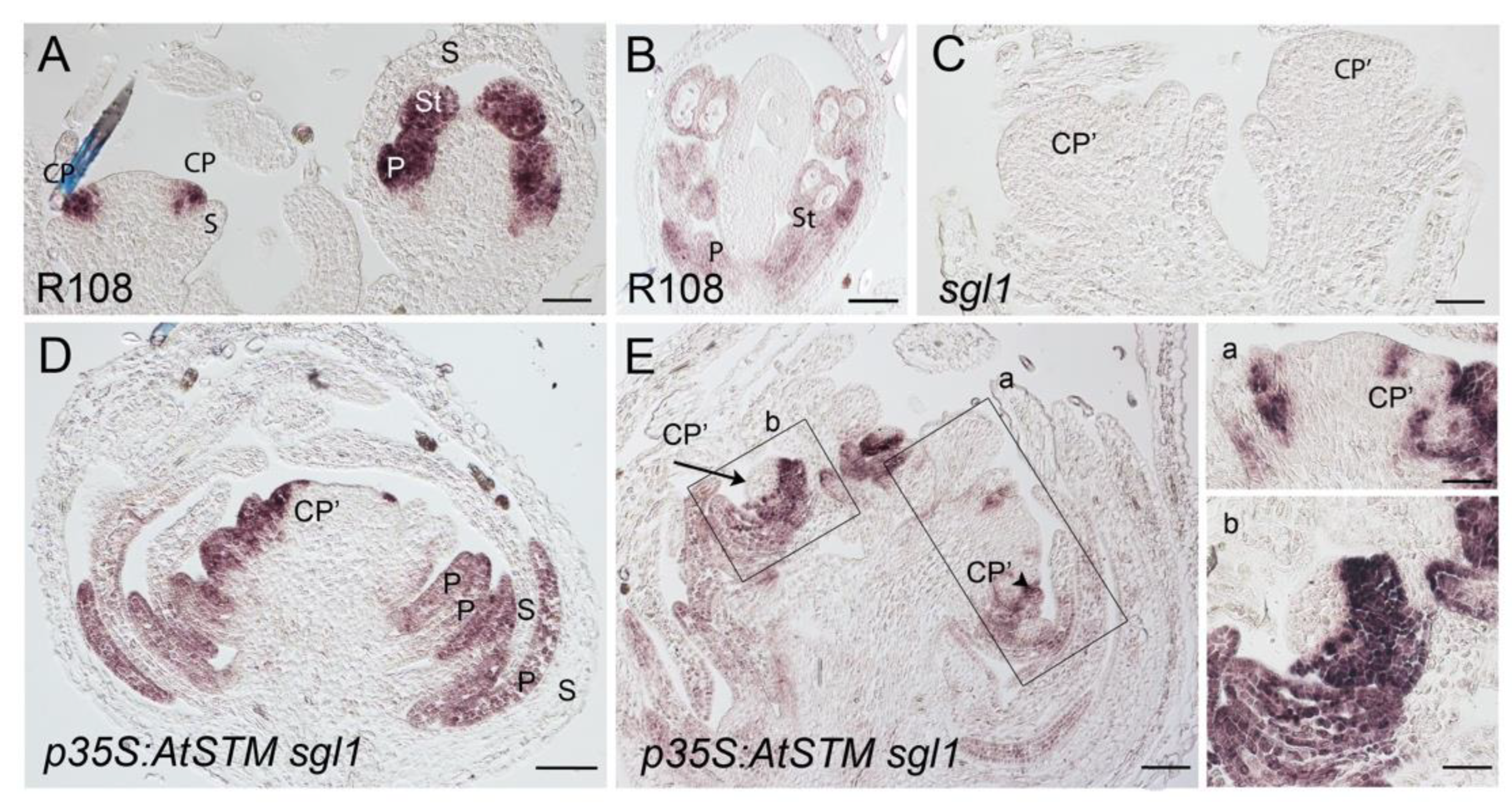
2.2. AtSTM Substitutes for SGL1 in M. truncatula in Specifying Petal Formation
2.3. AtSTM Substitutes for SGL1 to Promote Floral Organ Identity Gene Expression
3. Discussion
4. Materials and Methods
4.1. Plant Growth and Plant Material
4.2. Phenotypic Observations
4.3. Quantitative Analyses of Leaf Development
4.4. Scanning Electron Microscopy (SEM)
4.5. In Situ Localization of GUS Activity and In Situ Hybridization
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hepworth, S.R.; Pautot, V.A. Beyond the divide: Boundaries for patterning and stem cell regulation in Plants. Front. Plant Sci. 2015, 6, 1052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maugarny-Calès, A.; Gonçalves, B.; Jouannic, S.; Melkonian, M.; Ka-Shu Wong, G.; Laufs, P. Apparition of the NAC transcription factors predates the emergence of land plants. Mol. Plant 2016, 9, 1345–1348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Žádníková, P.; Simon, R. How boundaries control plant development. Curr. Opin. Plant Biol. 2014, 17, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Aida, M.; Ishida, T.; Tasaka, M. Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: Interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS Genes. Development 1999, 126, 1563–1570. [Google Scholar] [CrossRef]
- Endrizzi, K.; Moussian, B.; Haecker, A.; Levin, J.Z.; Laux, T. The SHOOT MERISTEMLESS gene is required for maintenance of undifferentiated cells in Arabidopsis shoot and floral meristems and acts at a different regulatory level than the meristem genes WUSCHEL and ZWILLE. Plant J. 1996, 10, 967–979. [Google Scholar] [CrossRef] [PubMed]
- Long, J.A.; Moan, E.I.; Medford, J.I.; Barton, M.K. A Member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 1996, 379, 66–69. [Google Scholar] [CrossRef] [PubMed]
- Takada, S.; Hibara, K.; Ishida, T.; Tasaka, M. The CUP-SHAPEDCOTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development 2001, 128, 1127–1135. [Google Scholar] [CrossRef]
- Belles-Boix, E.; Hamant, O.; Witiak, S.M.; Morin, H.; Traas, J.; Pautot, V. KNAT6: An Arabidopsis homeobox gene involved in meristem activity and organ separation. Plant Cell 2006, 18, 1900–1907. [Google Scholar] [CrossRef] [Green Version]
- Spinelli, S.V.; Martin, A.P.; Viola, I.L.; Gonzalez, D.H.; Palatnik, J.F. A Mechanistic link between STM and CUC1 during Arabidopsis development. Plant Physiol. 2011, 156, 1894–1904. [Google Scholar] [CrossRef] [Green Version]
- Scofield, S.; Murison, A.; Jones, A.; Fozard, J.; Aida, M.; Band, L.R.; Bennett, M.; Murray, J.A.H. Coordination of meristem and boundary functions by transcription factors in the SHOOT MERISTEMLESS Regulatory Network. Development 2018, 145, 157081. [Google Scholar] [CrossRef]
- Aida, M.; Tsubakimoto, Y.; Shimizu, S.; Ogisu, H.; Kamiya, M.; Iwamoto, R.; Takeda, S.; Karim, M.; Mizutani, M.; Lenhard, M.; et al. Establishment of the embryonic shoot meristem involves activation of two classes of genes with opposing functions for meristem activities. Int. J. Mol. Sci. 2020, 21, 5864. [Google Scholar] [CrossRef]
