Primary Breast Angiosarcoma: Comparative Transcriptome Analysis
Abstract
:1. Introduction
2. Results
2.1. Case Report
2.2. Differential Expression Genes
2.3. Functional Enrichment Analysis
2.4. Overlapped Genes among Angiosarcoma Case Report and Luminal A Subtype of Breast Cancer Sample
2.5. Overlapped Genes between the Case Report and the GSE163359 Dataset
3. Discussion
4. Conclusions
5. Materials and Methods
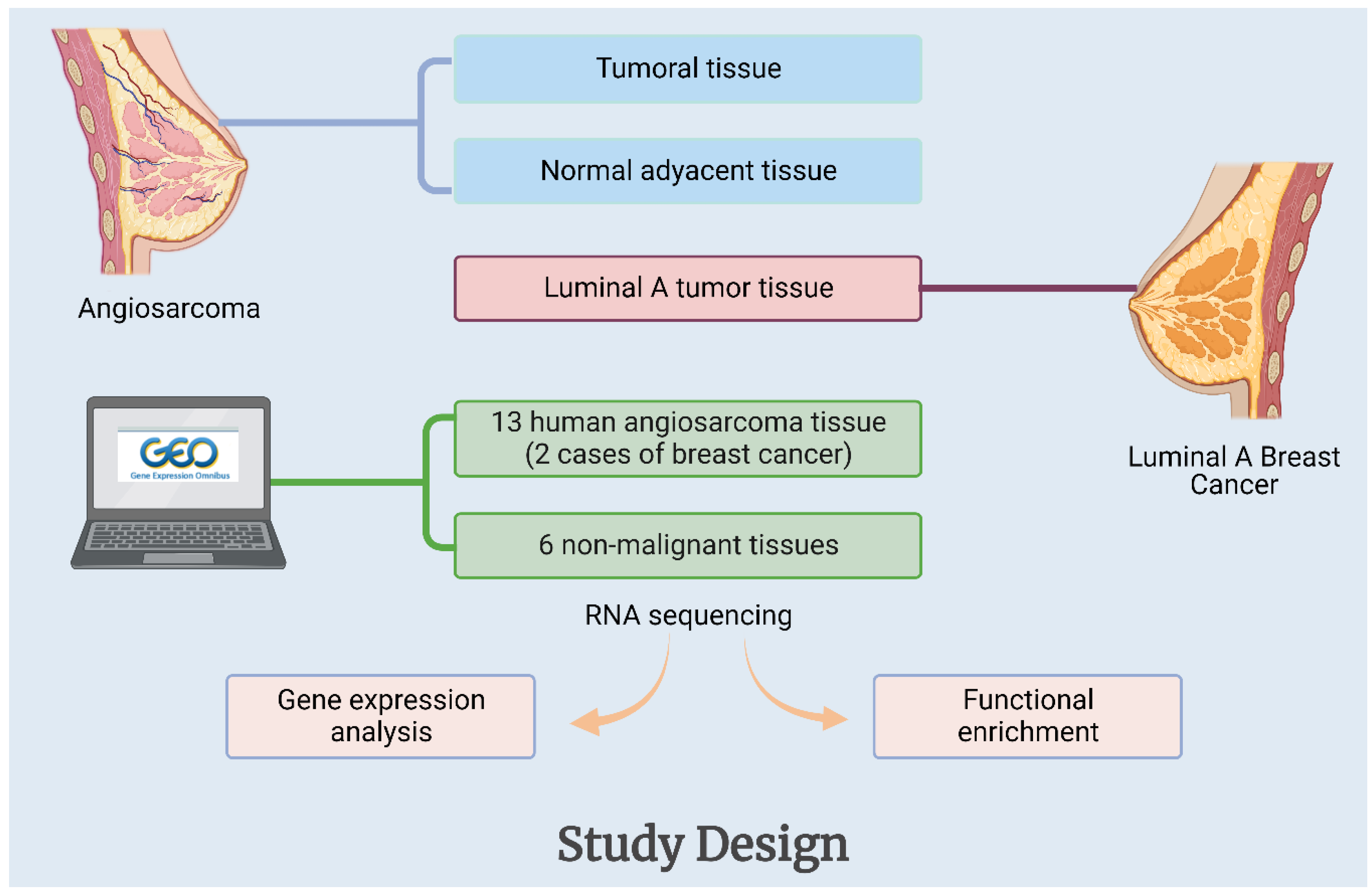
5.1. Samples
5.2. High-Throughput Gene Expression Data Retrieval
5.3. RNA Isolation
5.4. Angiosarcoma and Breast Cancer Tissues Whole Transcriptome Sequencing and GO-KEGG Enrichment
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alexandrova, E.; Sergieva, S.; Mihaylova, I.; Zarkova, A. Primary angiosarcoma of the breast complicated by the syndrome of disseminated intravascular coagulation (DIC): Case report and literature review. Rep. Pract. Oncol. Radiother. 2014, 19, 221–225. [Google Scholar] [CrossRef] [Green Version]
- Scow, J.S.; Reynolds, C.A.; Degnim, A.C.; Petersen, I.A.; Jakub, J.W.; Boughey, J.C. Primary and secondary angiosarcoma of the breast: The Mayo Clinic experience. J. Surg. Oncol. 2010, 101, 401–407. [Google Scholar] [CrossRef]
- Young, R.J.; Brown, N.J.; Reed, M.W.; Hughes, D.; Woll, P.J. Angiosarcoma. Lancet Oncol. 2010, 11, 983–991. [Google Scholar] [CrossRef]
- Babarović, E.; Zamolo, G.; Mustać, E.; Strčić, M. High grade angiosarcoma arising in fibroadenoma. Diagn. Pathol. 2011, 6, 125. [Google Scholar] [CrossRef] [Green Version]
- Esposito, E.; Avino, F.; di Giacomo, R.; Donzelli, I.; Marone, U.; Melucci, M.T.; Rinaldo, C.; Ruffolo, F.; Saponara, R.; Siani, C.; et al. Angiosarcoma of the breast, the unknown—A review of the current literature. Transl. Cancer Res. (Rare Tumors Breast) 2019, 8 (Suppl. S5), S510. [Google Scholar] [CrossRef]
- Arora, T.K.; Terracina, K.P.; Soong, J.; Idowu, M.O.; Takabe, K. Primary and secondary angiosarcoma of the breast. Gland Surg. 2014, 3, 28–34. [Google Scholar]
- Banks, J.; Ives, C.; Potter, S.; Holcombe, C. The BRASS (BReast Angiosarcoma Surveillance Study): Protocol for a retrospective multicentre cohort study to evaluate the management and outcomes of angiosarcoma of the breast and chest wall. Int. J. Surg. Protoc. 2017, 5, 5–10. [Google Scholar] [CrossRef] [Green Version]
- Bordoni, D.; Bolletta, E.; Falco, G.; Cadenelli, P.; Rocco, N.; Tessone, A.; Guarino, S.; Accurso, A.; Amato, B.; Magalotti, C. Primary angiosarcoma of the breast. Int. J. Surg. Case Rep. 2016, 20, 12–15. [Google Scholar] [CrossRef] [Green Version]
