NEAT1 Confers Radioresistance to Hepatocellular Carcinoma Cells by Inducing PINK1/Parkin-Mediated Mitophagy
Abstract
1. Introduction
2. Results
2.1. NEAT1v1 Suppresses Radiation-Induced Mitochondrial Damage
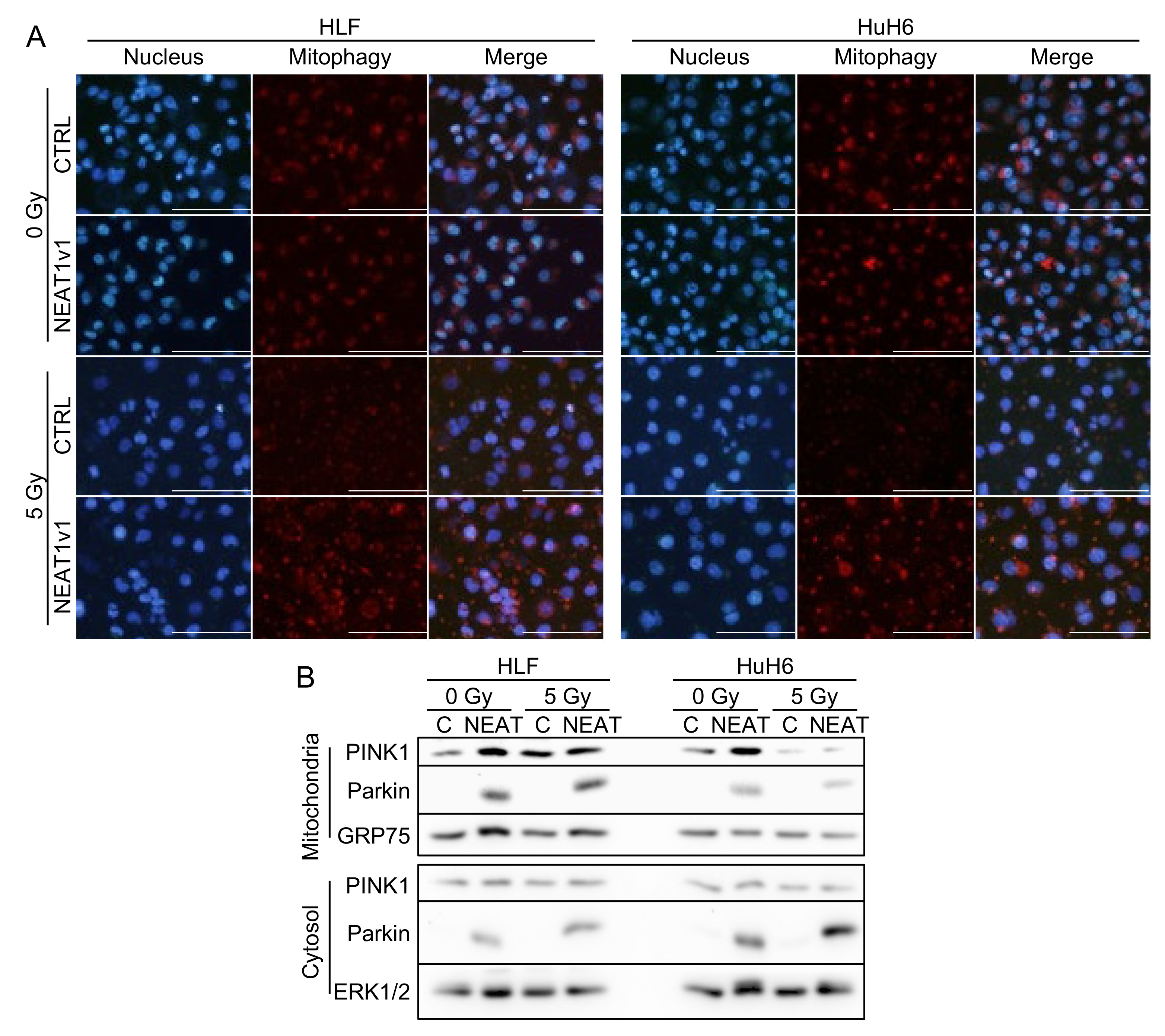
2.2. NEAT1v1 Promotes Mitophagy in Irradiated HCC Cells
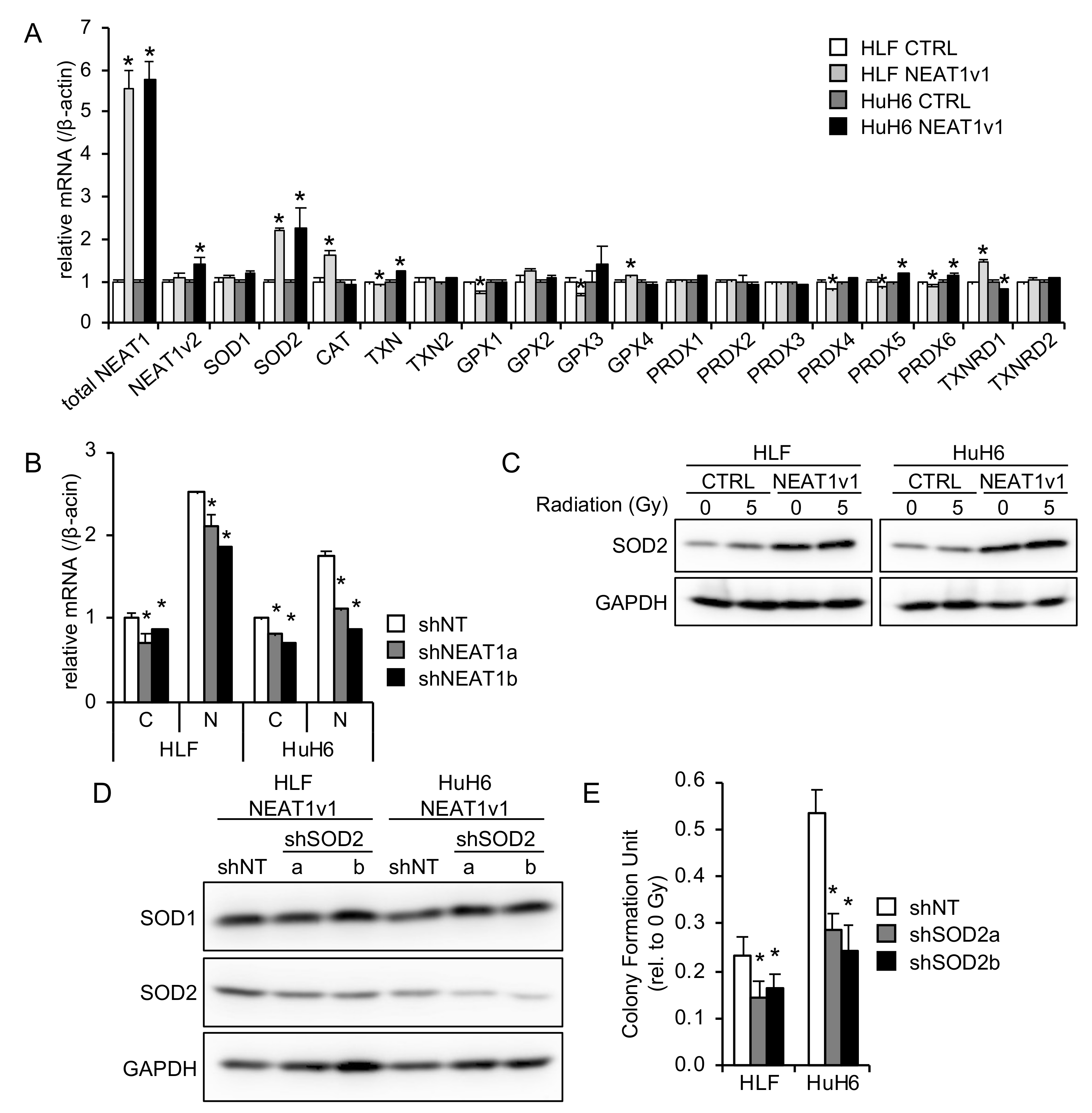
2.3. SOD2 Is Involved in NEAT1v1-Induced Radioresistance
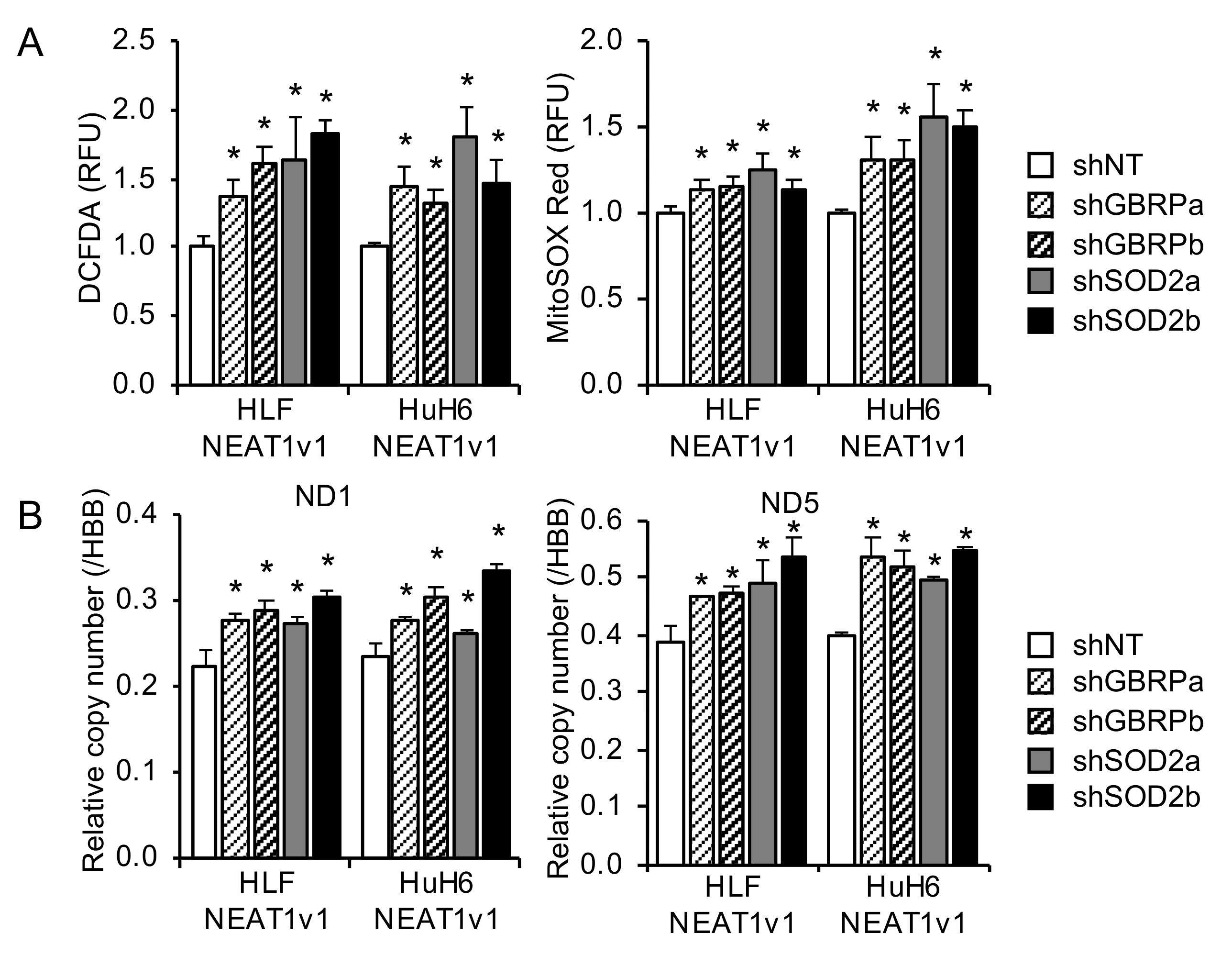
2.4. GABARAP and SOD2 Suppress Radiation-Induced Mitochondrial Oxidative Stress
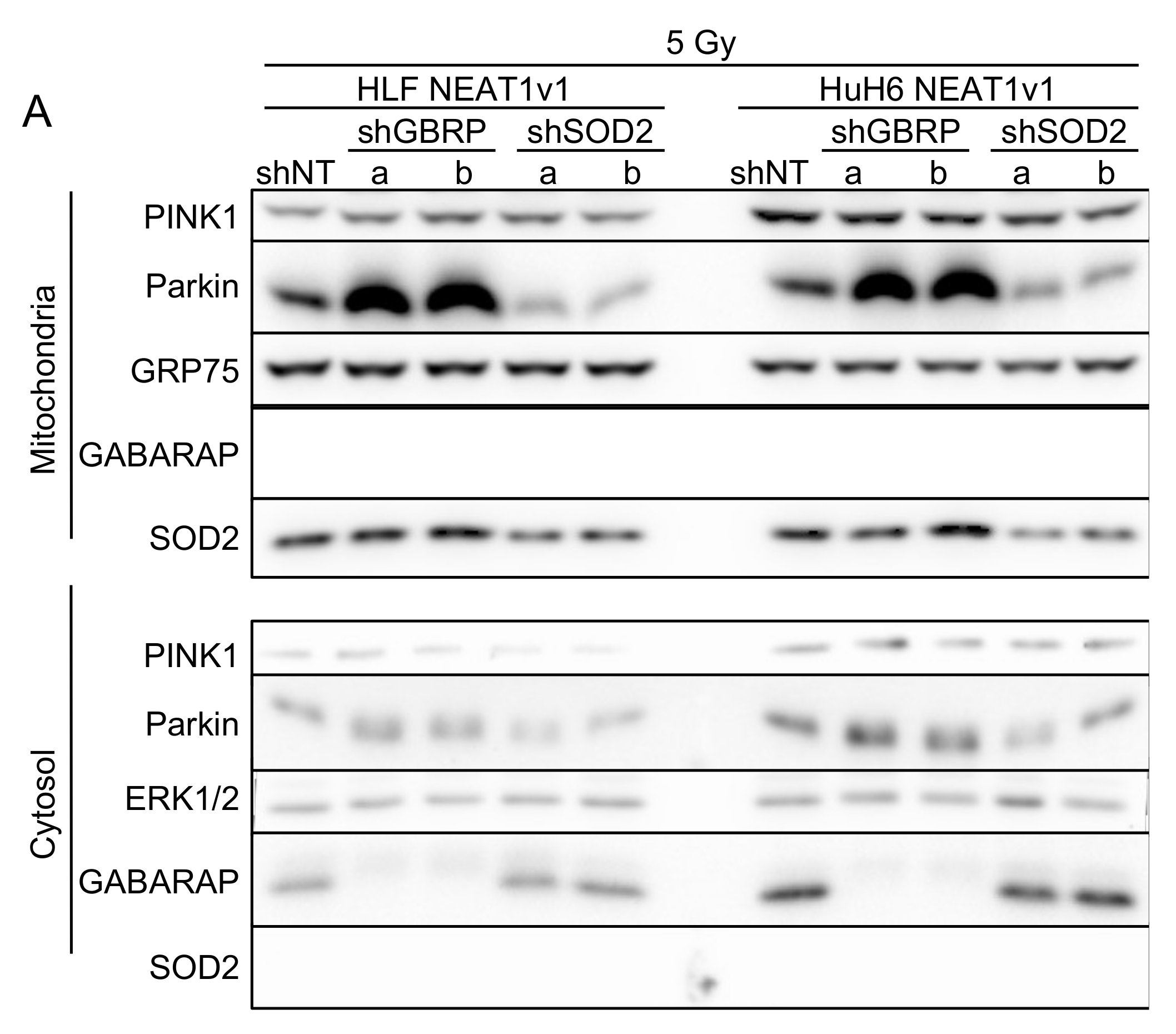
2.5. GABARAP and SOD2 Are Involved in NEAT1v1-Induced Mitophagy
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Adenovirus Vectors
4.3. Reverse Transcription-Quantitative Polymerase Chain Reaction (RT-qPCR) and Western Blot Analysis
4.4. Preparation of Cytosolic and Mitochondrial Fractions
4.5. Detemination of Mitochondrial DNA Content
4.6. Mesurement of Oxidative Stress
4.7. Mitophagy Detection
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M.; Pinyol, R.; Kelley, R.K.; El-Khoueiry, A.; Reeves, H.L.; Wang, X.W.; Gores, G.J.; Villanueva, A. Molecular pathogenesis and systemic therapies for hepatocellular carcinoma. Nat. Cancer 2022, 3, 386–401. [Google Scholar] [CrossRef] [PubMed]
- Kamimura, K.; Terai, S. The promise of radiotherapy for hepatocellular carcinoma. Hepatol. Res. 2021, 51, 837–838. [Google Scholar] [CrossRef]
- Su, T.S.; Liang, P.; Lu, H.Z.; Liang, J.; Gao, Y.C.; Zhou, Y.; Huang, Y.; Tang, M.Y.; Liang, J.N. Stereotactic body radiation therapy for small primary or recurrent hepatocellular carcinoma in 132 Chinese patients. J. Surg. Oncol. 2016, 113, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Shibuya, K.; Katoh, H.; Koyama, Y.; Shiba, S.; Okamoto, M.; Okazaki, S.; Araki, K.; Kakizaki, S.; Shirabe, K.; Ohno, T. Efficacy and Safety of 4 Fractions of Carbon-Ion Radiation Therapy for Hepatocellular Carcinoma: A Prospective Study. Liver Cancer 2021, 11, 61–74. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, S.; Naganuma, T.; Shioi, G.; Hirose, T. Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J. Cell Biol. 2011, 193, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Schmidt, B.F.; Bruchez, M.P.; McManus, C.J. Structural analyses of NEAT1 lncRNAs suggest long-range RNA interactions that may contribute to paraspeckle architecture. Nucleic Acids Res. 2018, 46, 3742–3752. [Google Scholar] [CrossRef]