- Laufs, P.; Peaucelle, A.; Morin, H.; Traas, J. MicroRNA Regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development 2004, 131, 4311–4322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mallory, A.C.; Reinhart, B.J.; Jones-Rhoades, M.W.; Tang, G.; Zamore, P.D.; Barton, M.K.; Bartel, D.P. MicroRNA Control of PHABULOSA in leaf development: Importance of pairing to the MicroRNA 5′ Region. EMBO J. 2004, 23, 3356–3364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parcy, F.; Nilsson, O.; Busch, M.A.; Lee, I.; Weigel, D. A Genetic framework for floral patterning. Nature 1998, 395, 561–566. [Google Scholar] [CrossRef] [PubMed]
- Wagner, D.; Sablowski, R.W.M.; Meyerowitz, E.M. Transcriptional activation of APETALA1 by LEAFY. Science 1999, 285, 582–584. [Google Scholar] [CrossRef] [PubMed]
- Lohmann, J.U.; Hong, R.L.; Hobe, M.; Busch, M.A.; Parcy, F.; Simon, R.; Weigel, D. A Molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 2001, 105, 793–803. [Google Scholar] [CrossRef] [Green Version]
- Chae, E.; Tan, Q.K.-G.; Hill, T.A.; Irish, V.F. An Arabidopsis F-Box Protein Acts as a transcriptional co-factor to regulate floral development. Development 2008, 135, 1235–1245. [Google Scholar] [CrossRef] [Green Version]
- Jin, R.; Klasfeld, S.; Zhu, Y.; Fernandez Garcia, M.; Xiao, J.; Han, S.-K.; Konkol, A.; Wagner, D. LEAFY is a pioneer transcription factor and licenses cell reprogramming to floral fate. Nat. Commun. 2021, 12, 626. [Google Scholar] [CrossRef]
- Lai, X.; Blanc-Mathieu, R.; GrandVuillemin, L.; Huang, Y.; Stigliani, A.; Lucas, J.; Thévenon, E.; Loue-Manifel, J.; Turchi, L.; Daher, H.; et al. The LEAFY floral regulator displays pioneer transcription factor properties. Mol. Plant 2021, 14, 829–837. [Google Scholar] [CrossRef]
- Moyroud, E.; Kusters, E.; Monniaux, M.; Koes, R.; Parcy, F. LEAFY blossoms. Trends Plant Sci. 2010, 15, 346–352. [Google Scholar] [CrossRef]
- Chahtane, H.; Vachon, G.; Le Masson, M.; Thévenon, E.; Périgon, S.; Mihajlovic, N.; Kalinina, A.; Michard, R.; Moyroud, E.; Monniaux, M.; et al. A Variant of LEAFY reveals its capacity to stimulate meristem development by inducing RAX1. Plant J. 2013, 74, 678–689. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhou, Y.; Liu, X.; Yu, P.; Cohen, J.D.; Meyerowitz, E.M. LEAFY controls auxin response pathways in floral primordium formation. Sci. Signal. 2013, 6, ra23. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, N.; Wu, M.-F.; Winter, C.M.; Berns, M.C.; Nole-Wilson, S.; Yamaguchi, A.; Coupland, G.; Krizek, B.A.; Wagner, D. A molecular framework for auxin-mediated initiation of flower primordia. Dev. Cell 2013, 24, 271–282. [Google Scholar] [CrossRef] [Green Version]
- Zhao, D. The ASK1 gene regulates B function gene expression in cooperation with UFO and LEAFY in Arabidopsis. Development 2001, 128, 2735–2748. [Google Scholar] [CrossRef]