- Masai, K.; Kinoshita, T.; Jimbo, K.; Asaga, S.; Hojo, T. Clinicopathological features of breast angiosarcoma. Breast Cancer 2016, 23, 718–723. [Google Scholar] [CrossRef]
- Hodgson, N.C.; Bowen-Wells, C.; Moffat, F.; Franceschi, D.; Avisar, E. Angiosarcomas of the Breast. Am. J. Clin. Oncol. 2007, 30, 570–573. [Google Scholar] [CrossRef]
- Martínez Pérez, F.; Álvarez Flórez, M.; Sánchez Burgos, R.; Martínez Gimeno, C. Metástasis cutánea facial de angiosarcoma de mama. Rev. Española Cirugía Oral y Maxilofac. 2015, 37, 53–54. [Google Scholar] [CrossRef]
- Rohan, V.; Hanji, A.; Patel, J.; Tankshali, R. Primary angiosarcoma of the breast in a postmenopausal patient. J. Cancer Res. Ther. 2010, 6, 120. [Google Scholar] [CrossRef]
- Itakura, E.; Yamamoto, H.; Oda, Y.; Tsuneyoshi, M. Detection and characterization of vascular endothelial growth factors and their receptors in a series of angiosarcomas. J. Surg. Oncol. 2008, 97, 74–81. [Google Scholar] [CrossRef]
- Dim, D.; Ravi, V.; Tan, J.; Hicks, D.; Wong, M. The actin-bundling motility protein fascin and vascular endothelial growth factor (VEGF) are universally over-expressed in human angiosarcoma. J. Clin. Oncol. 2007, 25, 10068. [Google Scholar] [CrossRef]
- Schmid, H.; Zietz, C. Human herpesvirus 8 and angiosarcoma: Analysis of 40 cases and review of the literature. Pathology 2005, 37, 284–287. [Google Scholar] [CrossRef]
- Naka, N.; Tomita, Y.; Nakanishi, H.; Araki, N.; Hongyo, T.; Ochi, T.; Aozasa, K. Mutations of p53 tumor-suppressor gene in angiosarcoma. Int. J. Cancer 1997, 71, 952–955. [Google Scholar] [CrossRef]
- Johnson, K.D.; Glinskii, O.V.; Mossine, V.V.; Turk, J.R.; Mawhinney, T.P.; Anthony, D.C.; Henry, C.J.; Huxley, V.H.; Glinsky, G.V.; Pienta, K.J.; et al. Galectin-3 as a potential therapeutic target in tumors arising from malignant endothelia. Neoplasia 2007, 9, 662–670. [Google Scholar] [CrossRef] [Green Version]
- Garcia, J.M.; Gonzalez, R.; Silva, J.M.; Dominguez, G.; Vegazo, I.S.; Gamallo, C.; Provencio, M.; España, P.; Bonilla, F. Mutational status of K- ras and TP53 genes in primary sarcomas of the heart. Br. J. Cancer 2000, 82, 1183–1185. [Google Scholar] [CrossRef] [Green Version]
- Weihrauch, M.; Bader, M.; Lehnert, G.; Koch, B.; Wittekind, C.; Wrbitzky, R.; Tannapfel, A. Mutation analysis of K-ras-2 in liver angiosarcoma and adjacent nonneoplastic liver tissue from patients occupationally exposed to vinyl chloride. Environ. Mol. Mutagen. 2002, 40, 36–40. [Google Scholar] [CrossRef] [PubMed]