- Koyama, S.; Tsuchiya, H.; Amisaki, M.; Sakaguchi, H.; Honjo, S.; Fujiwara, Y.; Shiota, G. NEAT1 is Required for the Expression of the Liver Cancer Stem Cell Marker CD44. Int. J. Mol. Sci. 2020, 21, 1927. [Google Scholar] [CrossRef]
- Tsuchiya, H.; Shiota, G. Clinical and Biological Implications of Cancer Stem Cells in Hepatocellular Carcinoma. Yonago Acta Med. 2021, 64, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Sakaguchi, H.; Tsuchiya, H.; Kitagawa, Y.; Tanino, T.; Yoshida, K.; Uchida, N.; Shiota, G. NEAT1 Confers Radioresistance to Hepatocellular Carcinoma Cells by Inducing Autophagy through GABARAP. Int. J. Mol. Sci. 2022, 23, 711. [Google Scholar] [CrossRef]
- Johansen, T.; Lamark, T. Selective Autophagy: ATG8 Family Proteins, LIR Motifs and Cargo Receptors. J. Mol. Biol. 2020, 432, 80–103. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.N.; Padman, B.S.; Usher, J.; Oorschot, V.; Ramm, G.; Lazarou, M. Atg8 family LC3/GABARAP proteins are crucial for autophagosome-lysosome fusion but not autophagosome formation during PINK1/Parkin mitophagy and starvation. J. Cell Biol. 2016, 215, 857–874. [Google Scholar] [CrossRef] [PubMed]
- Ichimura, Y.; Komatsu, M. Activation of p62/SQSTM1-Keap1-Nuclear Factor Erythroid 2-Related Factor 2 Pathway in Cancer. Front. Oncol. 2018, 8, 210. [Google Scholar] [CrossRef] [PubMed]
- Takamura, A.; Komatsu, M.; Hara, T.; Sakamoto, A.; Kishi, C.; Waguri, S.; Eishi, Y.; Hino, O.; Tanaka, K.; Mizushima, N. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011, 25, 795–800. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Niu, Y.; Wan, A.; Chen, D.; Liang, H.; Chen, X.; Sun, L.; Zhan, S.; Chen, L.; Cheng, C.; et al. RNA m6 A methylation regulates sorafenib resistance in liver cancer through FOXO3-mediated autophagy. EMBO J. 2020, 39, e103181. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, S.; Takehara, T.; Hikita, H.; Kodama, T.; Tsunematsu, H.; Miyagi, T.; Hosui, A.; Ishida, H.; Tatsumi, T.; Kanto, T.; et al. Inhibition of autophagy potentiates the antitumor effect of the multikinase inhibitor sorafenib in hepatocellular carcinoma. Int. J. Cancer 2012, 131, 548–557. [Google Scholar] [CrossRef] [PubMed]
