Improvement of Rice Agronomic Traits by Editing Type-B Response Regulators
Abstract
1. Introduction
2. Results
2.1. Domain and Phylogenetic Tree Analysis of Rice Cytokinin Response Regulators
2.2. Sequence Alignment and mRNA Expression Patterns of Type-B OsRRs in Rice
2.3. Gene Editing of Type-B OsRRs by CRISPR/Cas9 System
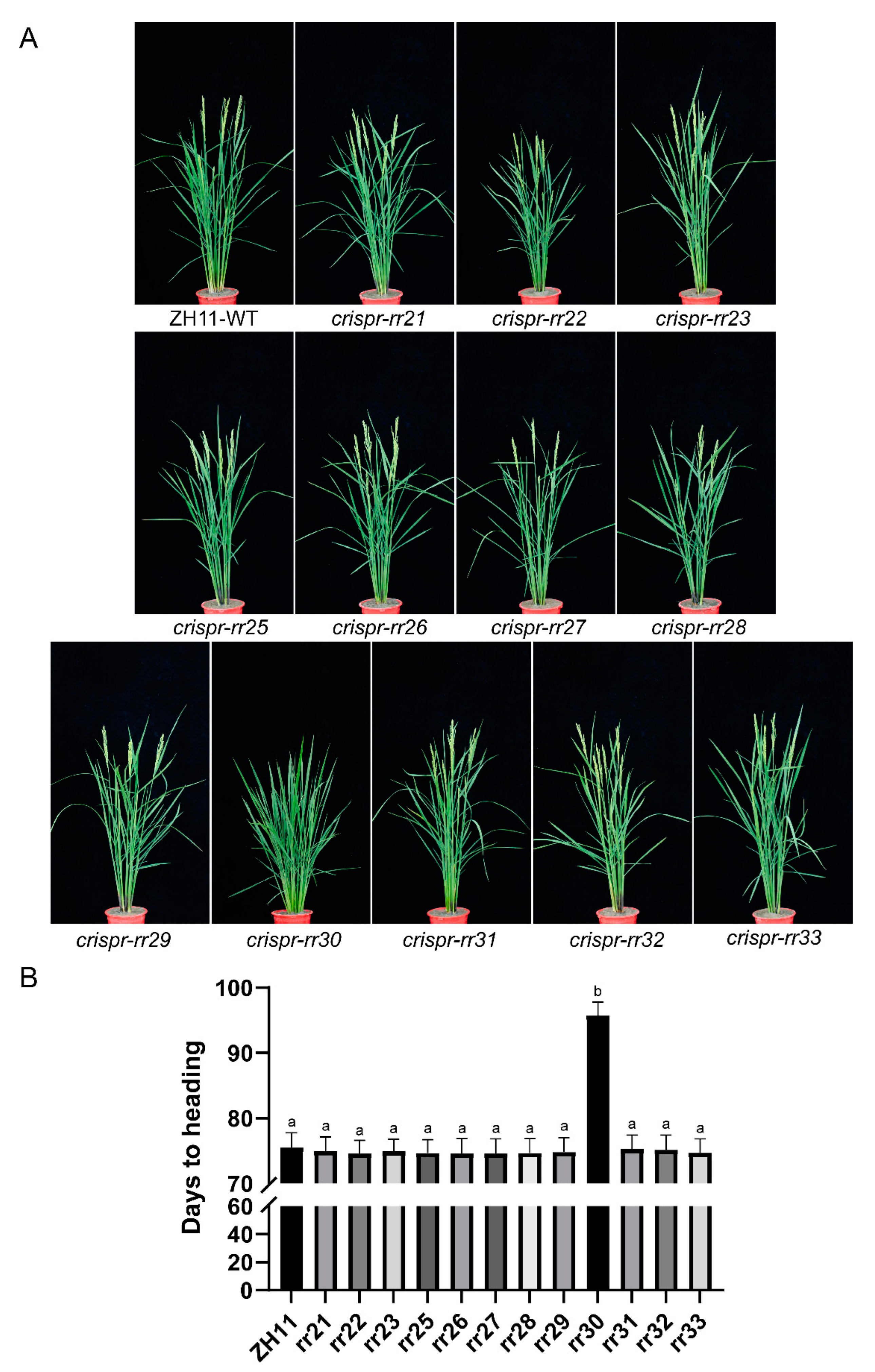
2.4. Field Agronomic Traits of T2 Generation Mutants of Type-B OsRRs
2.5. OsRR30 Mutation Delays Heading while Improves Rice Yield and Quality
3. Discussion
3.1. Characteristics of Type-B OsRRs
3.2. Application of Gene Editing in Rice Breeding
3.3. Application Potential of OsRR30 in Rice Breeding and Strategies for Further Improvement
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Vector Construction and Plant Transformation
4.3. Phenotype Investigation and Evaluation of Agronomic Traits
4.4. Molecular Characterization of the Mutant Plants
4.5. RNA Isolation and RNA Expression Analysis
4.6. Sequence and Expression Analysis of Type-B Response Regulators
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Capra, E.J.; Laub, M.T. Evolution of two-component signal transduction systems. Annu. Rev. Microbiol. 2012, 66, 325–347. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, T. Compilation of all genes encoding two-component phosphotransfer signal transducers in the genome of Escherichia coli. DNA Res. 1997, 4, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Nixon, B.T.; Ronson, C.W.; Ausubel, F.M. Two-component regulatory systems responsive to environmental stimuli share strongly conserved domains with the nitrogen assimilation regulatory genes ntrB and ntrC. Proc. Natl. Acad. Sci. USA 1986, 83, 7850–7854. [Google Scholar] [CrossRef] [PubMed]
- Hwang, I.; Sheen, J.; Muller, B. Cytokinin signaling networks. Annu. Rev. Plant Biol. 2012, 63, 353–380. [Google Scholar] [CrossRef] [PubMed]
- Schaller, G.E. Histidine Kinases and the Role of Two-Component Systems in Plants. Adv. Bot. Res. 2000, 32, 109–148. [Google Scholar]
- Hwang, I.; Chen, H.C.; Sheen, J. Two-component signal transduction pathways in Arabidopsis. Plant Physiol. 2002, 129, 500–515. [Google Scholar] [CrossRef]
- Hutchison, C.E.; Kieber, J.J. Cytokinin signaling in Arabidopsis. Plant Cell 2002, 14, S47–S59. [Google Scholar] [CrossRef]
- Bourret, R.B. Receiver domain structure and function in response regulator proteins. Curr. Opin. Microbiol. 2010, 13, 142–149. [Google Scholar] [CrossRef]