- Naito, S.; Shimizu, K.; Nakashima, M.; Nakayama, T.; Ito, T.; Ito, M.; Yamashita, S.; Sekine, I. Overexpression of Ets-1 Transcription Factor in Angiosarcoma of the Skin. Pathol.-Res. Pract. 2000, 196, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Behjati, S.; Tarpey, P.S.; Sheldon, H.; Martincorena, I.; Van Loo, P.; Gundem, G.; Wedge, D.C.; Ramakrishna, M.; Cooke, S.L.; Pillay, N.; et al. Recurrent PTPRB and PLCG1 mutations in angiosarcoma. Nat. Genet. 2014, 46, 376–379. [Google Scholar] [CrossRef] [PubMed]
- Painter, C.A.; Jain, E.; Tomson, B.N.; Dunphy, M.; Stoddard, R.E.; Thomas, B.S.; Damon, A.L.; Shah, S.; Kim, D.; Gómez Tejeda Zañudo, J.; et al. The Angiosarcoma Project: Enabling genomic and clinical discoveries in a rare cancer through patient-partnered research. Nat. Med. 2020, 26, 181–187. [Google Scholar] [CrossRef]
- Chan, J.Y.; Lim, J.Q.; Yeong, J.; Ravi, V.; Guan, P.; Boot, A.; Tay, T.K.Y.; Selvarajan, S.; Md Nasir, N.D.; Loh, J.H.; et al. Multiomic analysis and immunoprofiling reveal distinct subtypes of human angiosarcoma. J. Clin. Investig. 2020, 130, 5833–5846. [Google Scholar] [CrossRef] [PubMed]
- Espejo-Freire, A.P.; Elliott, A.; Rosenberg, A.; Costa, P.A.; Barreto-Coelho, P.; Jonczak, E.; D’Amato, G.; Subhawong, T.; Arshad, J.; Diaz-Perez, J.A.; et al. Genomic Landscape of Angiosarcoma: A Targeted and Immunotherapy Biomarker Analysis. Cancers 2021, 13, 4816. [Google Scholar] [CrossRef]
- Huang, S.-C.; Zhang, L.; Sung, Y.-S.; Chen, C.-L.; Kao, Y.-C.; Agaram, N.P.; Singer, S.; Tap, W.D.; D’Angelo, S.; Antonescu, C.R. Recurrent CIC gene abnormalities in angiosarcomas: A molecular study of 120 cases with concurrent investigation of PLCG1, KDR, MYC, and FLT4 gene alterations. Am. J. Surg. Pathol. 2016, 40, 645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antonescu, C.R.; Yoshida, A.; Guo, T.; Chang, N.-E.; Zhang, L.; Agaram, N.P.; Qin, L.-X.; Brennan, M.F.; Singer, S.; Maki, R.G. KDR Activating Mutations in Human Angiosarcomas Are Sensitive to Specific Kinase Inhibitors. Cancer Res. 2009, 69, 7175–7179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Segal, N.H.; Pavlidis, P.; Antonescu, C.R.; Maki, R.G.; Noble, W.S.; DeSantis, D.; Woodruff, J.M.; Lewis, J.J.; Brennan, M.F.; Houghton, A.N.; et al. Classification and subtype prediction of adult soft tissue sarcoma by functional genomics. Am. J. Pathol. 2003, 163, 691–700. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Zhang, C.; Zhang, Z.; Zhang, Z.; Ji, W.; Cao, S.; Cai, X.; Lei, D.; Pan, X. E26 Transformation-Specific Transcription Factor ETS2 as an Oncogene Promotes the Progression of Hypopharyngeal Cancer. Cancer Biother. Radiopharm. 2017, 32, 327–334. [Google Scholar] [CrossRef]