- Amaravadi, R.K.; Kimmelman, A.C.; Debnath, J. Targeting Autophagy in Cancer: Recent Advances and Future Directions. Cancer Discov. 2019, 9, 1167–1181. [Google Scholar] [CrossRef]
- Kawamura, K.; Qi, F.; Kobayashi, J. Potential relationship between the biological effects of low-dose irradiation and mitochondrial ROS production. J. Radiat. Res. 2018, 59, ii91–ii97. [Google Scholar] [CrossRef]
- Vara-Perez, M.; Felipe-Abrio, B.; Agostinis, P. Mitophagy in Cancer: A Tale of Adaptation. Cells 2019, 8, 493. [Google Scholar] [CrossRef] [PubMed]
- Narendra, D.P.; Jin, S.M.; Tanaka, A.; Suen, D.F.; Gautier, C.A.; Shen, J.; Cookson, M.R.; Youle, R.J. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010, 8, e1000298. [Google Scholar] [CrossRef]
- Narendra, D.; Tanaka, A.; Suen, D.F.; Youle, R.J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 2008, 183, 795–803. [Google Scholar] [CrossRef]
- Koyano, F.; Okatsu, K.; Kosako, H.; Tamura, Y.; Go, E.; Kimura, M.; Kimura, Y.; Tsuchiya, H.; Yoshihara, H.; Hirokawa, T.; et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature 2014, 510, 162–166. [Google Scholar] [CrossRef] [PubMed]
- Lazarou, M.; Sliter, D.A.; Kane, L.A.; Sarraf, S.A.; Wang, C.; Burman, J.L.; Sideris, D.P.; Fogel, A.I.; Youle, R.J. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 2015, 524, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Wong, Y.C.; Holzbaur, E.L. Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc. Natl. Acad. Sci. USA 2014, 111, E4439–E4448. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Liu, H.; Ikeda, Y.; Amiot, B.P.; Rinaldo, P.; Duncan, S.A.; Nyberg, S.L. Hepatocyte-like cells differentiated from human induced pluripotent stem cells: Relevance to cellular therapies. Stem Cell Res. 2012, 9, 196–207. [Google Scholar] [CrossRef]
- Kameyama, K.; Motoyama, K.; Tanaka, N.; Yamashita, Y.; Higashi, T.; Arima, H. Induction of mitophagy-mediated antitumor activity with folate-appended methyl-β-cyclodextrin. Int. J. Nanomed. 2017, 12, 3433–3446. [Google Scholar] [CrossRef] [PubMed]
