Abstract
ω-3 Polyunsaturated fatty acids (PUFAs) have been found to exert many actions, including neuroprotective effects. In this regard, the exact molecular mechanisms are not well understood. Parkinson’s disease (PD) is the second most common age-related neurodegenerative disease. Emerging evidence supports the hypothesis that PD is the result of complex interactions between genetic abnormalities, environmental toxins, mitochondrial dysfunction, and other cellular processes, such as DNA methylation. In this context, BDNF (brain-derived neurotrophic factor) and GDNF (glial cell line-derived neurotrophic factor) have a pivotal role because they are both involved in neuron differentiation, survival, and synaptogenesis. In this study, we aimed to elucidate the potential role of two PUFAs, eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA), and their effects on BDNF and GDNF expression in the SH-SY5Y cell line. Cell viability was determined using the MTT assay, and flow cytometry analysis was used to verify the level of apoptosis. Transmission electron microscopy was performed to observe the cell ultrastructure and mitochondria morphology. BDNF and GDNF protein levels and mRNA were assayed by Western blotting and RT-PCR, respectively. Finally, methylated and hydroxymethylated DNA immunoprecipitation were performed in the BDNF and GDNF promoter regions. EPA, but not DHA, is able (i) to reduce the neurotoxic effect of neurotoxin 6-hydroxydopamine (6-OHDA) in vitro, (ii) to re-establish mitochondrial function, and (iii) to increase BNDF and GDNF expression via epigenetic mechanisms.
Keywords:
PUFAs; EPA; DHA; Parkinson’s disease; neurodegeneration; apoptosis; mitochondrial damage; DNA methylation; BDNF; GDNF 1. Introduction
ω-3 Polyunsaturated fatty acids (PUFAs) are members of a large group of fatty acids (FA) possessing double bonds in their structure [1]. EPA (eicosapentaenoic acid; 20:5 ω-3) and DHA (docosahexaenoic acid; 22:6 ω-3) are very long-chain PUFAs that occur naturally in the highest quantities in fish [2]. In mammals, ω-3 PUFAs cannot be synthesized endogenously; therefore, the predominant source of EPA and DHA is dietary [3] and in a special kind of seafood.
ω-3 PUFAs have well-established anti-inflammatory properties [4] and have found clinical utility for cardiovascular disease prophylaxis and severe hypertriglyceridemia [5], with emerging evidence that they may be beneficial for the treatment of inflammatory diseases [6]. Recently, Tatsumi and co-workers reported that DHA and EPA induced antioxidant enzymes and protected Schwann cells from oxidative stress and, consequently, from diabetic neuropathy [7,8].
The dietary intake of ω-3 PUFAs has also been shown in human studies to inversely correlate with the overall risk of different types of cancer [9,10,11,12]. A substantial number of in vitro studies on cancer cell lines, as well as studies on animal models of cancers, have shown the antiproliferative, apoptotic, angiogenesis, cytotoxic, and antimetastatic properties of DHA and EPA [13,14,15]. Recent evidence also points to the potent and, at the same time, selective actions of EPA and DHA on multiple types of cancer cells [16]. Although ample data are available regarding the effects responsible for their actions on cancer, the exact mechanism is still ambiguous; however, recently, it was demonstrated that new mechanisms may involve ω-3 PUFAs as modulators of the aberrant epigenetic assembly in cancer cells [17,18].
PUFAs have become an emerging dietary medical intervention for health maintenance [19,20] and the treatment of neurodegenerative diseases such as Alzheimer’s disease (AD) and Parkinson’s disease (PD) [21]. Although their mechanism remains unclear, PUFAs seem to play a pivotal role in neural functions due to their abundance in neural tissues. Arachidonic acid (AA) and DHA are the two most abundant PUFAs within the nervous system and together comprise roughly 35% of the lipid content in the brain tissue [22].
PD is a chronic neurodegenerative disorder characterized by the progressive loss of dopaminergic (DA) neurons in the substantia nigra (SN), a subsequent decrease in dopamine in the striatum, and the presence of Lewy bodies (LBs), in which specific proteins, including α-synuclein, are deposited [23]. Dopaminergic degeneration is highly linked to the lack of neurotrophic factors, i.e., brain-derived neurotrophic factor (BDNF) and glial cell line-derived neurotrophic factor (GDNF), in neurons or the brain associated with PD [24]. The neurochemical features of PD include reactive oxygen species (ROS) generation, mitochondrial dysfunction, inflammation, the accumulation of misfolded proteins, nitric oxide production, and ubiquitin-proteasome system dysfunction [25]. Aging, as well as environmental and genetic factors, are considered disease risk factors for PD [25], whose symptoms include movement disorders such as resting tremors, postural abnormalities, rigidity, and akinesia, all of which develop as a result of the loss of 50–70% of DA neurons [26]. Despite its high incidence today, current pharmacotherapy for PD is lacking, and can only partially tackle symptoms of the disorder, but it is still unable to stop the progressive degeneration of neurons [27].
In addition to genetic changes, epigenetic mechanisms have been found to contribute significantly to PD [28,29]. The level of DNA methylation is one of the most studied epigenetic modifications in PD [30,31]. Epigenetic factors in general are chemical modifications of chromatin that do not change the underlying genomic sequence. These modifications can modulate gene expression, allowing differentiation into peculiar cellular phenotypes by driving tissue-specific expression patterns [32]. In particular, DNA methylation involves the conversion of cytosine to 5′-methyl cytosine (5mC) by at least five types of DNA methyltransferases (DNMTs), and PUFAs take part in this pathway [33]. Such methylation occurs in so-called “CpG islands” (cytosine-phosphate-guanine), which are regions of DNA rich in the dinucleotide formed by cytosine followed by guanine. This mechanism, together with the eventual deacetylation of histone lysine, is responsible for the compact conformation of chromatin, which inactivates the transcription of the genes of interest by preventing the access of transcription factors to promoter regions rich in CpG islands [34,35]. Discovered in 2009, DNA hydroxymethylation (5hmC) is a relatively new epigenetic modification occurring on cytosine [36] by the Ten-Eleven Translocation (TET) protein-mediated oxidative catalysis of 5mC [37,38]. While 5mC drives gene silencing, 5hmC plays some role in maintaining and/or promoting gene expression, but the exact mechanism is still to be elucidated [39].
Many PD-associated genes appear to be hypomethylated. α-Synuclein, one of the hallmark genes involved in PD, exhibits a hypomethylated island in intron 1, which determines a high expression level, whereas other genes are hypermethylated, such as MAPT (microtubule-associated protein tau), which exhibits a higher methylation level correlated with lower expression [31]. Consequently, the discovery of new compounds affecting DNMTs or TET expression might be promising tools for epigenetic therapy.
In this context, neurotrophic factors play a key role in the development, phenotypic maintenance, and survival of neurons in PD [40]. BDNF and GDNF are particularly relevant in this regard because they are involved in neuron differentiation and synaptogenesis [41,42]. Thus, the down-regulation of BDNF has been demonstrated in the SN in individuals with PD [43], but the reduced expression of GDNF in mice causes a marked reduction in DA neurons [44].
Elevated BDNF DNA methylation in exon IV down-regulates its expression in neurons [45], whereas hypermethylation in the promoter of GDNF influences its expression in brain tissue [46]. GDNF and BDNF expression levels may be, at least in part, under epigenetic control. As previously mentioned, PUFAs exert beneficial effects in the brain and could prevent or attenuate neurodegenerative diseases [47]. A correlation between a higher intake of ω-3 PUFAs from fish, rich in EPA and DHA, and a lower prevalence of PD was found [48,49]. They are able to reduce the risk of developing PD [50,51]. Some of the positive effects of ω-3 PUFAs on PD are probably due to their neuroprotective properties, since oxidative stress and neuroinflammation are ameliorated [21,52]. Apart from neuroprotective effects, ω-3 PUFAs may enhance DA signal transduction [53]. However, the role of each ω-3 PUFA has not been established, and in general, EPA and DHA are used as a mixture because their molecular mechanisms are not clear yet. Recently, we found that EPA modulates gene expression through epigenetic action in leukemia and in solid tumor cells [54,55].
In this study, we have analyzed the potential role of EPA and DHA, used individually, on cell viability, apoptosis, mitochondrial integrity, and BDNF and GDNF expression in the SH-SY5Y cell line. In this cell line, using neurotoxin 6-hydroxydopamine (6-OHDA), we mimicked cellular death and damage typical of neurodegenerative disorders, such as PD. The compound 5-aza-2′-deoxycytidine (5-AZA), an inhibitor of DNA methylation mainly employed in the treatment of several types of cancer, was used as a control for demethylation.