- Galperin, M.Y. Diversity of structure and function of response regulator output domains. Curr. Opin. Microbiol. 2010, 13, 150–159. [Google Scholar] [CrossRef]
- D’Agostino, I.B.; Deruere, J.; Kieber, J.J. Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol. 2000, 124, 1706–1717. [Google Scholar] [CrossRef]
- To, J.P.; Kieber, J.J. Cytokinin signaling: Two-components and more. Trends Plant Sci. 2008, 13, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Imamura, A.; Hanaki, N.; Nakamura, A.; Suzuki, T.; Taniguchi, M.; Kiba, T.; Ueguchi, C.; Sugiyama, T.; Mizuno, T. Compilation and characterization of Arabiopsis thaliana response regulators implicated in His-Asp phosphorelay signal transduction. Plant Cell Physiol. 1999, 40, 733–742. [Google Scholar] [CrossRef]
- Kakimoto, T. Perception and signal transduction of cytokinins. Annu. Rev. Plant Biol. 2003, 54, 605–627. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Bouillet, S.; Stock, A.M. Structural Basis of Response Regulator Function. Annu. Rev. Microbiol. 2019, 73, 175–197. [Google Scholar] [CrossRef] [PubMed]
- Tsai, Y.C.; Weir, N.R.; Hill, K.; Zhang, W.; Kim, H.J.; Shiu, S.H.; Schaller, G.E.; Kieber, J.J. Characterization of genes involved in cytokinin signaling and metabolism from rice. Plant Physiol. 2012, 158, 1666–1684. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Jiao, F.; Chu, J.; Jin, G.; Chen, M.; Wu, P. The two-component signal system in rice (Oryza sativa L.): A genome-wide study of cytokinin signal perception and transduction. Genomics 2007, 89, 697–707. [Google Scholar] [CrossRef] [PubMed]
- Ito, Y.; Kurata, N. Identification and characterization of cytokinin-signalling gene families in rice. Gene 2006, 382, 57–65. [Google Scholar] [CrossRef]
- Sharan, A.; Soni, P.; Nongpiur, R.C.; Singla-Pareek, S.L.; Pareek, A. Mapping the ‘Two-component system’ network in rice. Sci. Rep. 2017, 7, 9287. [Google Scholar] [CrossRef]
- Jain, M.; Tyagi, A.K.; Khurana, J.P. Molecular characterization and differential expression of cytokinin-responsive type-A response regulators in rice (Oryza sativa). BMC Plant Biol. 2006, 6, 1. [Google Scholar] [CrossRef]
- Cheng, X.; Jiang, H.; Zhang, J.; Qian, Y.; Zhu, S.; Cheng, B. Overexpression of type—A rice response regulators, OsRR3 and OsRR5, results in lower sensitivity to cytokinins. Genet. Mol. Res. 2010, 9, 348–359. [Google Scholar] [CrossRef]