- McWilliams, T.G.; Prescott, A.R.; Allen, G.F.; Tamjar, J.; Munson, M.J.; Thomson, C.; Muqit, M.M.; Ganley, I.G. mito-QC illuminates mitophagy and mitochondrial architecture in vivo. J. Cell Biol. 2016, 214, 333–345. [Google Scholar] [CrossRef]
- Katayama, H.; Kogure, T.; Mizushima, N.; Yoshimori, T.; Miyawaki, A. A sensitive and quantitative technique for detecting autophagic events based on lysosomal delivery. Chem. Biol. 2011, 18, 1042–1052. [Google Scholar] [CrossRef]
- Xie, Y.; Liu, J.; Kang, R.; Tang, D. Mitophagy Receptors in Tumor Biology. Front. Cell Dev. Biol. 2020, 8, 594203. [Google Scholar] [CrossRef]
- Jin, X.; Zheng, X.; Li, F.; Liu, B.; Li, H.; Hirayama, R.; Li, P.; Liu, X.; Shen, G.; Li, Q. Fragmentation level determines mitochondrial damage response and subsequently the fate of cancer cells exposed to carbon ions. Radiother. Oncol. 2018, 129, 75–83. [Google Scholar] [CrossRef]
- Sia, J.; Szmyd, R.; Hau, E.; Gee, H.E. Molecular Mechanisms of Radiation-Induced Cancer Cell Death: A Primer. Front. Cell Dev. Biol. 2020, 8, 41. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Ai, F.Y.; Zhang, D.C.; Tian, L.; Yang, Z.Y.; Liu, S.J. LncRNA NEAT1 knockdown attenuates autophagy to elevate 5-FU sensitivity in colorectal cancer via targeting miR-34a. Cancer Med. 2020, 9, 1079–1091. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhou, Y.; Yang, L.; Ma, Y.; Peng, X.; Yang, S.; Li, H.; Liu, J. LncRNA NEAT1 promotes autophagy via regulating miR-204/ATG3 and enhanced cell resistance to sorafenib in hepatocellular carcinoma. J. Cell Physiol. 2020, 235, 3402–3413. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Zhang, C.F.; Guo, J.L.; Su, J.L.; Guo, Z.K.; Li, H.Y. Involvement of NEAT1/PINK1-mediated mitophagy in chronic obstructive pulmonary disease induced by cigarette smoke or PM2.5. Ann. Transl. Med. 2022, 10, 277. [Google Scholar] [CrossRef]
- Huang, Z.; Zhao, J.; Wang, W.; Zhou, J.; Zhang, J. Depletion of LncRNA NEAT1 Rescues Mitochondrial Dysfunction Through NEDD4L-Dependent PINK1 Degradation in Animal Models of Alzheimer’s Disease. Front. Cell Neurosci. 2020, 14, 28. [Google Scholar] [CrossRef]