2. Results
2.1. Cell Viability by MTT with All Different Compounds Used Individually and in Combination
In our experiments, a neuronal cell line (SH-SY5Y) was used as an in vitro model. To mimic neurodegeneration, 6-OHDA, an analogue of the natural neurotransmitter dopamine, was utilized because its presence has been found in both rat and human brains [56]. Since its first description in 1959, 6-OHDA has played a fundamental role in preclinical research on PD [57]. First, to demonstrate the effects of 6-OHDA, EPA and DHA and their combination on the SH-SY5Y cell line, an MTT assay was performed after 24 h of treatment. For the above-mentioned test, a BSA/NaCl solution was used as a vehicle treatment because both EPA and DHA were prepared in this solution (Figure 1A). The concentrations used, from 1 to 20 µL, were safe. We observed a slight cytotoxic effect only after 50 µL of treatment. The toxin 6-OHDA diluted in PBS 1X, as previously reported [58,59], has a strong effect on cell viability. As shown in Figure 1B, SH-SY5Y viability decreased, and this cytotoxicity was dose-dependent. The first two concentrations, 10 µM (94.57 ± 3.26%) and 25 µM (87.35 ± 2.52%), had a moderate toxic effect. Significant cell death was observed with 100, 150, 200, and 500 µM. The dose chosen for inducing cell cytotoxicity and for evaluating the possible protective effects of EPA or DHA in all experiments was 50 µM 6-OHDA, which resulted in 71.37 ± 3.07% relative cell viability (p < 0.0001). Furthermore, 24 h of treatment with EPA (Figure 1C) and DHA (Figure 1D) alone from 10 up to 150 µM had no significant effect on cell viability, except for the two maximum doses tested, namely, 200 and 500 µM. As shown in Figure 1E,F, 6-OHDA was always present at a 50 µM concentration, whereas EPA and DHA were used from 10 to 500 µM. EPA incubation significantly prevented 6-OHDA-induced cell cytotoxicity; in particular, 100 µM EPA (Figure 1E) was able to return the cell viability approximatively to the control (CTR) value (95.09 ± 4.12%). DHA (Figure 1F) had a slight effect on cell viability, with best results at 50, 100, and 200 µM with viability around 87%. For this reason and because it was previously reported in the literature as the physiological dose [17], 100 µM of the two ω-3 PUFAs was selected as a standard dose in the following experiments.
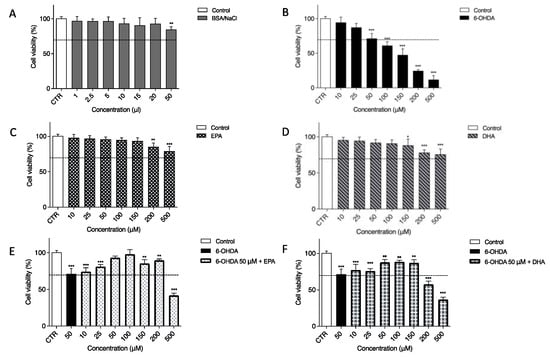
Figure 1.
Effect of 6-OHDA, EPA, and DHA (alone or in combination) on the viability of SH-SY5Y cell line. Cells were treated for 24 h using BSA/NaCl solution as vehicle treatment condition (A) and different concentrations of 6-OHDA (B), EPA (C), or DHA (D). Based on this result, 50 μM 6-OHDA was used to treat the cells together with EPA (E) or DHA (F). In the last two cases, the first column represents untreated cells (white), the second column is the viability after 50 μM 6-OHDA treatment (black) and all other columns show co-treatment with 50 μM 6-OHDA and two omega-3 fatty acids. Cell viability was assessed using MTT assay. Data from three independent experiments using four independent cultures per condition were analyzed by ANOVA using Graphpad Prism software. * p < 0.01, ** p < 0.001, and *** p < 0.0001.
2.2. Flow Cytometry Analysis Showed a Strong Induction of Apoptosis after 6-OHDA Treatment, Whereas EPA Had a Protective Effect
The apoptosis values were determined after 24 h in CTR and 6-OHDA-, EPA-, DHA-, and 5-AZA-treated cells, as shown in Figure 2, by flow cytometry. Using 50 µM 6-OHDA alone, we obtained a strong effect (p < 0.0001) on apoptosis (20.73 ± 2.48%) compared to untreated cells (1.43 ± 0.32%) (Figure 2), in accordance with MTT results. PUFA treatments alone (100 µM) did not induce SH-SY5Y apoptosis. Together with 6-OHDA, EPA treatment produced the greatest effect on reducing apoptosis (6.57 ± 1.37%) (Figure 2), but not DHA (17.37 ± 5.16%) (Figure 2). In addition, 5-AZA (1 µM) had a safe profile in terms of apoptosis (Figure 2), similar to PUFA treatments. The effects of all compounds on the percent distribution of the various phases of the cell cycle were studied, with no significant changes observed (data not shown).
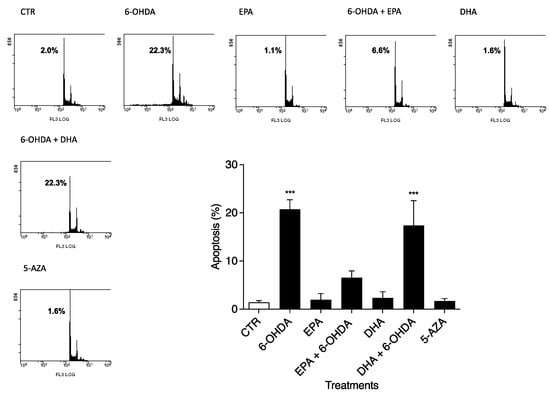
Figure 2.
Effects of 6-OHDA, EPA, DHA, and 5-AZA on the apoptosis of SH-SY5Y cell line. Cells were treated for 24 h using 50 μM 6-OHDA, 100 μM EPA, 100 μM DHA, and 1 μM 5-AZA. Cells were harvested, stained with PI, and analyzed using flow cytometry. Representative plots are reported together with means ± S.D. Data from three independent experiments using four independent cultures per condition were analyzed by ANOVA using Graphpad Prism software *** p < 0.0001.
Flow cytometry analysis was used before every experiment to ensure the correct cellular response.
2.3. Cell Morphology and Mitochondrial Organization Using Transmission Electron Microscopy (TEM) Analysis Were Compromised after 6-OHDA Treatment, but EPA Was Able to Reduce Damage
Transmission electron microscopy (TEM) was performed to observe the cell ultrastructure and mitochondrial morphology. The 50 µM 6-OHDA treatment after 24 h of incubation in vitro caused the swelling of mitochondria and damage to the cristae in the SH-SY5Y model (Figure 3B) compared to untreated cells, where mitochondria showed normal morphology with preserved cristae (Figure 3A). Electron-dense cytoplasmic inclusions and vacuoles of various sizes were also observed after 6-OHDA treatment (Figure 3B). Neither 100 µM EPA (Figure 3C) nor 100 µM DHA treatment (Figure 3E) altered the morphology of mitochondria, and no vacuoles or electron-dense cytoplasmic inclusions were observed. TEM showed that EPA reduces 6-OHDA-induced damage to mitochondrial membranes and cristae. No electron-dense cytoplasmic inclusions were found, and a few small cytoplasmic vacuoles were present (Figure 3D). DHA + 6-OHDA-treated cells showed partially swollen mitochondria, and small cytoplasmic vacuoles were visible in the cytoplasm (Figure 3F). This result shows that DHA has a less protective effect than EPA against 6-OHDA-induced toxicity. Treatment with 1 µM 5-AZA did not alter the morphology of mitochondria (Figure 3G).
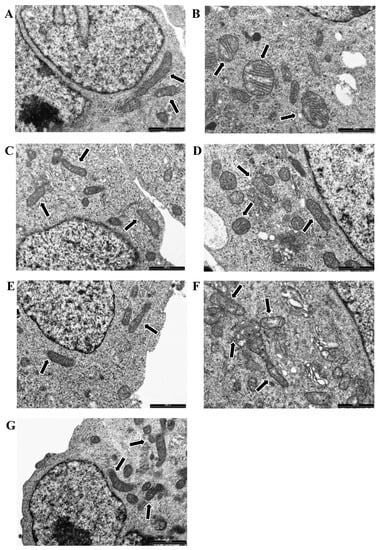
Figure 3.
Representative transmission electron microscopy (TEM) images of CTR (A) and 6-OHDA- (B), EPA- (C), EPA + 6-OHDA- (D), DHA- (E), DHA + 6-OHDA- (F), and 5-AZA-treated (G) cells are reported. Mitochondria are highlighted with black arrows. Bars = 1000 nm.
2.4. BDNF and GDNF Protein Levels by Western Blot Analysis Increased Significantly after EPA Treatment
Western blotting was used to investigate the roles of 6-OHDA, EPA, DHA, and 5-AZA on BDNF and GDNF protein levels in the SH-SY5Y neuronal cell line. Treatment with 50 µM 6-OHDA induced reductions in BDNF and GDNF protein expression of 0.59- (Figure 4A) and 0.53-fold (Figure 4B), respectively, compared with CTR (arbitrarily set to 1). Significant increments in BDNF (p < 0.0001) and GDNF (p < 0.0001) were observed after 100 µM EPA treatment (2.42 and 1.73 times, respectively, in comparison with CTR, as shown in Figure 4). This ability of EPA to up-regulate BDNF and GDNF protein expression was also confirmed in combination with 50 µM 6-OHDA: 2.18 times for BDNF (p < 0.0001) and 1.66 times for GDNF (p < 0.0001) vs. CTR. As shown in Figure 4, different results were obtained with DHA. In fact, DHA alone was unable to significantly change the neurotrophic factor levels. The combination (100 µM DHA + 50 µM 6-OHDA) seems to slightly improve the effect of 6-OHDA alone, returning BDNF and GNDF expression to levels similar to CTR. The 1 µM 5-AZA treatment significantly increased BDNF (p < 0.01) and GDNF (p < 0.0001) protein levels (Figure 4).
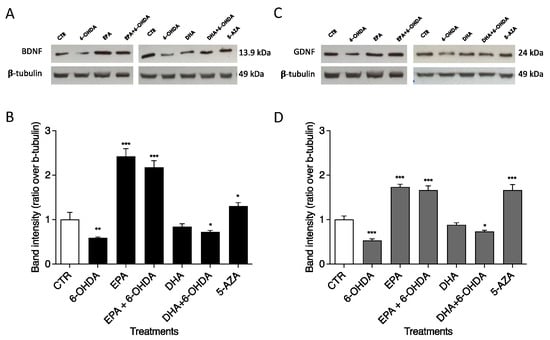
Figure 4.