- Zhang, A.; Liu, Y.; Wang, F.; Li, T.; Chen, Z.; Kong, D.; Bi, J.; Zhang, F.; Luo, X.; Wang, J. Enhanced rice salinity tolerance via CRISPR/Cas9-targeted mutagenesis of the OsRR22 gene. Mol. Breed. 2019, 39, 47. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.-C.; Lin, T.-C.; Kieber, J.; Tsai, Y.-C. Response Regulators 9 and 10 Negatively Regulate Salinity Tolerance in Rice. Plant Cell Physiol. 2019, 60, 2549–2563. [Google Scholar] [CrossRef] [PubMed]
- Takagi, H.; Tamiru, M.; Abe, A.; Yoshida, K.; Uemura, A.; Yaegashi, H.; Obara, T.; Oikawa, K.; Utsushi, H.; Kanzaki, E.; et al. MutMap accelerates breeding of a salt-tolerant rice cultivar. Nat. Biotechnol. 2015, 33, 445–449. [Google Scholar] [CrossRef]
- Worthen, J.M.; Yamburenko, M.V.; Lim, J.; Nimchuk, Z.L.; Kieber, J.J.; Schaller, G.E. Type-B response regulators of rice play key roles in growth, development and cytokinin signaling. Development 2019, 146, dev174870. [Google Scholar] [CrossRef] [PubMed]
- Hille, F.; Richter, H.; Wong, S.P.; Bratovic, M.; Ressel, S.; Charpentier, E. The Biology of CRISPR-Cas: Backward and Forward. Cell 2018, 172, 1239–1259. [Google Scholar] [CrossRef]
- Horvath, P.; Barrangou, R. CRISPR/Cas, the immune system of bacteria and archaea. Science 2010, 327, 167–170. [Google Scholar] [CrossRef]
- Sorek, R.; Kunin, V.; Hugenholtz, P. CRISPR—A widespread system that provides acquired resistance against phages in bacteria and archaea. Nat. Rev. Microbiol. 2008, 6, 181–186. [Google Scholar] [CrossRef]
- Bhaya, D.; Davison, M.; Barrangou, R. CRISPR-Cas systems in bacteria and archaea: Versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet. 2011, 45, 273–297. [Google Scholar] [CrossRef]
- Brouns, S.J.; Jore, M.M.; Lundgren, M.; Westra, E.R.; Slijkhuis, R.J.; Snijders, A.P.; Dickman, M.J.; Makarova, K.S.; Koonin, E.V.; Van Der Oost, J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 2008, 321, 960–964. [Google Scholar] [CrossRef]
- Marraffini, L.A.; Sontheimer, E.J. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 2008, 322, 1843–1845. [Google Scholar] [CrossRef]
- Gasiunas, G.; Barrangou, R.; Horvath, P.; Siksnys, V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. USA 2012, 109, E2579-2586. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; East, A.; Cheng, A.; Lin, S.; Ma, E.; Doudna, J. RNA-programmed genome editing in human cells. eLife 2013, 2, e00471. [Google Scholar] [CrossRef]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef] [PubMed]