- Yang, D.Y.; Zhou, X.; Liu, Z.W.; Xu, X.Q.; Liu, C. LncRNA NEAT1 accelerates renal tubular epithelial cell damage by modulating mitophagy via miR-150-5p-DRP1 axis in diabetic nephropathy. Exp. Physiol. 2021, 106, 1631–1642. [Google Scholar] [CrossRef]
- Salah, F.S.; Ebbinghaus, M.; Muley, V.Y.; Zhou, Z.; Al-Saadi, K.R.; Pacyna-Gengelbach, M.; O’Sullivan, G.A.; Betz, H.; König, R.; Wang, Z.Q.; et al. Tumor suppression in mice lacking GABARAP, an Atg8/LC3 family member implicated in autophagy, is associated with alterations in cytokine secretion and cell death. Cell Death Dis. 2016, 7, e2205. [Google Scholar] [CrossRef]
- Miao, Y.; Zhang, Y.; Chen, Y.; Chen, L.; Wang, F. GABARAP is overexpressed in colorectal carcinoma and correlates with shortened patient survival. Hepatogastroenterology 2010, 57, 257–261. [Google Scholar]
- Bortnik, S.; Tessier-Cloutier, B.; Leung, S.; Xu, J.; Asleh, K.; Burugu, S.; Magrill, J.; Greening, K.; Derakhshan, F.; Yip, S.; et al. Differential expression and prognostic relevance of autophagy-related markers ATG4B, GABARAP, and LC3B in breast cancer. Breast Cancer Res. Treat. 2020, 183, 525–547. [Google Scholar] [CrossRef]
- Moussay, E.; Kaoma, T.; Baginska, J.; Muller, A.; Van Moer, K.; Nicot, N.; Nazarov, P.V.; Vallar, L.; Chouaib, S.; Berchem, G.; et al. The acquisition of resistance to TNFα in breast cancer cells is associated with constitutive activation of autophagy as revealed by a transcriptome analysis using a custom microarray. Autophagy 2011, 7, 760–770. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, D.; Lei, M.; Gao, J.; Cui, Y.; Jin, X.; Yu, Q.; Jiang, Y.; Guo, Y.; Liu, Y.; et al. GABARAP suppresses EMT and breast cancer progression via the AKT/mTOR signaling pathway. Aging 2021, 13, 5858–5874. [Google Scholar] [CrossRef]
- Kim, E.H.; Sohn, S.; Kwon, H.J.; Kim, S.U.; Kim, M.J.; Lee, S.J.; Choi, K.S. Sodium selenite induces superoxide-mediated mitochondrial damage and subsequent autophagic cell death in malignant glioma cells. Cancer Res. 2007, 67, 6314–6324. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Yazdi, A.S.; Menu, P.; Tschopp, J. A role for mitochondria in NLRP3 inflammasome activation. Nature 2011, 469, 221–225. [Google Scholar] [CrossRef]
- Lee, J.; Giordano, S.; Zhang, J. Autophagy, mitochondria and oxidative stress: Cross-talk and redox signalling. Biochem. J. 2012, 441, 523–540. [Google Scholar] [CrossRef] [PubMed]