Effects of EPA and DHA alone and in combination with 6-OHDA on protein expression in SH-SY5Y cells. Cells were cultured and treated as reported in M&M. BDNF (A) and GDNF (C) levels were analyzed by Western blotting. The positions of proteins are indicated in relation to the positions of molecular size standards. β-Tubulin III (neuronal) was used to normalize protein levels. Area density was evaluated by Chemidoc Imagequant TL software. Values of BDNF (B) and GDNF (D) are expressed relative to the control, which was set to an arbitrary value (1.0). Experimental samples are reported as the mean ± SD. Data from three independent experiments using four independent cultures per condition were analyzed by ANOVA using Graphpad Prism software. * p < 0.01, ** p < 0.001, and *** p < 0.0001.
2.5. BDNF and GDNF Genes Were Up-Regulated after EPA Treatment
To confirm the Western blotting data, BDNF and GDNF mRNA levels were determined by RT-PCR using the TaqMan Assay. SH-SY5Y cells were treated with the same doses of chemical compounds used for the previous experiments. Similar results were recorded with 6-OHDA, EPA, and DHA treatments in terms of gene expression in comparison with CTR (Figure 5). The BDNF mRNA level significantly increased (p < 0.0001) by 2.46 and 2.12 times, respectively, with EPA and EPA + 6-OHDA treatments (Figure 5A). On the contrary, DHA treatment did not change BDNF expression. A similar trend was obtained for the GDNF level (Figure 5B). EPA significantly increased (p < 0.0001) GDNF expression by 1.72 and 1.54 times, respectively, alone and in combination with 6-OHDA, whereas after DHA treatment, the GDNF level remained unchanged or even slightly decreased in combination with 6-OHDA (p < 0.01). 5-AZA was able to increase the BDNF and GDNF levels by 1.58 and 1.39 times, respectively (p < 0.001).

Figure 5.
Effects of EPA and DHA alone and in combination with 6-OHDA on mRNA expression in SH-SY5Y cells. Cells were cultured and treated as reported in M&M. BDNF (A) and GDNF (B) gene expression levels were studied by RT-PCR. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as a housekeeping gene. Experimental samples are reported as the mean ± SD. Data from three independent experiments using four independent cultures per condition were analyzed by ANOVA using Graphpad Prism software. * p < 0.01, ** p < 0.001, and *** p < 0.0001.
2.6. Different Effects of EPA and DHA on Hypermethylated CpG Islands Located in Promoter Regions of BDNF and GDNF Genes by DNA Immunoprecipitation (DNA IP)
Firstly, we sought CpG islands in the GDNF and BDNF promoter regions. In BDNF, a hypermethylated sequence (−4721/−5265) was identified; it is 400 bp long and contains 27 CpG. A hypermethylated region consisting of 712 bp was found in the GDNF promoter sequence (−7321/−8033) containing 59 CpG. To evaluate the methylation levels of these two CpG islands after 24 h EPA and DHA treatments, we performed a DNA IP assay using 5mC and 5hmC antibodies. The methylation and hydroxymethylation levels are approximatively the same in the control cells for both genes. Notably, we observed different effects in the treatment with the two PUFAs independently. In particular, the MeDIP assay demonstrated that 5mC levels decreased significantly (p < 0.0001) after 24 h EPA conditioning (Figure 6) in both promoter regions compared to CTR. Conversely, methylation in the BDNF promoter region (Figure 6A) remained almost stable, with a slight drop after DHA treatment compared to CTR, but decreased considerably (p < 0.0001) in the GDNF promoter region (Figure 6B). The hMeDIP assay revealed an important increase (p < 0.0001) in 5hmC content with 24 h EPA treatment, particularly for GDNF, showing the active transcription of both genes, in agreement with Western blot analysis and RT-PCR analysis data. The 5hmC levels did not change significantly after DHA conditioning in either gene (Figure 6) in comparison to untreated cells.
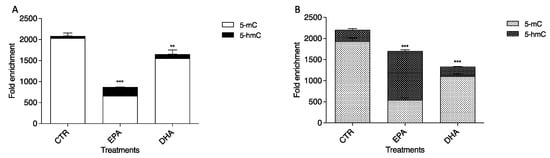
Figure 6.
Effects of EPA and DHA on BDNF (A) and GDNF (B) methylation and hydroxymethylation levels. Experimental samples are reported as the mean ± SD. Data from three independent experiments using four independent cultures per condition were analyzed by ANOVA using Graphpad Prism software. ** p < 0.001, and *** p < 0.0001.
3. Discussion
This is the first study that reports a unique effect of EPA on a human neuronal cell line, that is, suppressing apoptosis via mitochondrial integrity and neurotrophin over-expression. In this context, several studies have shown the protective effects of dietary enrichment with PUFAs in different animal models of neurodegenerative diseases and clinical/epidemiological studies [60,61]. These studies showed that the prevalence of AD and PD negatively correlates with fish consumption. EPA has a central role in neurodegenerative disorders [62] but right now is used in combination with other PUFAs, particularly with DHA, because the exact molecular mechanisms are still not elucidated.
The aim of the present study was to understand which PUFAs (EPA or DHA) are able to prevent cell apoptosis, to reduce mitochondrial damage, and to re-establish the abnormal epigenetic modifications involved in the pathogenesis of neurodegenerative disorders, particularly PD.
Neurotoxin 6-OHDA, an analogue of the natural neurotransmitter dopamine, has a dose-dependent toxicity [58,59] and causes mitochondrial dysfunction [63] mimicking neurodegenerative disorders, such as PD [57]. To investigate this mitochondrial dysfunction, we used transmission electron microscopy on SH-SY5Y cells treated with 50 µM 6-OHDA. Treatment with 50 µM 6-OHDA caused the swelling of mitochondria and damage to the cristae, whereas PUFAs were safe, and treated cells looked like CTR cells. In this regard, it was recently demonstrated that the toxic effects induced by the herbicide Paraquat (1,1′-dimethyl-4-4′-bipyridinium dichloride) in Drosophila melanogaster were prevented by the concomitant dietary ingestion of an EPA/DHA mixture, and more interestingly, this neuroprotective effect involves mitochondrial protection [64]. In our study, we demonstrated, for the first time, that EPA was able to reduce 6-OHDA-induced damage to mitochondrial membranes and cristae better than DHA. In fact, TEM images indicate that EPA had a protective role against mitochondria swelling and damage to the cristae.
Recent studies suggest that neurodegeneration is due to abnormal DNA methylation, one of the most studied epigenetic modifications in PD [31,65,66]. Epigenetic factors do not change the underlying genomic sequence but may modulate gene expression. In this study, using the DNA IP technique, we were able to discriminate 5mC and 5hmC in two crucial neurotrophic factors, BDNF and GDNF genes, both important for the survival, maintenance, and regeneration of specific neuronal populations in the adult brain [67].
Our results show that EPA but not DHA is able to modify the methylation and hydroxymethylation levels of BDNF and GDNF and, as a consequence, their mRNA levels and protein expression. In particular, EPA considerably decreased 5mC and increased 5hmC in the promoter regions of these two important genes, crucial for neuron differentiation and synaptogenesis. The first one, a member of the neurotrophin family of growth factors, has been shown to modulate the survival, differentiation, synaptic plasticity, and activity of neurons. In PD, a reduction in BDNF expression in SN might contribute to the death of DA neurons because inhibiting BDNF expression in the SN causes parkinsonism in the rat [68]. A similar role was supposed for GDNF, but recent preclinical data suggest that GDNF can rescue DA neurons, and a possible therapy was evaluated for PD patients in clinical trials [69].
Replacement strategies and the administration of a therapy involving these neurotrophic proteins were considered in the past as potential therapeutics for PD [67], but the major problems found are the blood–brain barrier and other secondary side effects, such as anorexia and weight loss [70]. So, novel therapeutic approaches are needed. In this scenario, Nam and co-workers demonstrated that the human ras homolog enriched in the brain, which has an S16H mutation [hRheb(S16H)], protects the nigrostriatal DA projection in the 6-OHDA-treated animal PD model thanks to BDNF and GDNF activation [71]. Animal studies also revealed that physical activity promotes angiogenesis and neuroplasticity and may activate some extracellular signals, including BDNF [72].
Recently, Che and co-workers [73] demonstrated that EPA-enriched ethanolamine plasmalogen (EPA-pPE) enhanced BDNF/TrkB/CREB signaling and inhibited neuronal apoptosis in vitro and in vivo better than EPA-enriched phosphatidylethanolamine (EPA-PE). They proposed this molecule as a potential therapeutic agent in long-term AD therapy. Moreover, ethyl-eicosapentaenoate (E-EPA) can protect against motor impairments, neurodegeneration, and inflammation [74] and partially attenuated the striatal DA turnover [75] in a 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)-probenecid (MPTP-P) mouse model of PD.
Since in mammals, both DHA and EPA cannot be synthesized de novo, their plasma levels primarily reflect dietary intake and a small amount of transformation from the precursor alpha-linolenic acid (ALA, 18:3 ω-3) [76,77]. As previously described, PUFAs are essential for normal brain functioning, and their reduction in plasma has been associated with major depression and other neurological diseases [78,79,80].
In animal models, ω-3 PUFA deficiency induced in offspring via the manipulation of the rat maternal diet causes the marked over-expression of many genes that encode neurotransmitter receptors, particularly those for dopamine, glutamate, and acetylcholine [81].
In this context, Sublette and co-workers suggested that the dietary intake of PUFAs (both absolute and relative to the omega-6 status) may also contribute to the regulation of brain DA functioning in humans [82]. Potential mechanisms whereby ω-3 PUFAs may regulate dopamine include reducing cortical MAO-B activity, thereby decreasing the degradation of monoamines [83]; improving dopamine neuron survival [84]; and influencing lipid rafts [85].