- Gagne, J.M.; Downes, B.P.; Shiu, S.-H.; Durski, A.M.; Vierstra, R.D. The F-Box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc. Natl. Acad. Sci. USA 2002, 99, 11519–11524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Risseeuw, E.; Venglat, P.; Xiang, D.; Komendant, K.; Daskalchuk, T.; Babic, V.; Crosby, W.; Datla, R. An activated form of UFO alters leaf development and produces ectopic floral and inflorescence meristems. PLoS ONE 2013, 8, e83807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamaguchi, N.; Wu, M.-F.; Winter, C.; Wagner, D. LEAFY and polar auxin transport coordinately regulate Arabidopsis flower development. Plants 2014, 3, 251–265. [Google Scholar] [CrossRef] [Green Version]
- Running, M.P.; Fletcher, J.C.; Meyerowitz, E.M. The WIGGUM gene is required for proper regulation of floral meristem size in Arabidopsis. Development 1998, 125, 2545–2553. [Google Scholar] [CrossRef]
- Chen, Q.; Atkinson, A.; Otsuga, D.; Christensen, T.; Reynolds, L.; Drews, G.N. The Arabidopsis FILAMENTOUS FLOWER gene is required for flower formation. Development 1999, 126, 2715–2726. [Google Scholar] [CrossRef]
- Sawa, S.; Watanabe, K.; Goto, K.; Kanaya, E.; Morita, E.H.; Okada, K. FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and hmg-related domains. Genes Dev. 1999, 13, 1079–1088. [Google Scholar] [CrossRef]
- Norberg, M.; Holmlund, M.; Nilsson, O. The BLADE ON PETIOLE genes act redundantly to control the growth and development of lateral organs. Development 2005, 132, 2203–2213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanrar, S.; Onguka, O.; Smith, H.M.S. Arabidopsis inflorescence architecture requires the activities of KNOX-BELL homeodomain heterodimers. Planta 2006, 224, 1163–1173. [Google Scholar] [CrossRef] [PubMed]
- Kanrar, S.; Bhattacharya, M.; Arthur, B.; Courtier, J.; Smith, H.M.S. Regulatory networks that function to specify flower meristems require the function of homeobox genes PENNYWISE and POUND-FOOLISH in Arabidopsis. Plant J. 2008, 54, 924–937. [Google Scholar] [CrossRef] [PubMed]
- Keller, T.; Abbott, J.; Moritz, T.; Doerner, P. Arabidopsis REGULATOR OF AXILLARY MERISTEMS1 controls a leaf axil stem cell niche and modulates vegetative development. Plant Cell 2006, 18, 598–611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Müller, D.; Schmitz, G.; Theres, K. Blind homologous R2R3 myb genes control the pattern of lateral meristem initiation in Arabidopsis. Plant Cell 2006, 18, 586–597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, D.J.; Park, J.Y.; Ku, S.-J.; Ha, Y.-M.; Kim, S.; Kim, M.D.; Oh, M.-H.; Kim, J. Genome-wide expression profiling of ARABIDOPSIS RESPONSE REGULATOR 7(ARR7) overexpression in cytokinin response. Mol. Genet. Genomics. 2007, 277, 115–137. [Google Scholar] [CrossRef]
- Hareven, D.; Gutfinger, T.; Parnis, A.; Eshed, Y.; Lifschitz, E. The making of a compound leaf: Genetic manipulation of leaf architecture in tomato. Cell 1996, 84, 735–744. [Google Scholar] [CrossRef] [Green Version]
- Bharathan, G.; Goliber, T.E.; Moore, C.; Kessler, S.; Pham, T.; Sinha, N.R. Homologies in leaf form inferred from KNOXI gene expression during development. Science 2002, 296, 1858–1860. [Google Scholar] [CrossRef]