- Yao, N.; Wang, C.; Hu, N.; Li, Y.; Liu, M.; Lei, Y.; Chen, M.; Chen, L.; Chen, C.; Lan, P.; et al. Inhibition of PINK1/Parkin-dependent mitophagy sensitizes multidrug-resistant cancer cells to B5G1, a new betulinic acid analog. Cell Death Dis. 2019, 10, 232. [Google Scholar] [CrossRef] [PubMed]
- Rubio, N.; Coupienne, I.; Di Valentin, E.; Heirman, I.; Grooten, J.; Piette, J.; Agostinis, P. Spatiotemporal autophagic degradation of oxidatively damaged organelles after photodynamic stress is amplified by mitochondrial reactive oxygen species. Autophagy 2012, 8, 1312–1324. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.D.; Zhang, J.L.; Chen, Y.T.; Zhang, J.X.; Wang, T.; Zeng, Q.Y. Insulin alleviates mitochondrial oxidative stress involving upregulation of superoxide dismutase 2 and uncoupling protein 2 in septic acute kidney injury. Exp. Ther. Med. 2018, 15, 3967–3975. [Google Scholar] [CrossRef]
- Dhar, S.K.; Batinic-Haberle, I.; St Clair, D.K. UVB-induced inactivation of manganese-containing superoxide dismutase promotes mitophagy via ROS-mediated mTORC2 pathway activation. J. Biol. Chem. 2019, 294, 6831–6842. [Google Scholar] [CrossRef]
- Li, Y.; Ma, Y.; Song, L.; Yu, L.; Zhang, L.; Zhang, Y.; Xing, Y.; Yin, Y.; Ma, H. SIRT3 deficiency exacerbates p53/Parkin-mediated mitophagy inhibition and promotes mitochondrial dysfunction: Implication for aged hearts. Int. J. Mol. Med. 2018, 41, 3517–3526. [Google Scholar] [CrossRef]
- Dongil, P.; Pérez-García, A.; Hurtado-Carneiro, V.; Herrero-de-Dios, C.; Blazquez, E.; Alvarez, E.; Sanz, C. Pas Kinase Deficiency Triggers Antioxidant Mechanisms in the Liver. Sci. Rep. 2018, 8, 13810. [Google Scholar] [CrossRef]
- Xi, J.; Rong, Y.; Zhao, Z.; Huang, Y.; Wang, P.; Luan, H.; Xing, Y.; Li, S.; Liao, J.; Dai, Y.; et al. Scutellarin ameliorates high glucose-induced vascular endothelial cells injury by activating PINK1/Parkin-mediated mitophagy. J. Ethnopharmacol. 2021, 271, 113855. [Google Scholar] [CrossRef]
- Ryoo, I.G.; Kwak, M.K. Regulatory crosstalk between the oxidative stress-related transcription factor Nfe2l2/Nrf2 and mitochondria. Toxicol. Appl. Pharmacol. 2018, 359, 24–33. [Google Scholar] [CrossRef]