- Sun, X.; Zhao, X.; Xu, S.; Zhou, Y.; Jia, Z.; Li, Y. CircSRSF4 Enhances Proliferation, Invasion, and Migration to Promote the Progression of Osteosarcoma via Rac1. Int. J. Mol. Sci. 2022, 23, 6200. [Google Scholar] [CrossRef]
- Truong, A.H.; Ben-David, Y. The role of Fli-1 in normal cell function and malignant transformation. Oncogene 2000, 19, 6482–6489. [Google Scholar] [CrossRef] [Green Version]
- Ben-David, Y.; Gajendran, B.; Sample, K.M.; Zacksenhaus, E. Current insights into the role of Fli-1 in hematopoiesis and malignant transformation. Cell. Mol. Life Sci. 2022, 79, 163. [Google Scholar] [CrossRef]
- Park, S.-W.; Do, H.-J.; Choi, W.; Kim, J.-H. Fli-1 promotes proliferation and upregulates NANOGP8 expression in T-lymphocyte leukemia cells. Biochimie 2020, 168, 1–9. [Google Scholar] [CrossRef]
- Sato, F.; Yamamoto, T. Breast angiosarcoma after primary breast cancer surgery: A systematic review. J. Plast. Reconstr. Aesthet. Surg. 2022, 75, 2882–2889. [Google Scholar] [CrossRef]
- Sanders, M.E.; Simpson, J.F.; Cates, J.M. Vascular Lesions of the Breast. In A Comprehensive Guide to Core Needle Biopsies of the Breast; Springer International Publishing: Cham, Switzerland, 2016; pp. 667–685. [Google Scholar]
- Kim, J.-Y.; Bang, S.I.; Lee, S.D. α-Casein Changes Gene Expression Profiles and Promotes Tumorigenesis of Prostate Cancer Cells. Nutr. Cancer 2020, 72, 239–251. [Google Scholar] [CrossRef]
- Bessrour, I.; Kouidhi, S.; Belhassen, S.; Elgaaïed, A.B. Discrimination by RT-Nested PCR of Alpha-Lactalbumin Transcript Expression in Mammary Tumors. J. Environ. Pathol. Toxicol. Oncol. Off. Organ Int. Soc. Environ. Toxicol. Cancer 2019, 38, 313–327. [Google Scholar] [CrossRef]
- Nakato, R.; Wada, Y.; Nakaki, R.; Nagae, G.; Katou, Y.; Tsutsumi, S.; Nakajima, N.; Fukuhara, H.; Iguchi, A.; Kohro, T.; et al. Comprehensive epigenome characterization reveals diverse transcriptional regulation across human vascular endothelial cells. Epigenetics Chromatin 2019, 12, 77. [Google Scholar] [CrossRef] [Green Version]
- Yusof, K.M.; Groen, K.; Rosli, R.; Abdullah, M.; Mahmud, R.; Avery-Kiejda, K.A. Evaluation of Circulating MicroRNAs and Adipokines in Breast Cancer Survivors with Arm Lymphedema. Int. J. Mol. Sci. 2022, 23, 11359. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.-J.; Yin, Y.; Chua, B.T.; Li, P. CIDE family proteins control lipid homeostasis and the development of metabolic diseases. Traffic 2020, 21, 94–105. [Google Scholar] [CrossRef]