- Hay, A.; Tsiantis, M. The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta. Nat. Genet. 2006, 38, 942–947. [Google Scholar] [CrossRef]
- Shani, E.; Burko, Y.; Ben-Yaakov, L.; Berger, Y.; Amsellem, Z.; Goldshmidt, A.; Sharon, E.; Ori, N. Stage-specific regulation of Solanum lycopersicum leaf maturation by class 1 KNOTTED1-LIKE HOMEOBOX proteins. Plant Cell 2009, 21, 3078–3092. [Google Scholar] [CrossRef]
- Hofer, J.; Turner, L.; Hellens, R.; Ambrose, M.; Matthews, P.; Michael, A.; Ellis, N. UNIFOLIATA regulates leaf and flower morphogenesis in Pea. Curr. Biol. 1997, 7, 581–587. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Chen, J.; Wen, J.; Tadege, M.; Li, G.; Liu, Y.; Mysore, K.S.; Ratet, P.; Chen, R. Control of compound leaf development by FLORICAULA/LEAFY ortholog SINGLE LEAFLET1 in Medicago truncatula. Plant Physiol. 2008, 146, 1759–1772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hofer, J.; Gourlay, C.; Michael, A.; Ellis, T.H.N. Expression of a Class 1 Knotted1-like homeobox gene is down-regulated in pea compound leaf primordia. Plant Mol. Biol. 2001, 45, 387–398. [Google Scholar] [CrossRef]
- Champagne, C.E.M.; Goliber, T.E.; Wojciechowski, M.F.; Mei, R.W.; Townsley, B.T.; Wang, K.; Paz, M.M.; Geeta, R.; Sinha, N.R. Compound leaf development and evolution in the legumes. Plant Cell 2007, 19, 3369–3378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Busch, A.; Gleissberg, S. EcFLO, a FLORICAULA-like gene from Eschscholzia californica is expressed during organogenesis at the vegetative shoot apex. Planta 2003, 217, 841–848. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Han, L.; Li, G.; Chai, M.; Fu, C.; Cheng, X.; Wen, J.; Tang, Y.; Wang, Z.-Y. STM/BP-like KNOXI is uncoupled from ARP in the regulation of compound leaf development in Medicago truncatula. Plant Cell 2014, 26, 1464–1479. [Google Scholar] [CrossRef] [Green Version]
- Taylor, S.; Hofer, J.; Murfet, I. Stamina pistilloida, the pea ortholog of Fim and UFO, is required for normal development of flowers, inflorescences, and leaves. Plant Cell 2001, 13, 31–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, Z.; Zhao, Z.; Liu, C.; Luo, J.; Yang, J.; Huang, W.; Hu, X.; Wang, T.L.; Luo, D. Floral patterning in Lotus japonicus. Plant Physiol. 2005, 137, 1272–1282. [Google Scholar] [CrossRef] [Green Version]
- Nikolov, L.A.; Runions, A.; Das Gupta, M.; Tsiantis, M. Leaf development and evolution. Curr. Top. Dev. Biol. 2019, 131, 109–139. [Google Scholar]
- Challa, K.R.; Rath, M.; Sharma, A.N.; Bajpai, A.K.; Davuluri, S.; Acharya, K.K.; Nath, U. Active suppression of leaflet emergence as a mechanism of simple leaf development. Nat. Plants 2021, 7, 1264–1275. [Google Scholar] [CrossRef]
- Lincoln, C.; Long, J. A Knottedl-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell 1994, 6, 1859–1876. [Google Scholar] [PubMed]