- Sheik Abdul, N.; Nagiah, S.; Chuturgoon, A.A. Fusaric acid induces NRF2 as a cytoprotective response to prevent NLRP3 activation in the liver derived HepG2 cell line. Toxicol. In Vitro 2019, 55, 151–159. [Google Scholar] [CrossRef]
- Bin, J.; Bai, T.; Zhao, Q.; Duan, X.; Deng, S.; Xu, Y. Parkin overexpression reduces inflammation-mediated cardiomyocyte apoptosis through activating Nrf2/ARE signaling pathway. J. Recept. Signal Transduct. Res. 2021, 41, 451–456. [Google Scholar] [CrossRef] [PubMed]
- Alves, A.F.; Moura, A.C.; Andreolla, H.F.; Veiga, A.B.G.D.; Fiegenbaum, M.; Giovenardi, M.; Almeida, S. Gene expression evaluation of antioxidant enzymes in patients with hepatocellular carcinoma: RT-qPCR and bioinformatic analyses. Genet. Mol. Biol. 2021, 44, e20190373. [Google Scholar] [CrossRef]
- Hempel, N.; Carrico, P.M.; Melendez, J.A. Manganese superoxide dismutase (Sod2) and redox-control of signaling events that drive metastasis. Anticancer Agents Med. Chem. 2011, 11, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Cao, W.; Zhou, H.; Qian, H.; Wang, H. CLTRN, Regulated by NRF1/RAN/DLD Protein Complex, Enhances Radiation Sensitivity of Hepatocellular Carcinoma Cells Through Ferroptosis Pathway. Int. J. Radiat. Oncol. Biol. Phys. 2021, 110, 859–871. [Google Scholar] [CrossRef] [PubMed]
- Lan, X.; Wu, Y.Z.; Wang, Y.; Wu, F.R.; Zang, C.B.; Tang, C.; Cao, S.; Li, S.L. CD133 silencing inhibits stemness properties and enhances chemoradiosensitivity in CD133-positive liver cancer stem cells. Int. J. Mol. Med. 2013, 31, 315–324. [Google Scholar] [CrossRef]
- Chen, C.C.; Chen, C.Y.; Wang, S.H.; Yeh, C.T.; Su, S.C.; Ueng, S.H.; Chuang, W.Y.; Hsueh, C.; Wang, T.H. Melatonin Sensitizes Hepatocellular Carcinoma Cells to Chemotherapy Through Long Non-Coding RNA RAD51-AS1-Mediated Suppression of DNA Repair. Cancers 2018, 10, 320. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhao, H.; Yu, J.; Xu, X.; Jing, H.; Li, N.; Tang, Y.; Wang, S.; Li, Y.; Cai, J.; et al. MiR-320b/RAD21 axis affects hepatocellular carcinoma radiosensitivity to ionizing radiation treatment through DNA damage repair signaling. Cancer Sci. 2021, 112, 575–588. [Google Scholar] [CrossRef]