A possibility may be that an endogenous molecule and/or a molecule derived from the diet is able to impart the important ability to continuously produce BDNF and GDNF as therapeutic agents against neurodegeneration in DA neurons. We already know that ω-3 PUFA deficiency can reduce the nigrostriatal system’s ability to maintain homeostasis under oxidative conditions, which may enhance the risk of PD. In particular, in rodents fed a diet deficient in ω-3 PUFAs, a dramatic decrease in brain PUFA contents has been observed with an important reduction in BDNF expression in SN [86].
Our findings suggest that EPA could be much more effective than DHA as an epigenetic modulator in preventing the loss of DA neurons via BDNF and GDNF expression. Our previous studies showed that only EPA wielded an important function in DNA demethylation in solid tumor cells [46,47]. For this reason, we aimed to investigate whether it could be a neuroprotective agent in the SH-SY5Y cell line through epigenetic mechanisms. In fact, it was previously reported that the entire molecular process behind DNA demethylation involved two stages: increased 5hmC levels, due to greater TET1 binding to DNA [17], and decreased DNA methyltransferase 1 (DNMT1) mRNA and protein levels [18].
In conclusion, the present study strengthens the evidence that EPA is able to reduce the neurotoxic effect induced by the neurotoxin 6-OHDA on the SH-SY5Y cell line via methylation reduction and the hydroxymethylation activation of two characteristic genes: BDNF and GDNF. Additional in vivo and clinical studies are needed to define its therapeutic use in humans. In any case, we speculate that EPA supplementation may prevent neurodegeneration, and it could be administered before clinical manifestations.
Future research directions should consider PUFA administration as a single treatment and not as a complex mixture, particularly for neurodegenerative disorder treatment.
4. Materials and Methods
4.1. Materials
The human bone marrow neuroblastoma SH-SY5Y cell line was purchased from Istituto Zooprofilattico Sperimentale della Lombardia e dell’Emilia Romagna ‘Bruno Ubertini’ (Brescia, Italy). RPMI 1640, L-glutamine, trypsin, and ethylenediaminetetraacetic acid disodium and tetrasodium salt (EDTA) were from Microtech Srl (Pozzuoli, NA, Italy).
Fetal Bovine Serum (FBS), penicillin–streptomycin, Master Mix TaqMan®Gene Expression and all primers for RT-PCR were from Thermo Fisher Scientific (Waltham, MA, USA). Antibiotics and Dulbecco’s phosphate-buffered saline pH 7.4 (PBS) were from Invitrogen Srl (Milan, Italy). Dimethyl sulfoxide (DMSO), ethanol, hydrochloric acid, sodium chloride, sodium deoxycholate, and SDS (Sodium Dodecyl Sulfate) were purchased from Carlo Erba (Reagenti Srl, Milan, Italy). 6-Hydroxydopamine hydrochloride (6-OHDA), eicosapentaenoic acid (20:5, ω-3; EPA), docosahexaenoic acid (22:6, ω-3; DHA), bovine serum albumin fraction V (BSA; fatty acid-free), 5-aza-2′-deoxycytidine, and monoclonal anti-β-tubulin III (neuronal) (T8578) were purchased from Sigma (Saint Louis, MO 63178 USA). Anti-BDNF (TA328615) rabbit polyclonal antibody was from Origene (Rockville, MD 20850), whereas anti-GDNF (ab176564) rabbit monoclonal antibody, was from Abcam (Cambridge, MA, USA). Anti-β-tubulin antibody (Sigma-Aldrich) was used to normalize. Monoclonal horseradish peroxidase-conjugated goat anti-rabbit secondary antibodies were from Santa Cruz Biotechnology (Bergheimer Str. 89-2, 69115 Heidelberg, Germany). Immunoreactive bands were visualized using the SuperSignal West Pico Chemiluminescent Substrate (ThermoFisher Scientific, Via Morolense 5, 03013 Ferentino FR, Italy). The EpiQuik Hydroxymethylated DNA Immunoprecipitation (hMeDIP/P-1038-24) Kit and Methylamp Methylated DNA Capture (MeDIP/P1015-24) Kit were from TEMA ricerca Srl (Via XXI Ottobre 1944, 11/240055 Castenaso, Bologna, Italy).
4.2. Albumin-Bound Fatty Acid
As previously described [87], EPA and DHA were prepared as stock solutions (10 mM). In brief, EPA and DHA were diluted in ethanol and precipitated by adding NaOH (final concentration: 0.25 M). The precipitate was dried under nitrogen, reconstituted with 0.9% (w/v) NaCl, and stirred at RT for 10 min with defatted BSA fraction V (fatty acid free) (final concentration: 10% w/v) in 0.15 M NaCl. Each solution was adjusted to pH 7.4 with NaOH and stored in the dark in aliquots in liquid nitrogen. The fatty acid/BSA molar ratio was 4:1 [17].
4.3. Cell Culture and Treatment
SH-SY5Y neuroblastoma cells, a subclone of the original SK-N-SH bone-marrow-derived cells, were grown in monolayer cultures with RPMI 1640 supplemented with 10% heat-inactivated FBS, 1 mmol/L sodium pyruvate, 2 mM L-glutamine, and antibiotics (100 U/mL penicillin and 100 μg/mL streptomycin). The cells were maintained in a cell incubator at 37 °C in a humidified atmosphere containing 5% CO2. When the cells reached 80–90% confluence, the routine culture medium was aspirated, and SH-SY5Y cells were washed with PBS 1X. The cells were then harvested by 0.05% trypsin in 0.02% Na4EDTA for 1–2 min at 37 °C and suspended in 1:8 supplemented growth medium to be maintained in the exponential growth phase. For experiments, the cells were counted using a trypan blue dye exclusion assay and seeded at a standard concentration for different assays.
For all experiments, the cells were cultured in the presence or absence of different substances: 6-OHDA, EPA, DHA, and 5-AZA. Stock solutions of 6-OHDA (2 mM) and 5-AZA (1 mM) were prepared before using in PBS 1X, whereas stock solutions of EPA (10 mM) and DHA (10 mM) were in BSA/NaCl stored in liquid nitrogen, as described in Section 4.2. The final concentrations for TEM, Western blotting, and RT-PCR analysis were 50 µM for 6-OHDA, 100 µM for EPA, 100 µM for DHA, and 1 µM for 5-AZA.
4.4. Viability by MTT Assay
Cellular viability was assessed by the reduction of MTT to formazan [88]. SH-SY5Y cells were seeded onto a 96-well plate at a density of 1 × 104 cells/well with RPMI 1640 complete medium. After 24 h, fresh complete medium was replaced for treatment with different concentrations (from 10 to 500 µM) of 6-OHDA, EPA, and DHA for 24 h. Then, MTT reagent was dissolved in PBS 1X and added to the culture at a 0.5 mg/mL final concentration. After 3 h incubation at 37 °C, the supernatant was carefully removed, and formazan salt crystals were dissolved in 200 μL of DMSO added to each well [89]. The absorbance (OD) values were measured spectrophotometrically at 540 nm using an automatic microplate reader (Eliza MAT 2000, DRG Instruments, GmbH). Each experiment was performed twice in quadruplicate, and cell viability was expressed as a relative percentage, as previously described [90,91].
4.5. Flow Cytometry Analysis of Apoptosis and Cell Cycle
SH-SY5Y cells treated for 24 h with 50 µM 6-OHDA, 100 μM fatty acids (EPA and DHA), and 1 μM 5-aza were recovered, washed, and centrifuged at 200× g, then the cell pellets were resuspended in 1 mL of hypotonic propidium iodide (PI) solution (50 μg/mL in 1% sodium citrate plus 0.1% Triton X-100; Sigma), and the samples were incubated at 4 °C in the dark for 2 h. The fluorescence of PI of single nuclei was evaluated with an EPICS XL-MCL flow cytometer (Beckman Coulter, Brea, CA, USA), and the data were processed with an Intercomp computer and analyzed with EXPO32 software (Beckman Coulter) [92,93,94]. The percentage of apoptosis (hypodiploid DNA content) was determined with EXPO32 software (Beckman Coulter) as previously described [93,94,95]. The cell cycle was examined by measuring DNA-bound PI fluorescence in the orange-red fluorescence channel (FL2) with linear amplification. The percentage of cells in each cell-cycle phase was evaluated with ModFit software (Verity Software House, Topsham, ME, USA) [96].
Flow cytometry analyses were repeated three times in independent experiments. DNA fluorescence flow cytometric profiles of one experiment are representative of three, and graphs showing the mean ± standard deviation of percentage hypodiploid obtained in three different experiments are shown. The data were analyzed as described in the Statistical Analysis section.
4.6. Transmission Electron Microscopy (TEM) Analysis
Cells were fixed with 2% glutaraldehyde in 0.1 M phosphate buffer for 2 h at 20 °C. The fixative was washed off twice with PBS and post-fixed with 1% OsO4 in PBS for 1 h.
Samples were dehydrated through ethanol gradients and embedded in Epon.
Ultrathin sections were cut with a diamond knife on a Reichert ultramicrotome; the sections were mounted on Formvar single-hole grids or 150-mesh grids, stained with uranyl acetate and lead citrate, and then examined at 80 kV under a Philips EM 208 microscope equipped with a digital camera (University Centre for Electron Microscopy, CUME, Perugia, Italy).