- Pautot, V.; Dockx, J.; Hamant, O.; Kronenberger, J.; Grandjean, O.; Jublot, D.; Traas, J. KNAT2: Evidence for a link between Knotted-like genes and carpel development. Plant Cell 2001, 13, 1719–1734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piazza, P.; Bailey, C.D.; Cartolano, M.; Krieger, J.; Cao, J.; Ossowski, S.; Schneeberger, K.; He, F.; de Meaux, J.; Hall, N.; et al. Arabidopsis thaliana leaf form evolved via loss of KNOX expression in leaves in association with a selective sweep. Curr. Biol. 2010, 20, 2223–2228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, I.; Wolfe, D.S.; Nilsson, O.; Weigel, D. A LEAFY co-regulator encoded by UNUSUAL FLORAL ORGANS. Curr. Biol. 1997, 7, 95–104. [Google Scholar] [CrossRef] [Green Version]
- Blein, T.; Pulido, A.; Vialette-Guiraud, A.; Nikovics, K.; Morin, H.; Hay, A.; Johansen, I.E.; Tsiantis, M.; Laufs, P. A conserved molecular framework for compound leaf development. Science 2008, 322, 1835–1839. [Google Scholar] [CrossRef] [Green Version]
- Bilsborough, G.D.; Runions, A.; Barkoulas, M.; Jenkins, H.W.; Hasson, A.; Galinha, C.; Laufs, P.; Hay, A.; Prusinkiewicz, P.; Tsiantis, M. Model for the regulation of Arabidopsis thaliana leaf margin development. Proc. Natl. Acad. Sci. USA 2011, 108, 3424–3429. [Google Scholar] [CrossRef] [Green Version]
- Vialette-Guiraud, A.C.M.; Chauvet, A.; Gutierrez-Mazariegos, J.; Eschstruth, A.; Ratet, P.; Scutt, C.P. A conserved role for the NAM/MiR164 developmental module reveals a common mechanism underlying carpel margin fusion in monocarpous and syncarpous eurosids. Front. Plant Sci. 2016, 6, 1239. [Google Scholar] [CrossRef] [Green Version]
- Cheng, X.; Peng, J.; Ma, J.; Tang, Y.; Chen, R.; Mysore, K.S.; Wen, J. NO APICAL MERISTEM (MtNAM) regulates floral organ identity and lateral organ separation in Medicago truncatula. New Phytol. 2012, 195, 71–84. [Google Scholar] [CrossRef]
- Benlloch, R.; Navarro, C.; Beltrán, J.; Cañas, L.A. Floral development of the model legume Medicago truncatula: Ontogeny studies as a tool to better characterize homeotic mutations. Sex. Plant Reprod. 2003, 15, 231–241. [Google Scholar] [CrossRef]
- Benlloch, R.; Berbel, A.; Ali, L.; Gohari, G.; Millán, T.; Madueño, F. Genetic control of inflorescence architecture in legumes. Front. Plant Sci. 2015, 6, 543. [Google Scholar] [CrossRef] [Green Version]
- Cheng, X.; Li, G.; Tang, Y.; Wen, J. Dissection of genetic regulation of compound inflorescence development in Medicago truncatula. Development 2018, 145, 158766. [Google Scholar] [CrossRef] [PubMed]
- Hecht, V.; Foucher, F.; Ferrándiz, C.; Macknight, R.; Navarro, C.; Morin, J.; Vardy, M.E.; Ellis, N.; Beltrán, J.P.; Rameau, C.; et al. Conservation of Arabidopsis flowering genes in model legumes. Plant Physiol. 2005, 137, 1420–1434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weigel, D.; Alvarez, J.; Smyth, D.R.; Yanofsky, M.F.; Meyerowitz, E.M. LEAFY controls floral meristem identity in Arabidopsis. Cell 1992, 69, 843–859. [Google Scholar] [CrossRef] [Green Version]
- Benlloch, R.; d’Erfurth, I.; Ferrandiz, C.; Cosson, V.; Beltrán, J.P.; Cañas, L.A.; Kondorosi, A.; Madueño, F.; Ratet, P. Isolation of mtpim proves Tnt1 a useful reverse genetics tool in Medicago truncatula and uncovers new aspects of AP1-like functions in legumes. Plant Physiol. 2006, 142, 972–983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roque, E.; Serwatowska, J.; Cruz Rochina, M.; Wen, J.; Mysore, K.S.; Yenush, L.; Beltrán, J.P.; Cañas, L.A. Functional specialization of duplicated AP3-like genes in Medicago truncatula. Plant J. 2013, 73, 663–675. [Google Scholar] [CrossRef] [Green Version]