- Murugan, K.; Babu, K.; Sundaresan, R.; Rajan, R.; Sashital, D.G. The Revolution Continues: Newly Discovered Systems Expand the CRISPR-Cas Toolkit. Mol. Cell 2017, 68, 15–25. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Koblan, L.W.; Liu, D.R. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 2020, 38, 824–844. [Google Scholar] [CrossRef]
- Gao, C. Genome engineering for crop improvement and future agriculture. Cell 2021, 184, 1621–1635. [Google Scholar] [CrossRef]
- Hickey, L.T.; Hafee, N.A.; Robinson, H.; Jackson, S.A.; Leal-Bertioli, S.C.M.; Tester, M.; Gao, C.; Godwin, I.D.; Hayes, B.J.; Wulff, B.B.H. Breeding crops to feed 10 billion. Nat. Biotechnol. 2019, 37, 744–754. [Google Scholar] [CrossRef]
- Chen, K.; Wang, Y.; Zhang, R.; Zhang, H.; Gao, C. CRISPR/Cas Genome Editing and Precision Plant Breeding in Agriculture. Annu. Rev. Plant Biol. 2019, 70, 667–697. [Google Scholar] [CrossRef]
- Zhu, H.; Li, C.; Gao, C. Applications of CRISPR-Cas in agriculture and plant biotechnology. Nat. Rev. Mol. Cell Biol. 2020, 21, 661–677. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, Y.; de la Bastide, M.; Hamilton, J.P.; Kanamori, H.; McCombie, W.R.; Ouyang, S.; Schwartz, D.C.; Tanaka, T.; Wu, J.; Zhou, S.; et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 2013, 6, 4. [Google Scholar] [CrossRef] [PubMed]
- Sakai, H.; Lee, S.S.; Tanaka, T.; Numa, H.; Kim, J.; Kawahara, Y.; Wakimoto, H.; Yang, C.C.; Iwamoto, M.; Abe, T.; et al. Rice Annotation Project Database (RAP-DB): An integrative and interactive database for rice genomics. Plant Cell Physiol. 2013, 54, e6. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Stock, A.M.; Robinson, V.L.; Goudreau, P.N. Two-component signal transduction. Annu. Rev. Biochem. 2000, 69, 183–215. [Google Scholar] [CrossRef]
- Sato, Y.; Takehisa, H.; Kamatsuki, K.; Minami, H.; Namiki, N.; Ikawa, H.; Ohyanagi, H.; Sugimoto, K.; Antonio, B.A.; Nagamura, Y. RiceXPro version 3.0: Expanding the informatics resource for rice transcriptome. Nucleic Acids Res. 2013, 41, D1206–D1213. [Google Scholar] [CrossRef]
- Zhao, T.; Ren, L.; Chen, X.; Yu, H.; Liu, C.; Shen, Y.; Shi, W.; Tang, D.; Du, G.; Li, Y.; et al. The OsRR24/LEPTO1 Type-B Response Regulator is Essential for the Organization of Leptotene Chromosomes in Rice Meiosis. Plant Cell 2018, 30, 3024–3037. [Google Scholar] [CrossRef]