- Hu, L.; Wang, H.; Huang, L.; Zhao, Y.; Wang, J. The Protective Roles of ROS-Mediated Mitophagy on 125I Seeds Radiation Induced Cell Death in HCT116 Cells. Oxidative Med. Cell Longev. 2016, 2016, 9460462. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.; Song, X.; Geng, X.; Wang, X.; Wang, W.; Chen, T.C.; Xie, L.; Song, X. Temozolomide-Perillyl alcohol conjugate impairs Mitophagy flux by inducing lysosomal dysfunction in non-small cell lung Cancer cells and sensitizes them to irradiation. J. Exp. Clin. Cancer Res. 2018, 37, 250. [Google Scholar] [CrossRef] [PubMed]
- Gallaher, S.D.; Berk, A.J. A rapid Q-PCR titration protocol for adenovirus and helper-dependent adenovirus vectors that produces biologically relevant results. J. Virol. Methods 2013, 192, 28–38. [Google Scholar] [CrossRef] [PubMed]
- Miki, T.; Awa, M.; Nishikawa, Y.; Kiyonaka, S.; Wakabayashi, M.; Ishihama, Y.; Hamachi, I. A conditional proteomics approach to identify proteins involved in zinc homeostasis. Nat. Methods 2016, 13, 931–937. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsuchiya, H.; Shinonaga, R.; Sakaguchi, H.; Kitagawa, Y.; Yoshida, K.; Shiota, G. NEAT1 Confers Radioresistance to Hepatocellular Carcinoma Cells by Inducing PINK1/Parkin-Mediated Mitophagy. Int. J. Mol. Sci. 2022, 23, 14397. https://doi.org/10.3390/ijms232214397
Tsuchiya H, Shinonaga R, Sakaguchi H, Kitagawa Y, Yoshida K, Shiota G. NEAT1 Confers Radioresistance to Hepatocellular Carcinoma Cells by Inducing PINK1/Parkin-Mediated Mitophagy. International Journal of Molecular Sciences. 2022; 23(22):14397. https://doi.org/10.3390/ijms232214397
Chicago/Turabian StyleTsuchiya, Hiroyuki, Ririko Shinonaga, Hiromi Sakaguchi, Yutaka Kitagawa, Kenji Yoshida, and Goshi Shiota. 2022. "NEAT1 Confers Radioresistance to Hepatocellular Carcinoma Cells by Inducing PINK1/Parkin-Mediated Mitophagy" International Journal of Molecular Sciences 23, no. 22: 14397. https://doi.org/10.3390/ijms232214397
APA StyleTsuchiya, H., Shinonaga, R., Sakaguchi, H., Kitagawa, Y., Yoshida, K., & Shiota, G. (2022). NEAT1 Confers Radioresistance to Hepatocellular Carcinoma Cells by Inducing PINK1/Parkin-Mediated Mitophagy. International Journal of Molecular Sciences, 23(22), 14397. https://doi.org/10.3390/ijms232214397