- Benlloch, R.; Roque, E.; Ferrándiz, C.; Cosson, V.; Caballero, T.; Penmetsa, R.V.; Beltrán, J.P.; Cañas, L.A.; Ratet, P.; Madueño, F. Analysis of B function in legumes: PISTILLATA proteins do not require the PI motif for floral organ development in Medicago truncatula. Plant J. 2009, 60, 102–111. [Google Scholar] [CrossRef]
- Roque, E.; Fares, M.A.; Yenush, L.; Rochina, M.C.; Wen, J.; Mysore, K.S.; Gómez-Mena, C.; Beltrán, J.P.; Cañas, L.A. Evolution by gene duplication of Medicago truncatula PISTILLATA-like transcription factors. J. Exp. Bot. 2016, 67, 1805–1817. [Google Scholar] [CrossRef] [Green Version]
- Serwatowska, J.; Roque, E.; Gómez-Mena, C.; Constantin, G.D.; Wen, J.; Mysore, K.S.; Lund, O.S.; Johansen, E.; Beltrán, J.P.; Cañas, L.A. Two euAGAMOUS genes control C-function in Medicago truncatula. PLoS ONE 2014, 9, e103770. [Google Scholar] [CrossRef] [Green Version]
- Zhu, B.; Li, H.; Hou, Y.; Zhang, P.; Xia, X.; Wang, N.; Wang, H.; Mysore, K.S.; Wen, J.; Pei, Y.; et al. AGAMOUS and TERMINAL FLOWER controls floral organ identity and inflorescence development in Medicago truncatula. J. Integr. Plant Biol. 2019, 61, 917–923. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Liu, R.; Xu, Y.; Wang, M.; Zhang, J.; Bai, M.; Han, C.; Xiang, F.; Wang, Z.-Y.; Mysore, K.S.; et al. AGLF provides C-function in floral organ identity through transcriptional regulation of AGAMOUS in Medicago truncatula. Proc. Natl. Acad. Sci. USA 2019, 116, 5176–5181. [Google Scholar] [CrossRef] [Green Version]
- Clark, S.E.; Jacobsen, S.E.; Levin, J.Z.; Meyerowitz, E.M. The CLAVATA and SHOOT MERISTEMLESS loci competitively regulate meristem activity in Arabidopsis. Development 1996, 122, 1567–1575. [Google Scholar] [CrossRef] [PubMed]
- Scofield, S.; Dewitte, W.; Murray, J.A.H. The KNOX Gene SHOOT MERISTEMLESS is required for the development of reproductive meristematic tissues in Arabidopsis. Plant J. 2007, 50, 767–781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schultz, E.A.; Haughn, G.W. LEAFY, a homeotic gene that regulates inflorescence development in Arabidopsis. Plant Cell 1991, 3, 771–781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, C.; Han, L.; Hou, C.; Metelli, A.; Qi, L.; Tadege, M.; Mysore, K.S.; Wang, Z.-Y. Developmental analysis of a Medicago truncatula smooth leaf margin1 mutant reveals context-dependent effects on compound leaf development. Plant Cell 2011, 23, 2106–2124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, Y.; Liu, H.; Wang, H.; Liu, Y.; Zhu, B.; Wang, Z.; Hou, Y.; Zhang, P.; Wen, J.; Yang, H.; et al. HEADLESS, a WUSCHEL homolog, uncovers novel aspects of shoot meristem regulation and leaf blade development in Medicago truncatula. J. Exp. Bot. 2019, 70, 149–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Xu, Y.; Hong, L.; Zhang, X.; Wang, X.; Zhang, J.; Ding, Z.; Meng, Z.; Wang, Z.