- West, A.H.; Stock, A.M. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 2001, 26, 369–376. [Google Scholar] [CrossRef]
- Werner, T.; Motyka, V.; Laucou, V.; Smets, R.; Van Onckelen, H.; Schmulling, T. Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 2003, 15, 2532–2550. [Google Scholar] [CrossRef]
- Yamburenko, M.V.; Worthen, J.M.; Zeenat, A.; Azhar, B.J.; Swain, S.; Couitt, A.R.; Shakeel, S.N.; Kieber, J.J.; Schaller, G.E. Functional Analysis of the Rice Type-B Response Regulator RR22. Front. Plant Sci. 2020, 11, 577676. [Google Scholar] [CrossRef]
- Godfray, H.C.J.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M.; Toulmin, C. Food security: The challenge of feeding 9 billion people. Science 2010, 327, 812–818. [Google Scholar] [CrossRef] [PubMed]
- Tilman, D.; Balzer, C.; Hill, J.; Befort, B.L. Global food demand and the sustainable intensification of agriculture. Proc. Natl. Acad. Sci. USA 2011, 108, 20260–20264. [Google Scholar] [CrossRef] [PubMed]
- Tester, M.; Langridge, P. Breeding technologies to increase crop production in a changing world. Science 2010, 327, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Prado, J.R.; Segers, G.; Voelker, T.; Carson, D.; Dobert, R.; Phillips, J.; Cook, K.; Cornejo, C.; Monken, J.; Grapes, L.; et al. Genetically engineered crops: From idea to product. Annu. Rev. Plant Biol. 2014, 65, 769–790. [Google Scholar] [CrossRef] [PubMed]
- Eshed, Y.; Lippman, Z.B. Revolutions in agriculture chart a course for targeted breeding of old and new crops. Science 2019, 366, eaax0025. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Yang, X.; Yu, Y.; Si, X.; Zhai, X.; Zhang, H.; Dong, W.; Gao, C.; Xu, C. Domestication of wild tomato is accelerated by genome editing. Nat. Biotechnol. 2018, 36, 1160–1163. [Google Scholar] [CrossRef]
- Yu, H.; Lin, T.; Meng, X.; Du, H.; Zhang, J.; Liu, G.; Chen, M.; Jing, Y.; Kou, L.; Li, X.; et al. A route to de novo domestication of wild allotetraploid rice. Cell 2021, 184, 1156–1170.e1114. [Google Scholar] [CrossRef]
- Zsogon, A.; Cermak, T.; Naves, E.R.; Notini, M.M.; Edel, K.H.; Weinl, S.; Freschi, L.; Voytas, D.F.; Kudla, J.; Peres, L.E.P. De novo domestication of wild tomato using genome editing. Nat. Biotechnol. 2018, 36, 1211–1216. [Google Scholar] [CrossRef]