4.7. Western Blotting
After treatment, SH-SY5Y cells were harvested and lysed in ice-cold RIPA lysis buffer containing 150 mM sodium chloride, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS (Sodium Dodecyl Sulfate), and 50 mM Tris pH 8.0. Protein concentration was determined according to the method of Bradford [97]. About 60 µg of protein was used to perform 10% SDS–polyacrylamide gel electrophoresis at 200 V for 1 h, as previously described [98]. Proteins were transferred onto 0.45 µm cellulose nitrate strip membranes (Sartorius Stedim Biotech S.A.) in transfer buffer for 1 h at 100 V at 4 °C. Membranes were blocked with 5% (w/v) non-fat dry milk in PBS, pH 7.5, for 1 h at RT. The blot was incubated overnight at 4 °C with anti-BDNF, anti-GDNF, and anti-β-tubulin III specific antibodies (1:1000). The blot was treated with horseradish peroxidase-conjugated goat anti-rabbit secondary antibodies (1:5000). SuperSignal West Pico Chemiluminescent Substrate (ThermoFisher Scientific) was used to detect the chemiluminescent (ECL) HRP substrate. The apparent molecular weights of BDNF and GDNF were calculated according to the migration of molecular size standards. Images were acquired using the VersaDoc Imaging System (Bio-Rad, Hercules, CA, USA), and signals were quantified using Quantity One software (Bio-Rad). The area densities of the bands were evaluated by densitometry scanning and analyzed with Scion Image.
4.8. Reverse Transcription Quantitative PCR (RTq-PCR)
SH-SY5Y cells were used for total RNA extraction, which was performed by using the RNAqueous ®-4PCR kit (Ambion Inc., Austin, TX, USA) as previously reported [99]. Before cDNA synthesis, the integrity of RNA was evaluated by denaturing electrophoresis in TAE 1.2% agarose gel. cDNA was synthesized using 1 μg of total RNA for all samples using a High-Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA, USA) under the following conditions: 50 °C for 2 min, 95 °C for 10 min, 95 °C for 15 s, and 60 °C for 1 min for 40 cycles. RTq-PCR was performed using Master Mix TaqMan®Gene Expression and the 7.500 RT-PCR instrument (Applied Biosystems), targeting genes in the Taqman Array 96-Well Plate (P/N: 4414292): brain-derived neurotrophic factor (BDNF, Hs02718934_s1), glial cell-derived neurotrophic factor (GDNF, Hs01931883_s1), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH, Hs99999905_m1).
4.9. Methylated DNA Immunoprecipitation (MeDIP) and Hydroxymethylated DNA Immunoprecipitation (hMeDIP) Assays
Fragmented DNA (0.5 mg/sample; 200–1000 bp in size) was denatured for 10 min at 95 °C. Hydroxymethylated DNA immunoprecipitation (hMeDIP; P-1038-24; Epigentek) and methylated DNA immunoprecipitation (MeDIP; P-2019-24; Epigentek) assays were performed according to the manufacturer’s instructions.
Antibody buffer (100 µL) was added to the following antibodies: 1 µL of non-immune IgG (negative controls), 1 µL of anti-5hmC, and 1 µL of anti-5mC and incubated at room temperature for 60 min. After removing the antibody buffer from the wells and washing with diluted washing buffer, 0.5 µg of fragmented DNA was added to each well. After incubation for 90 min at room temperature on an orbital shaker, the reaction wells were washed with diluted washing buffer. Proteinase K was added for 15 min at 60 °C, and the reaction wells were incubated at 95 °C for 3 min to inactivate the enzyme.
DNA samples were subjected to qRT-PCR using Brilliant SYBR Green qPCR Master Mix.
The following primers were used to amplify the BDNF promoter region (−4721, −5265):
Forward, 5′-GGAAAGTTGTTGGGCTGGTT-3′;
Reverse, 5′-GTTTCCTAGGGCTGCCTTCT-3′.
The following primers were used to amplify the GDNF proximal promoter sequence (NG_011675):
Forward, 5′-TTCGGTGGTTTAGTGGGGTG-3′;
Reverse, 5′-CAGTGGAAAACTCCCTGCCT-3′.
Fold enrichment was calculated by means of the amplification efficiency ratio of hMeDIP or MeDIP DNA samples over non-immune IgG by using 2Δ(IgG Ct−sample Ct) [17].
4.10. Statistical Analysis
Data from three independent experiments using at least four independent cultures per condition were analyzed by ANOVA. GraphPad Prism version 9.2.0.332 was used (GraphPad software, San Diego, CA, USA) for MTT, flow cytometry, RT-PCR, Western blotting, and immunoprecipitation analyses. Data are expressed as mean ± S.D. A one-way ANOVA test was performed, and the significance thresholds were set as * p < 0.01, ** p < 0.001, and *** p < 0.0001. The nonparametric Mann–Whitney test or Kruskal–Wallis with post hoc Dunn’s test was used.
Author Contributions
Experiments and analysis of data, M.R.C. and V.C.; conceptualization, A.V.; analysis of data and revision of the manuscript, M.R.C. and M.C. (Michela Codini); methodology, M.R.C. and S.C.; software, M.R.C., K.F. and M.C. (Mario Calvitti); writing—original draft preparation, M.R.C. and T.B.; supervision, M.R.C. and V.C.; project administration, E.A.; funding acquisition, A.V. and T.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author: Tommaso Beccari, email tommaso.beccari@unipg.it.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Calder, P.C. Mechanisms of action of (n-3) fatty acids. J. Nutr. 2012, 142, 592S–599S. [Google Scholar] [CrossRef] [PubMed]
- Calder, P.C.; Yaqoob, P. Omega-3 polyunsaturated fatty acids and human health outcomes. Biofactors 2009, 35, 266–272. [Google Scholar] [CrossRef] [PubMed]
- Baker, E.J.; Miles, E.A.; Burdge, G.C.; Yaqoob, P.; Calder, P.C. Metabolism and functional effects of plant-derived omega-3 fatty acids in humans. Prog. Lipid Res. 2016, 64, 30–56. [Google Scholar] [CrossRef] [PubMed]
- Im, D.S. Omega-3 fatty acids in anti-inflammation (pro-resolution) and GPCRs. Prog. Lipid Res. 2012, 51, 232–237. [Google Scholar] [CrossRef] [PubMed]
- Kwak, S.M.; Myung, S.K.; Lee, Y.J.; Seo, H.G.; Korean Meta-analysis Study Group. Efficacy of omega-3 fatty acid supplements (eicosapentaenoic acid and docosahexaenoic acid) in the secondary prevention of cardiovascular disease: A meta-analysis of randomized, double-blind, placebo-controlled trials. Arch. Intern. Med. 2012, 172, 686–694. [Google Scholar] [CrossRef]
- Michalak, A.; Mosińska, P.; Fichna, J. Polyunsaturated Fatty Acids and Their Derivatives: Therapeutic Value for Inflammatory, Functional Gastrointestinal Disorders, and Colorectal Cancer. Front. Pharmacol. 2016, 7, 459. [Google Scholar] [CrossRef]
- Tatsumi, Y.; Kato, A.; Sango, K.; Himeno, T.; Kondo, M.; Kato, Y.; Kamiya, H.; Nakamura, J.; Kato, K. Omega-3 polyunsaturated fatty acids exert anti-oxidant effects through the nuclear factor (erythroid-derived 2)-related factor 2 pathway in immortalized mouse Schwann cells. J. Diabetes Investig. 2019, 10, 602–612. [Google Scholar] [CrossRef]
- Tatsumi, Y.; Kato, A.; Niimi, N.; Yako, H.; Himeno, T.; Kondo, M.; Tsunekawa, S.; Kato, Y.; Kamiya, H.; Nakamura, J.; et al. Docosahexaenoic Acid Suppresses Oxidative Stress-Induced Autophagy and Cell Death via the AMPK-Dependent Signaling Pathway in Immortalized Fischer Rat Schwann Cells 1. Int. J. Mol. Sci. 2022, 23, 4405. [Google Scholar] [CrossRef]
- Slagsvold, J.E.; Pettersen, C.H.; Follestad, T.; Krokan, H.E.; Schønberg, S.A. The antiproliferative effect of EPA in HL60 cells is mediated by alterations in calcium homeostasis. Lipids 2009, 44, 103–113. [Google Scholar] [CrossRef]
- Song, E.A.; Kim, H. Docosahexaenoic Acid Induces Oxidative DNA Damage and Apoptosis, and Enhances the Chemosensitivity of Cancer Cells. Int. J. Mol. Sci. 2016, 17, 1257. [Google Scholar] [CrossRef]
- Cockbain, A.J.; Toogood, G.J.; Hull, M.A. Omega-3 polyunsaturated fatty acids for the treatment and prevention of colorectal cancer. Gut 2012, 61, 135–149. [Google Scholar] [CrossRef] [PubMed]