-Y.; Long, R.; et al. HEADLESS regulates auxin response and compound leaf morphogenesis in Medicago truncatula. Front. Plant Sci. 2019, 10, 1024. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Sandal, N.; Polowick, P.L.; Stiller, J.; Stougaard, J.; Fobert, P.R. Proliferating Floral Organs (Pfo), a Lotus japonicus gene required for specifying floral meristem determinacy and organ identity, encodes an F-Box Protein. Plant J. 2003, 33, 607–619. [Google Scholar] [CrossRef]
- Roth, O.; Alvarez, J.P.; Levy, M.; Bowman, J.L.; Ori, N.; Shani, E. The KNOXI transcription factor SHOOT MERISTEMLESS regulates floral fate in Arabidopsis. Plant Cell 2018, 30, 1309–1321. [Google Scholar] [CrossRef] [Green Version]
- Cosson, V.; Eschstruth, A.; Ratet, P. Medicago truncatula transformation using leaf explants. Methods Mol. Biol. 2015, 1223, 43–56. [Google Scholar] [CrossRef]
- Kakar, K.; Wandrey, M.; Czechowski, T.; Gaertner, T.; Scheible, W.-R.; Stitt, M.; Torres-Jerez, I.; Xiao, Y.; Redman, J.C.; Wu, H.C.; et al. A community resource for high-throughput quantitative RT-PCR analysis of transcription factor gene expression in Medicago truncatula. Plant Methods 2008, 4, 18. [Google Scholar] [CrossRef] [Green Version]
- Nikovics, K.; Blein, T.; Peaucelle, A.; Ishida, T.; Morin, H.; Aida, M.; Laufs, P. The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell 2006, 18, 2929–2945. [Google Scholar] [CrossRef] [PubMed]
- Ferrándiz, C.; Liljegren, S.J.; Yanofsky, M.F. Negative Regulation of the SHATTERPROOF genes by FRUITFULL during Arabidopsis fruit development. Science 2000, 289, 436–438. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pautot, V.; Berbel, A.; Cayla, T.; Eschstruth, A.; Adroher, B.; Ratet, P.; Madueño, F.; Laufs, P. Arabidopsis thaliana SHOOT MERISTEMLESS Substitutes for Medicago truncatula SINGLE LEAFLET1 to Form Complex Leaves and Petals. Int. J. Mol. Sci. 2022, 23, 14114. https://doi.org/10.3390/ijms232214114
Pautot V, Berbel A, Cayla T, Eschstruth A, Adroher B, Ratet P, Madueño F, Laufs P. Arabidopsis thaliana SHOOT MERISTEMLESS Substitutes for Medicago truncatula SINGLE LEAFLET1 to Form Complex Leaves and Petals. International Journal of Molecular Sciences. 2022; 23(22):14114. https://doi.org/10.3390/ijms232214114
Chicago/Turabian StylePautot, Véronique, Ana Berbel, Thibaud Cayla, Alexis Eschstruth, Bernard Adroher, Pascal Ratet, Francisco Madueño, and Patrick Laufs. 2022. "Arabidopsis thaliana SHOOT MERISTEMLESS Substitutes for Medicago truncatula SINGLE LEAFLET1 to Form Complex Leaves and Petals" International Journal of Molecular Sciences 23, no. 22: 14114. https://doi.org/10.3390/ijms232214114
APA StylePautot, V., Berbel, A., Cayla, T., Eschstruth, A., Adroher, B., Ratet, P., Madueño, F., & Laufs, P. (2022). Arabidopsis thaliana SHOOT MERISTEMLESS Substitutes for Medicago truncatula SINGLE LEAFLET1 to Form Complex Leaves and Petals. International Journal of Molecular Sciences, 23(22), 14114. https://doi.org/10.3390/ijms232214114