- Wei, Y.; Yang, X.; Gao, L.; Xu, Y.; Yi, C. Differences in potential key genes and pathways between primary and radiation-associated angiosarcoma of the breast. Transl. Oncol. 2022, 19, 101385. [Google Scholar] [CrossRef]
- Ruh, M.; Stemmler, M.P.; Frisch, I.; Fuchs, K.; van Roey, R.; Kleemann, J.; Roas, M.; Schuhwerk, H.; Eccles, R.L.; Agaimy, A.; et al. The EMT transcription factor ZEB1 blocks osteoblastic differentiation in bone development and osteosarcoma. J. Pathol. 2021, 254, 199–211. [Google Scholar] [CrossRef]
- Rosenbaum, E.; Antonescu, C.R.; Smith, S.; Bradic, M.; Kashani, D.; Richards, A.L.; Donoghue, M.; Kelly, C.M.; Nacev, B.; Chan, J.E.; et al. Clinical, genomic, and transcriptomic correlates of response to immune checkpoint blockade-based therapy in a cohort of patients with angiosarcoma treated at a single center. J. Immunother. Cancer 2022, 10, e004149. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. ClusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. Omi. J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]






| References | Sample | Angiogenic Markers in Patients | Additional Notes |
|---|---|---|---|
| Itakura et al., 2008 [13] | 34 angiosarcomas | 32 (94%) VEGF-A positive. 4 (12%) VEGF-C positive. 32 (94%) VEGFR-1 positive. 22 (65%) VEGFR-2 positive. 27 (79%) VEGFR-3 positive. | VEGF-A expression correlates with VEGFR-1 expression. Loss of VEGFR-2 expression is associated with poor prognosis. VEGFR-1 and VEGFR-3 were not associated with prognosis. |
| Dim et al., 2007 [14] | 20 angiosarcomas 5 hemangioma | All angiosarcomas were VEGF-A positive. | |
| Zietz et al., 2005 [15] | 19 angiosarcomas 10 benign vascular lesions | 18 angiosarcomas (95%) VEGF-A positive. 1 benign vascular lesion (10%) VEGF-A positive. | Abnormal MDM2 expression in 68% of angiosarcomas, abnormal P53 expression in 53%; abnormal P53 and MDM2 expression correlates with VEGF expression, which correlates with tumor grade. |
| Methodology | Origin | Genes Altered | Additional Notes | References |
|---|---|---|---|---|
| Whole-exome sequencing (GES) | Primary breast, cardiac, skin, bladder, lung and others | TP53, KDR, PIK3CA, POT1, PLCG1, NF1, FAT1, LRP1B, FLT4, NOTCH2, PTPRB, GRIN2A, MYC | The leading mutated gene in primary breast angiosarcoma was PIK3CA. | Painter, C.A. et al. [22] |
| Multiomic- sequencing and immuno-oncology profiling | Head and neck, breast, liver, lung, and others | AURKA, AURKB, PLK1, PLK4, CHEK1, and CDK4. | TMB was determined high, and there was significant tumor infiltration by immune system cells. | Chan, J.Y. et al. [23] |