- Sawada, N.; Inoue, M.; Iwasaki, M.; Sasazuki, S.; Shimazu, T.; Yamaji, T.; Takachi, R.; Tanaka, Y.; Mizokami, M.; Tsugane, S.; et al. Consumption of n-3 fatty acids and fish reduces risk of hepatocellular carcinoma. Gastroenterology 2012, 142, 1468–1475. [Google Scholar] [CrossRef] [PubMed]
- Betiati Dda, S.; de Oliveira, P.F.; Camargo Cde, Q.; Nunes, E.A.; Trindade, E.B. Effects of omega-3 fatty acids on regulatory T cells in hematologic neoplasms. Rev. Bras. Hematol. Hemoter. 2013, 35, 119–125. [Google Scholar] [CrossRef][Green Version]
- Azrad, M.; Turgeon, C.; Demark-Wahnefried, W. Current evidence linking polyunsaturated Fatty acids with cancer risk and progression. Front. Oncol. 2013, 3, 224. [Google Scholar] [CrossRef]
- Huang, Q.; Wen, J.; Chen, G.; Ge, M.; Gao, Y.; Ye, X.; Liu, C.; Cai, C. Omega-3 Polyunsaturated Fatty Acids Inhibited Tumor Growth via Preventing the Decrease of Genomic DNA Methylation in Colorectal Cancer Rats. Nutr. Cancer 2016, 68, 113–119. [Google Scholar] [CrossRef]
- Abdi, J.; Garssen, J.; Faber, J.; Redegeld, F.A. Omega-3 fatty acids, EPA and DHA induce apoptosis and enhance drug sensitivity in multiple myeloma cells but not in normal peripheral mononuclear cells. J. Nutr. Biochem. 2014, 25, 1254–1262. [Google Scholar] [CrossRef] [PubMed]
- Ceccarelli, V.; Valentini, V.; Ronchetti, S.; Cannarile, L.; Billi, M.; Riccardi, C.; Ottini, L.; Talesa, V.N.; Grignani, F.; Vecchini, A. Eicosapentaenoic acid induces DNA demethylation in carcinoma cells through a TET1-dependent mechanism. FASEB J. 2018, 32, 5990–6001. [Google Scholar] [CrossRef]
- Ceccarelli, V.; Ronchetti, S.; Marchetti, M.C.; Calvitti, M.; Riccardi, C.; Grignani, F.; Vecchini, A. Molecular mechanisms underlying eicosapentaenoic acid inhibition of HDAC1 and DNMT expression and activity in carcinoma cells. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194481. [Google Scholar] [CrossRef]
- Wall, R.; Ross, R.P.; Fitzgerald, G.F.; Stanton, C. Fatty acids from fish: The anti-inflammatory potential of long-chain omega-3 fatty acids. Nutr. Rev. 2010, 68, 280–289. [Google Scholar] [CrossRef]
- Yang, L.; Liang, J.; Lam, S.M.; Yavuz, A.; Shui, G.; Ding, M.; Huang, X. Neuronal lipolysis participates in PUFA-mediated neural function and neurodegeneration. EMBO Rep. 2020, 21, e50214. [Google Scholar] [CrossRef]
- Burckhardt, M.; Herke, M.; Wustmann, T.; Watzke, S.; Langer, G.; Fink, A. Omega-3 fatty acids for the treatment of dementia. Cochrane Database Syst. Rev. 2016, 4, CD009002. [Google Scholar] [CrossRef] [PubMed]
- Bishop-Bailey, D.; Thomson, S.; Askari, A.; Faulkner, A.; Wheeler-Jones, C. Lipid-metabolizing CYPs in the regulation and dysregulation of metabolism. Annu. Rev. Nutr. 2014, 34, 261–279. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, S.; Wood, N.W. Molecular pathogenesis of Parkinson’s disease. Hum. Mol. Genet. 2005, 14, 2749–2755. [Google Scholar] [CrossRef] [PubMed]
- Siegel, G.J.; Chauhan, N.B. Neurotrophic factors in Alzheimer’s and Parkinson’s disease brain. Brain Res. Rev. 2000, 33, 199–227. [Google Scholar] [CrossRef]
- Renani, P.G.; Taheri, F.; Rostami, D.; Farahani, N.; Abdolkarimi, H.; Abdollahi, E.; Taghizadeh, E.; Gheibi Hayat, S.M. Involvment of aberrant regulation of epigenetic machanisms in the pathogenesis of Parkinson’s disease and epigenetic-based therapies. J. Cell Physiol. 2019, 234, 19307–19319. [Google Scholar] [CrossRef]
- Iranzo, A.; Valldeoriola, F.; Lomeña, F.; Molinuevo, J.L.; Serradell, M.; Salamero, M.; Cot, A.; Ros, D.; Pavía, J.; Santamaria, J.; et al. Serial dopamine transporter imaging of nigrostriatal function in patients with idiopathic rapid-eye-movement sleep behaviour disorder: A prospective study. Lancet Neurol. 2011, 10, 797–805. [Google Scholar] [CrossRef] [PubMed]
- Cheon, S.M.; Chan, L.; Chan, D.K.; Kim, J.W. Genetics of Parkinson’s disease—A clinical perspective. J. Mov. Disord. 2012, 5, 33–41. [Google Scholar] [CrossRef]
- Harrison, I.F.; Dexter, D.T. Epigenetic targeting of histone deacetylase: Therapeutic potential in Parkinson’s disease? Pharmacol. Ther. 2013, 140, 34–52. [Google Scholar] [CrossRef]
- Coppedè, F. Genetics and epigenetics of Parkinson’s disease. Sci. World J. 2012, 2012, 489830. [Google Scholar] [CrossRef]
- Miranda-Morales, E.; Meier, K.; Sandoval-Carrillo, A.; Salas-Pacheco, J.; Vazquez-Cardenas, P.; Arias-Carrion, O. Implications of DNA methylation in Parkinson’s disease. Front. Mol. Neurosci. 2017, 10, 225. [Google Scholar] [CrossRef]
- Labbé, C.; Lorenzo-Betancor, O.; Ross, O.A. Epigenetic regulation in Parkinson’s disease. Acta Neurophatol. 2016, 132, 515–530. [Google Scholar] [CrossRef] [PubMed]
- Jun, W. Characterization of tissue- specific differential DNA methylation suggests distinct modes of positive and negative gene expression regulation. BMC Genom. 2015, 16, 49. [Google Scholar] [CrossRef]
- Sarabi, M.M.; Naghibalhossaini, F. The impact of polyunsaturated fatty acids on DNA methylation and expression of DNMTs in human colorectal cancer cells. Biomed. Pharmacother. 2018, 101, 94–99. [Google Scholar] [CrossRef] [PubMed]
- Maunakea, A.K. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 2010, 466, 253–258. [Google Scholar] [CrossRef] [PubMed]
- Peter, A.J. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012, 13, 484. [Google Scholar] [CrossRef]
- Kriaucionis, S.; Heintz, N. The Nuclear DNA Base 5-Hydroxymethylcytosine Is Present in Purkinje Neurons and the Brain. Science 2009, 324, 929–930. [Google Scholar] [CrossRef] [PubMed]
- Tahiliani, M.; Koh, K.P.; Shen, Y.; Pastor, W.A.; Bandukwala, H.; Brudno, Y.; Agarwal, S.; Iyer, L.M.; Liu, D.R.; Aravind, L.; et al. Conversion of 5-Methylcytosine to 5-Hydroxymethylcytosine in Mammalian DNA by MLL Partner TET1. Science 2009, 324, 930–935. [Google Scholar] [CrossRef]
- Xu, Y.; Wu, F.; Tan, L.; Kong, L.; Xiong, L.; Deng, J.; Barbera, A.J.; Zheng, L.; Zhang, H.; Huang, S.; et al. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol. Cell 2011, 42, 451–464. [Google Scholar] [CrossRef]
- Haffner, M.C.; Chaux, A.; Meeker, A.K.; Esopi, D.M.; Gerber, J.; Pellakuru, L.G.; Toubaji, A.; Argani, P.; Iacobuzio-Donahue, C.; Nelson, W.G.; et al. Global 5-hydroxymethylcytosine content is significantly reduced in tissue stem/progenitor cell compartments and in human cancers. Oncotarget 2011, 2, 627–637. [Google Scholar] [CrossRef]
- Chauhan, N.B.; Siegel, G.J.; Lee, J.M. Depletion of glial cell line-derived neurotrophic factor in substantia nigra neurons of Parkinson’s disease brain. J. Chem. Neuroanat. 2001, 21, 277–288. [Google Scholar] [CrossRef]
- Lin, L.F.; Doherty, D.H.; Lile, J.D.; Bektesh, S.; Collins, F. GDNF: A glial cell line-derived neutrophic factor for midbrain dopaminergic neurons. Science 1993, 260, 1130–1132. [Google Scholar] [CrossRef] [PubMed]
- Hyman, C.; Hofer, M.; Barde, Y.A.; Juhasz, M.; Yancopoulos, G.D.; Squinto, S.P.; Lindsay, R.M. BDNF is a neutrophic factor for dopaminergic neurons of the substantia nigra. Nature 1991, 350, 230–232. [Google Scholar] [CrossRef] [PubMed]
- Mogi, M.; Togari, A.; Kondo, T.; Mizuno, Y.; Komure, O.; Kuno, S.; Ichinose, H.; Nagatsu, T. Brain-derived growth factor and nerve growth factor concentrations are decreased in the substantia nigra in Parkinson’s disease. Neurosci. Lett. 1999, 270, 45–48. [Google Scholar] [CrossRef] [PubMed]
- Pascual, A.; Hidalgo-Figueroa, M.; Piruat, J.I.; Pindado, C.O.; Gomez-Diaz, R.; Lopez-Barneo, J. Absolute requirement of GDNF for adult catecholaminergic neuron survival. Nat. Neurosci. 2008, 11, 755–761. [Google Scholar] [CrossRef] [PubMed]