- Lemmon, Z.H.; Reem, N.T.; Dalrymple, J.; Soyk, S.; Swartwood, K.E.; Rodriguez-Leal, D.; Van Eck, J.; Lippman, Z.B. Rapid improvement of domestication traits in an orphan crop by genome editing. Nat. Plants 2018, 4, 766–770. [Google Scholar] [CrossRef]
- Doi, K.; Izawa, T.; Fuse, T.; Yamanouchi, U.; Kubo, T.; Shimatani, Z.; Yano, M.; Yoshimura, A. Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 2004, 18, 926–936. [Google Scholar] [CrossRef]
- Song, X.; Meng, X.; Guo, H.; Cheng, Q.; Jing, Y.; Chen, M.; Liu, G.; Wang, B.; Wang, Y.; Li, J.; et al. Targeting a gene regulatory element enhances rice grain yield by decoupling panicle number and size. Nat. Biotechnol. 2022, 40, 1403–1411. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Ding, Y.; Zhou, Y.; Jin, W.; Xie, K.; Chen, L.L. CRISPR-P 2.0: An Improved CRISPR-Cas9 Tool for Genome Editing in Plants. Mol. Plant 2017, 10, 530–532. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y.; et al. A Robust CRISPR/Cas9 System for Convenient, High-Efficiency Multiplex Genome Editing in Monocot and Dicot Plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef] [PubMed]
- Hiei, Y.; Ohta, S.; Komari, T.; Kumashiro, T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994, 6, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Sanger, F.; Coulson, A.R. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J. Mol. Biol. 1975, 94, 441–448. [Google Scholar] [CrossRef]
- Wang, L.; Xie, W.; Chen, Y.; Tang, W.; Yang, J.; Ye, R.; Liu, L.; Lin, Y.; Xu, C.; Xiao, J.; et al. A dynamic gene expression atlas covering the entire life cycle of rice. Plant J. 2010, 61, 752–766. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]


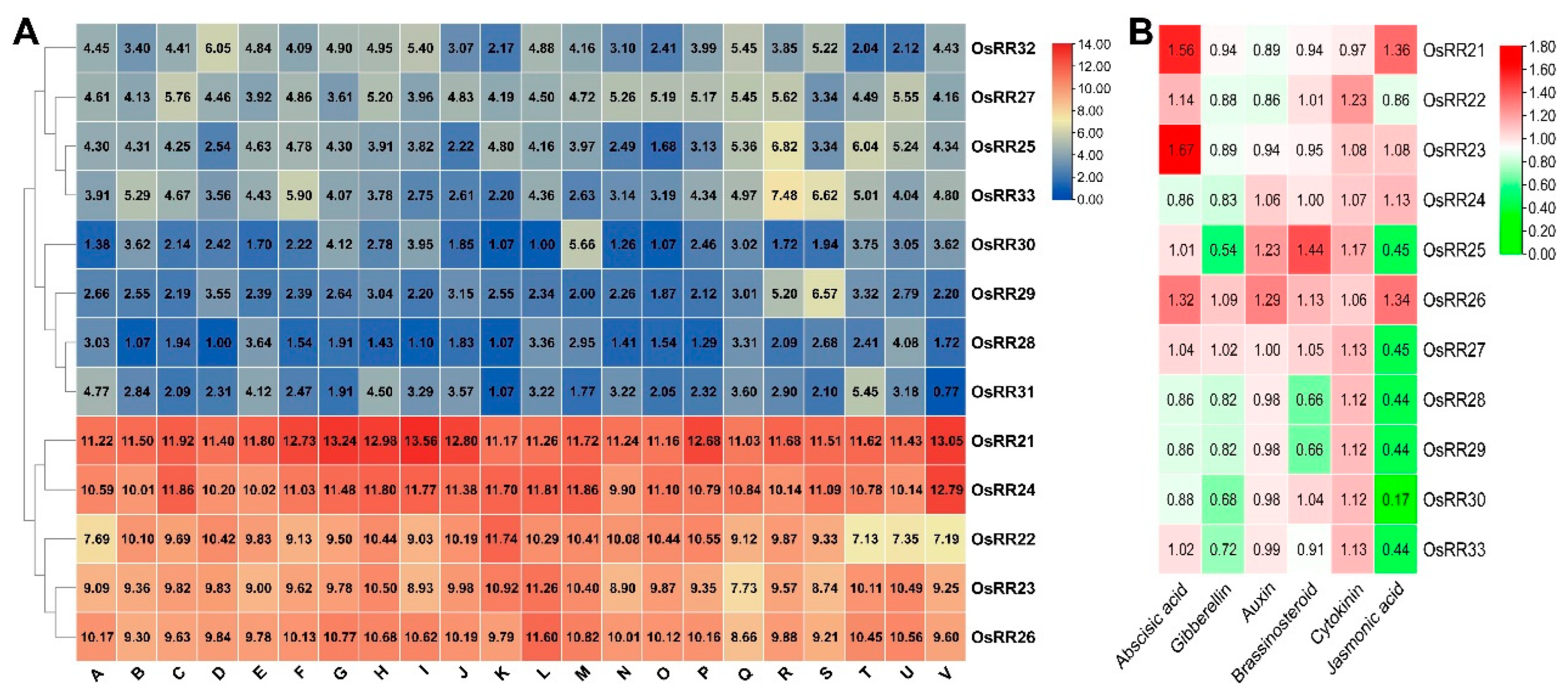



| Gene Name | MSU ID | RAP ID | Chromosome | Length (CDS) | Exon |
|---|---|---|---|---|---|
| OsRR21 | LOC_Os03g12350 | Os03g0224200 | 3 | 2073 | 5 |
| OsRR22 | LOC_Os06g08440 | Os06g0183100 | 6 | 2088 | 6 |
| OsRR23 | LOC_Os02g55320 | Os02g0796500 | 2 | 2064 | 6 |
| OsRR24 | LOC_Os02g08500 | Os02g0182100 | 2 | 1878 | 6 |
| OsRR25 | LOC_Os06g43910 | Os06g0647200 | 6 | 2070 | 6 |
| OsRR26 | LOC_Os01g67770 | Os01g0904700 | 1 | 1746 | 5 |
| OsRR27 | LOC_Os05g32880 | Os05g0395600 | 5 | 1860 | 5 |
| OsRR28 | LOC_Os04g28160 | Os04g0349100 | 4 | 1140 | 7 |
| OsRR29 | LOC_Os04g28130 | Os04g0348800 | 4 | 1170 | 7 |
| OsRR30 | LOC_Os10g32600 | Os10g0463400 | 10 | 1023 | 5 |