| Retrospective analysis (WES, NGS) | Head and neck, breast, víscera, skin and extremities | TP53, MYC, ARID1A, POT1, ATRX, HRAS, PIK3CA | The main changes in breast angiosarcoma were observed in genes such as MYC, HRAS, and PIK3CA. | Espejo-Freire. et al. [24] |
| Whole-exome sequencing (GES) and RNAseq | Breast, head and neck, chest, víscera, trunk and others | CIC, ETV1, ETV4, ETV5, PLCG1, KDR, MYC, FLT4 | In 50% of angiosarcoma cases, CIC was reported to be altered. | Huang SC et al. [25] |
| Experiment | Up Regulated Genes | Down Regulated Genes |
|---|---|---|
| Angiosarcoma Case Report | CSN2, LALBA, BTN1A1 CEL, CSN1S1, CSN3 CA6, XDH, SLC30A2 SHISA8, OLAH, LPO GC, TMEM171, NPFFR2 FABP3, ATP13A4, ALB | LEP, CIDEC |
| GSE163359 | FSCN1, MAP4K4, TIE1, SHC1, MARCKSL1, RHOJ, SMAD1, PPM1F, NID1, TSPAN18, FKBP10, ADAM19, NAV1, DBN1 | SCARA5, DHCR24, ZDHHC11B, HLF, ABCA8, IFNLR1 |
| Overlapped Genes | Angiosarcoma | Breast Cancer |
|---|---|---|
| CSN2 | 11.61 | −11.29 |
| LALBA | 10.73 | −11.30 |
| CSN1S1 | 9.97 | −5.28 |
| CSN3 | 9.71 | −5.73 |
| Comparison | Overlapped Genes | Count |
|---|---|---|
| Case report vs different human angiosarcoma dataset | CFD, ADH1B, ALDH2, MAOA, TMEM132C, GREM2, CLEC3B, PTPRT, CES4A, ADORA1, DLK1, DIO3, ENTPD3, EDN3, C1QTNF7, FRMD1 | 16 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rincón-Riveros, A.; De la Peña, J.; Rubiano, W.; Olivella, F.; Martinez-Agüero, M.; Villegas, V.E. Primary Breast Angiosarcoma: Comparative Transcriptome Analysis. Int. J. Mol. Sci. 2022, 23, 16032. https://doi.org/10.3390/ijms232416032
Rincón-Riveros A, De la Peña J, Rubiano W, Olivella F, Martinez-Agüero M, Villegas VE. Primary Breast Angiosarcoma: Comparative Transcriptome Analysis. International Journal of Molecular Sciences. 2022; 23(24):16032. https://doi.org/10.3390/ijms232416032
Chicago/Turabian StyleRincón-Riveros, Andrés, Jairo De la Peña, Wilson Rubiano, Fabio Olivella, María Martinez-Agüero, and Victoria E. Villegas. 2022. "Primary Breast Angiosarcoma: Comparative Transcriptome Analysis" International Journal of Molecular Sciences 23, no. 24: 16032. https://doi.org/10.3390/ijms232416032
APA StyleRincón-Riveros, A., De la Peña, J., Rubiano, W., Olivella, F., Martinez-Agüero, M., & Villegas, V. E. (2022). Primary Breast Angiosarcoma: Comparative Transcriptome Analysis. International Journal of Molecular Sciences, 23(24), 16032. https://doi.org/10.3390/ijms232416032