- Martinowich, K.; Hattori, D.; Wu, H.; Fouse, S.; He, F.; Hu, Y.; Fan, G.; Sun, Y.E. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science 2003, 302, 890–893. [Google Scholar] [CrossRef]
- Chen, M.; Liu, Z.; Deng, Y.; Chen, X.; Zhang, J. Methylation status of promoter 1 region of GDNF gene in human glioma cells. Int. J. Clin. Exp. Med. 2014, 7, 1735–1740. [Google Scholar]
- Lo Van, A.; Sakayori, N.; Hachem, M.; Belkouch, M.; Picq, M.; Fourmaux, B.; Lagarde, M.; Osumi, N.; Bernoud-Hubac, N. Targeting the brain with a Neuroprotective omega-3 fatty acid to enhance neurogenesis in hypoxic condition in culture. Mol. Neurobiol. 2019, 56, 986–999. [Google Scholar] [CrossRef]
- Gao, X.; Chen, H.; Fung, T.T.; Logroscino, G.; Schwarzschild, M.A.; Hu, F.B.; Ascherio, A. Prospective study of dietary pattern and risk of Parkinson disease. Am. J. Clin. Nutr. 2007, 86, 1486–1494. [Google Scholar] [CrossRef]
- Okubo, H.; Miyake, Y.; Sasaki, S.; Murakami, K.; Tanaka, K.; Fukushima, W.; Kiyohara, C.; Tsuboi, Y.; Yamada, T.; Oeda, T.; et al. Dietary patterns and risk of Parkinson’s disease: A case-control study in Japan. Eur. J. Neurol. 2012, 19, 681–688. [Google Scholar] [CrossRef]
- De Lau, L.M.; Bornebroek, M.; Witteman, J.C.; Hofman, A.; Koundstaal, P.J.; Breteler, M.M. Dietary fatty acids and the risk of Parkinson disease: The Rotterdam study. Neurology 2005, 64, 2040–2045. [Google Scholar] [CrossRef]
- Taghizadeh, M.; Tamtaji, O.R.; Dadgostar, E.; Daneshvar Kakhaki, R.; Bahmani, F.; Abolhassani, J.; Aarabi, M.H.; Kouchaki, E.; Memarzadeh, M.R.; Asemi, Z. The effects of omega-3 fatty acids and vitamin E co-supplementation on clinical and metabolic status in patients with Parkinson’s disease: A randomized, double-blind, placebo-controlled trials. Neurochem. Int. 2017, 108, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Orr, S.K.; Trepanier, M.O.; Bazinet, R.P. N-3 polyunsaturated fatty acids in animal models with neuroinflammation. Prostaglandins Leukot. Essent. Fatty Acids 2013, 88, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Healy-Stoffel, M.; Levant, B. N-3 (omega-3) fatty acids: Effects on brain dopamine systems and potential role in the etiology and treatment of neuropsychiatric disorder. CNS Neurol. Disord. Drug Targets 2018, 17, 216–232. [Google Scholar] [CrossRef] [PubMed]
- Ceccarelli, V.; Racanicchi, S.; Martelli, M.P.; Nocentini, G.; Fettucciari, K.; Riccardi, C.; Marconi, P.; Di Nardo, P.; Grignani, F.; Binaglia, L.; et al. Eicosapentaenoic acid demethylates a single CpG that mediates expression of tumor suppressor CCAAT/enhancer-binding protein delta in U937 leukemia cells. J. Biol. Chem. 2011, 286, 27092–27102. [Google Scholar] [CrossRef] [PubMed]
- Ceccarelli, V.; Nocentini, G.; Billi, M.; Racanicchi, S.; Riccardi, C.; Roberti, R.; Grignani, F.; Binaglia, L.; Vecchini, A. Eicosapentaenoic acid activates RAS/ERK/C/EBPb pathway through H-Ras intron 1 CpG Island demethylation in U937 leukemia cells. PLoS ONE 2014, 9, e85025. [Google Scholar] [CrossRef] [PubMed]
- Jellinger, K.; Linert, L.; Kienzl, E.; Herlinger, E.; Youdim, M.B. Chemical evidence for 6-hydroxydopamine to be an endogenous toxic factor in the pathogenesis of Parkinson’s disease. J. Neural Transm. Suppl. 1995, 46, 297–314. [Google Scholar]
- Simola, N.; Morelli, M.; Carta, A.R. The 6-hydroxydopamine model of Parkinson’s disease. Neurotox. Res. 2007, 1, 151–167. [Google Scholar] [CrossRef]
- Meesarapee, B.; Thampithak, A.; Jaisin, Y.; Sanvarinda, P.; Suksamrarn, A.; Tuchinda, P.; Morales, N.P.; Sanvarinda, Y. Curcumin I mediates neuroprotective effect through attenuation of quinoprotein formation, p-p38 MAPK expression, and caspase-3 activation in 6-hydroxydopamine treated SH-SY5Y cells. Phytother. Res. 2014, 28, 611–616. [Google Scholar] [CrossRef]
- Elyasi, L.; Eftekhar-Vaghefi, S.H.; Esmaeili-Mahani, S. Morphine protects SH-SY5Y human neuroblastoma cells against 6-hydroxydopamine-induced cell damage: Involvement of anti-oxidant, calcium blocking, and anti-apoptotic properties. Rejuv. Res. 2014, 17, 255–263. [Google Scholar] [CrossRef]
- Gonzalo-Gobernado, R.; Ayuso, M.I.; Sansone, L.; Bernal-Jiménez, J.J.; Ramos-Herrero, V.D.; Sánchez-García, E.; Ramos, T.L.; Abia, R.; Muriana, F.J.G.; Bermúdez, B.; et al. Neuroprotective Effects of Diets Containing Olive Oil and DHA/EPA in a Mouse Model of Cerebral Ischemia. Nutrients 2019, 11, 1109. [Google Scholar] [CrossRef]
- Zhang, Y.P.; Brown, R.E.; Zhang, P.C.; Zhao, Y.T.; Ju, X.H.; Song, C. DHA, EPA and their combination at various ratios differently modulated Aβ25-35-induced neurotoxicity in SH-SY5Y cells. Prostaglandins Leukot. Essent. Fatty Acids 2018, 136, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Kou, W.; Luchtman, D.; Song, C. Eicosapentaenoic acid (EPA) increases cell viability and expression of neurotrophin receptors in retinoic acid and brain-derived neurotrophic factor differentiated SH-SY5Y cells. Eur. J. Nutr. 2008, 47, 104–113. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Lazaro, M.; Bonekamp, N.A.; Galindo, M.F.; Jordán, J.; Schrader, M. 6-Hydroxydopamine (6-OHDA) induces Drp1-dependent mitochondrial fragmentation in SH-SY5Y cells. Free Radic. Biol. Med. 2008, 44, 1960–1969. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira-Souza, A.; Couto-Lima, C.A.; Catalão, C.H.R.; Santos-Júnior, N.N.; Dos Santos, J.F.; da Rocha, M.J.A.; Alberici, L.C. Neuroprotective action of Eicosapentaenoic (EPA) and Docosahexaenoic (DHA) acids on Paraquat intoxication in Drosophila melanogaster. Neurotoxicology 2019, 70, 154–160. [Google Scholar] [CrossRef]
- Kia, D.A.; Zhang, D.; Guelfi, S.; Manzoni, C.; Hubbard, L.; Reynolds, R.H.; Botía, J.; Ryten, M.; Ferrari, R.; Lewis, P.A.; et al. Identification of Candidate Parkinson Disease Genes by Integrating Genome-Wide Association Study, Expression, and Epigenetic Data Sets. JAMA Neurol. 2021, 78, 464–472. [Google Scholar] [CrossRef]
- Allen, S.J.; Watson, J.J.; Shoemark, D.K.; Barua, N.U.; Patel, N.K. GDNF, NGF and BDNF as therapeutic options for neurodegeneration. Pharmacol. Ther. 2013, 138, 155–175. [Google Scholar] [CrossRef]
- Masliah, E.; Dumaop, W.; Galasko, D.; Desplats, P. Distinctive patterns of DNA methylation associated with Parkinson disease: Identification of concordant epigenetic changes in brain and peripheral blood leukocytes. Epigenetics 2013, 8, 1030–1038. [Google Scholar] [CrossRef]
- Hernandez-Chan, N.G.; Bannon, M.J.; Orozco-Barrios, C.E.; Escobedo, L.; Zamudio, S.; De la Cruz, F.; Gongora-Alfaro, J.L.; Armendáriz-Borunda, J.; Reyes-Corona, D.; Espadas-Alvarez, A.J.; et al. Neurotensin-polyplex-mediated brain-derived neurotrophic factor gene delivery into nigral dopamine neurons prevents nigrostriatal degeneration in a rat model of early Parkinson’s disease. J. Biomed. Sci. 2015, 22, 59. [Google Scholar] [CrossRef]
- Barker, R.A.; Björklund, A.; Gash, D.M.; Whone, A.; Van Laar, A.; Kordower, J.H.; Bankiewicz, K.; Kieburtz, K.; Saarma, M.; Booms, S.; et al. GDNF and Parkinson’s Disease: Where Next? A Summary from a Recent Workshop. J. Parkinson’s Dis. 2020, 10, 875–891. [Google Scholar] [CrossRef]
- Nutt, J.G.; Burchiel, K.J.; Comella, C.L.; Jankovic, J.; Lang, A.E.; Laws, E.R., Jr.; Lozano, A.M.; Penn, R.D.; Simpson, R.K., Jr.; Stacy, M.; et al. Implanted intra-cerebroventricular. Glial cell line-derived neurotrophic factor Randomized, double-blind trial of glial cell line-derived neurotrophic factor (GDNF) in PD. Neurology 2003, 60, 69–73. [Google Scholar] [CrossRef]
- Nam, J.H.; Leem, E.; Jeon, M.T.; Jeong, K.H.; Park, J.W.; Jung, U.J.; Kholodilov, N.; Burke, R.E.; Jin, B.K.; Kim, S.R. Induction of GDNF and BDNF by hRheb(S16H) transduction of SNpc neurons: Neuroprotective mechanisms of hRheb(S16H) in a model of Parkinson’s disease. Mol. Neurobiol. 2015, 51, 487–499. [Google Scholar] [CrossRef] [PubMed]