| OsRR31 | LOC_Os08g35650 | Os08g0458400 | 8 | 1455 | 6 |
| OsRR32 | LOC_Os08g17760 | Os08g0279900 | 8 | 1710 | 6 |
| OsRR33 | LOC_Os08g35670 | Os08g0458600 | 8 | 1842 | 5 |
| Gene | No. of T0 Plants | Targeted Deletion | |
|---|---|---|---|
| Homozygous (No.%) | Heterozygous (No.%) | ||
| OsRR21 | 75 | 24 (32%) | 27 (36%) |
| OsRR22 | 35 | 1 (3%) | 12 (34%) |
| OsRR23 | 48 | 0 (0%) | 7 (15%) |
| OsRR24 | 54 | 0 (0%) | 14 (26%) |
| OsRR25 | 49 | 0 (0%) | 6 (12%) |
| OsRR26 | 51 | 1 (2%) | 16 (31%) |
| OsRR27 | 43 | 1 (2%) | 13 (30%) |
| OsRR28 | 64 | 6 (9%) | 16 (25%) |
| OsRR29 | 40 | 0 (0%) | 6 (15%) |
| OsRR30 | 78 | 6 (8%) | 25 (32%) |
| OsRR31 | 45 | 3 (7%) | 6 (13%) |
| OsRR32 | 39 | 2 (5%) | 5 (13%) |
| OsRR33 | 33 | 1 (3%) | 5 (15%) |
| Lines | Heading Date (d) | Plant Height (cm) | Tillers per Plant | Panicle Length (cm) | Filled Grain Rate (%) | 1000-Grain Weight (g) | Grains per Panicle | Yield per Plant (g) |
|---|---|---|---|---|---|---|---|---|
| WT | 80.4 ± 2.0 | 101.8 ± 2.0 | 9.7 ± 0.8 | 22.74 ± 0.33 | 79.74 ± 1.61 | 23.45 ± 0.43 | 145.3 ± 5.9 | 26.26 ± 1.05 |
| osrr30 | 92.2 ± 2.2 *** | 105.8 ± 1.9 *** | 10.0 ± 0.8 ns | 25.51 ± 0.50 *** | 82.26 ± 1.48 ** | 24.04 ± 0.98 ns | 170.5 ± 7.3 *** | 33.58 ± 0.9 *** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, C.; Gong, C.; Wu, J.; Yang, L.; Zhou, L.; Wu, B.; Gao, L.; Ling, F.; You, A.; Li, C.; et al. Improvement of Rice Agronomic Traits by Editing Type-B Response Regulators. Int. J. Mol. Sci. 2022, 23, 14165. https://doi.org/10.3390/ijms232214165
Li C, Gong C, Wu J, Yang L, Zhou L, Wu B, Gao L, Ling F, You A, Li C, et al. Improvement of Rice Agronomic Traits by Editing Type-B Response Regulators. International Journal of Molecular Sciences. 2022; 23(22):14165. https://doi.org/10.3390/ijms232214165
Chicago/Turabian StyleLi, Chuanhong, Chenbo Gong, Jiemin Wu, Linfeng Yang, Lei Zhou, Bian Wu, Liang Gao, Fei Ling, Aiqing You, Changyan Li, and et al. 2022. "Improvement of Rice Agronomic Traits by Editing Type-B Response Regulators" International Journal of Molecular Sciences 23, no. 22: 14165. https://doi.org/10.3390/ijms232214165
APA StyleLi, C., Gong, C., Wu, J., Yang, L., Zhou, L., Wu, B., Gao, L., Ling, F., You, A., Li, C., & Lin, Y. (2022). Improvement of Rice Agronomic Traits by Editing Type-B Response Regulators. International Journal of Molecular Sciences, 23(22), 14165. https://doi.org/10.3390/ijms232214165