- Palasz, E.; Wysocka, A.; Gasiorowska, A.; Chalimoniuk, M.; Niewiadomski, W.; Niewiadomska, G. BDNF as a Promising Therapeutic Agent in Parkinson’s Disease. Int. J. Mol. Sci. 2020, 21, 1170. [Google Scholar] [CrossRef] [PubMed]
- Che, H.; Zhang, L.; Ding, L.; Xie, W.; Jiang, X.; Xue, C.; Zhang, T.; Wang, Y. EPA-enriched ethanolamine plasmalogen and EPA-enriched phosphatidylethanolamine enhance BDNF/TrkB/CREB signaling and inhibit neuronal apoptosis in vitro and in vivo. Food Funct. 2020, 11, 1729–1739. [Google Scholar] [CrossRef] [PubMed]
- Luchtman, D.W.; Meng, Q.; Song, C. Ethyl-eicosapentaenoate (E-EPA) attenuates motor impairments and inflammation in the MPTP-probenecid mouse model of Parkinson’s disease. Behav. Brain Res. 2012, 226, 386–396. [Google Scholar] [CrossRef] [PubMed]
- Meng, Q.; Luchtman, D.W.; El Bahh, B.; Zidichouski, J.A.; Yang, J.; Song, C. Ethyl-eicosapentaenoate modulates changes in neurochemistry and brain lipids induced by parkinsonian neurotoxin 1-methyl-4-phenylpyridinium in mouse brain slices. Eur. J. Pharmacol. 2010, 649, 127–134. [Google Scholar] [CrossRef]
- Bezard, J.; Blond, J.P.; Bernard, A.; Clouet, P. The metabolism and availability of essential fatty acids in animal and human tissues. Reprod. Nutr. Dev. 1994, 34, 539–568. [Google Scholar] [CrossRef]
- Goyens, P.L.; Spilker, M.E.; Zock, P.L.; Katan, M.B.; Mensink, R.P. Compartmental modeling to quantify alpha-linolenic acid conversion after longer term intake of multiple tracer boluses. J. Lipid Res. 2005, 46, 1474–1483. [Google Scholar] [CrossRef]
- Peet, M.; Murphy, B.; Shay, J.; Horrobin, D. Depletion of omega-3 fatty acid levels in red blood cell membranes of depressive patients. Biol. Psychiatry 1998, 43, 315–319. [Google Scholar] [CrossRef]
- Lin, P.Y.; Huang, S.Y.; Su, K.P. A Meta-Analytic Review of Polyunsaturated Fatty Acid Compositions in Patients with Depression. Biol. Psychiatry 2010, 68, 140–147. [Google Scholar] [CrossRef]
- Avallone, R.; Vitale, G.; Bertolotti, M. Omega-3 fatty acids and neurodegenerative diseases: New evidence in clinical trials. Int. J. Mol. Sci. 2019, 20, 4256. [Google Scholar] [CrossRef]
- Kuperstein, F.; Yakubov, E.; Dinerman, P.; Gil, S.; Eylam, R.; Salem, N., Jr.; Yavin, E. Overexpression of dopamine receptor genes and their products in the postnatal rat brain following maternal n-3 fatty acid dietary deficiency. J. Neurochem. 2005, 95, 1550–1562. [Google Scholar] [CrossRef] [PubMed]
- Sublette, M.E.; Galfalvy, H.C.; Hibbeln, J.R.; Keilp, J.G.; Malone, K.M.; Oquendo, M.A.; Mann, J.J. Polyunsaturated fatty acid associations with dopaminergic indices in major depressive disorder. Int. J. Neuropsychopharmacol. 2014, 17, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Chalon, S.; Delion-Vancassel, S.; Belzung, C.; Guilloteau, D.; Leguisquet, A.M.; Besnard, J.C.; Durand, G. Dietary fish oil affects monoaminergic neurotransmission and behavior in rats. J. Nutr. 1998, 128, 2512–2519. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.O.; Park, J.H.; Radel, J.D.; Levant, B. Reduced numbers of dopamine neurons in the substantia nigra pars compacta and ventral tegmental area of rats fed an n-3 polyunsaturated fatty acid-deficient diet: A stereological study. Neurosci. Lett. 2008, 438, 303–307. [Google Scholar] [CrossRef]
- Rockett, B.D.; Franklin, A.; Harris, M.; Teague, H.; Rockett, A.; Shaikh, S.R. Membrane raft organization is more sensitive to disruption by (n-3) PUFA than nonraft organization in EL4 and B cells. J. Nutr. 2011, 141, 1041–1048. [Google Scholar] [CrossRef]
- Cardoso, H.D.; dos Santos Junior, E.F.; de Santana, D.F.; Gonçalves-Pimentel, C.; Angelim, M.K.; Isaac, A.R.; Lagranha, C.J.; Guedes, R.C.A.; Beltrão, E.I.; Morya, E.; et al. Omega-3 deficiency and neurodegeneration in the substantia nigra: Involvement of increased nitric oxide production and reduced BDNF expression. Biochim. Biophys. Acta. 2014, 1840, 1902–1912. [Google Scholar] [CrossRef]
- Hannah, V.C.; Ou, J.; Luong, A.; Goldstein, J.L.; Brown, M.S. Unsaturated fatty acids down-regulate srebp isoforms 1a and 1c by two mechanisms in HEK-293 cells. J. Biol. Chem. 2001, 276, 4365–4372. [Google Scholar] [CrossRef]
- Denizot, F.; Lang, R. Rapid colorimetric assay for cell growth and survival. Modifications to the tetrazolium dye procedure giving improved sensitivity and reliability. J. Immunol. Methods 1986, 89, 271–277. [Google Scholar] [CrossRef]
- Ceccarini, M.R.; Codini, M.; Cataldi, S.; Vannini, S.; Lazzarini, A.; Floridi, A.; Moretti, M.; Villarini, M.; Fioretti, B.; Beccari, T.; et al. Acid sphingomyelinase as target of Lycium Chinense: Promising new action for cell health. Lipids Health Dis. 2016, 15, 183–192. [Google Scholar] [CrossRef]
- Ceccarini, M.R.; Vannini, S.; Cataldi, S.; Moretti, M.; Villarini, M.; Fioretti, B.; Albi, E.; Beccari, T.; Codini, M. In Vitro Protective Effects of Lyciumbarbarum Berries Cultivated in Umbria (Italy) on Human Hepatocellular Carcinoma Cells. Biomed Res. Int. 2016, 2016, 7529521. [Google Scholar] [CrossRef]
- Ceccarini, M.R.; Puccetti, M.; Pagano, C.; Nocchetti, M.; Beccari, T.; di Michele, A.; Ricci, M.; Perioli, L. MgAl and ZnAl-Hydrotalcites as Materials for Cosmetic and Pharmaceutical Formulations: Study of Their Cytotoxicity on Different Cell Lines. Pharmaceuticals 2022, 15, 784. [Google Scholar] [CrossRef] [PubMed]
- Fettucciari, K.; Ponsini, P.; Gioè, D.; Macchioni, L.; Palumbo, C.; Antonelli, E.; Coaccioli, S.; Villanacci, V.; Corazzi, L.; Marconi, P.; et al. Enteric glial cells are susceptible to Clostridium difficile toxin B. Cell Mol. Life Sci. 2017, 74, 1527–1551. [Google Scholar] [CrossRef] [PubMed]
- Fettucciari, K.; Macchioni, L.; Davidescu, M.; Scarpelli, P.; Palumbo, C.; Corazzi, L.; Marchegiani, A.; Cerquetella, M.; Spaterna, A.; Marconi, P.; et al. Clostridium difficile toxin B induces senescence in enteric glial cells: A potential new mechanism of Clostridium difficile pathogenesis. Biochim. Biophys. Acta Mol. Cell Res. 2018, 1865, 1945–1958. [Google Scholar] [CrossRef] [PubMed]
- Fettucciari, K.; Marguerie, F.; Fruganti, A.; Marchegiani, A.; Spaterna, A.; Brancorsini, S.; Marconi, P.; Bassotti, G. Clostridioides difficile toxin B alone and with pro-inflammatory cytokines induces apoptosis in enteric glial cells by activating three different signalling pathways mediated by caspases, calpains and cathepsin B. Cell Mol. Life Sci. 2022, 79, 442. [Google Scholar] [CrossRef] [PubMed]
- Nicoletti, I.; Migliorati, G.; Pagliacci, M.C.; Grignani, F.; Riccardi, C. A rapid and simple method for measuring thymocyte apoptosis by propidium iodide staining and flow cytometry. J. Immunol. Methods 1991, 139, 271–279. [Google Scholar] [CrossRef] [PubMed]
- Gerashchenko, B.I.; Azzam, E.I.; Howell, R.W. Characterization of cell-cycle progression and growth of WB-F344 normal rat liver epithelial cells following gamma-ray exposure. Cytom. A 2004, 61, 134–141. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Cataldi, S.; Ceccarini, M.R.; Patria, F.; Beccari, T.; Mandarano, M.; Ferri, I.; Lazzarini, A.; Curcio, F.; Albi, E. The Effect of Vitamin D3 and Silver Nanoparticles on HaCaT Cell Viability. Int. J. Mol. Sci. 2022, 23, 1410. [Google Scholar] [CrossRef]
- Codini, M.; Cataldi, S.; Lazzarini, A.; Tasegian, A.; Ceccarini, M.R.; Floridi, A.; Lazzarini, R.; Ambesi-Impiombato, F.S.; Curcio, F.; Beccari, T.; et al. Why high cholesterol levels help hematological malignancies: Role of nuclear lipid microdomains. Lipids Health Dis. 2016, 15, 4. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).