Rapid, Label-Free Prediction of Antibiotic Resistance in Salmonella typhimurium by Surface-Enhanced Raman Spectroscopy
Abstract
:1. Introduction
2. Results and Discussion
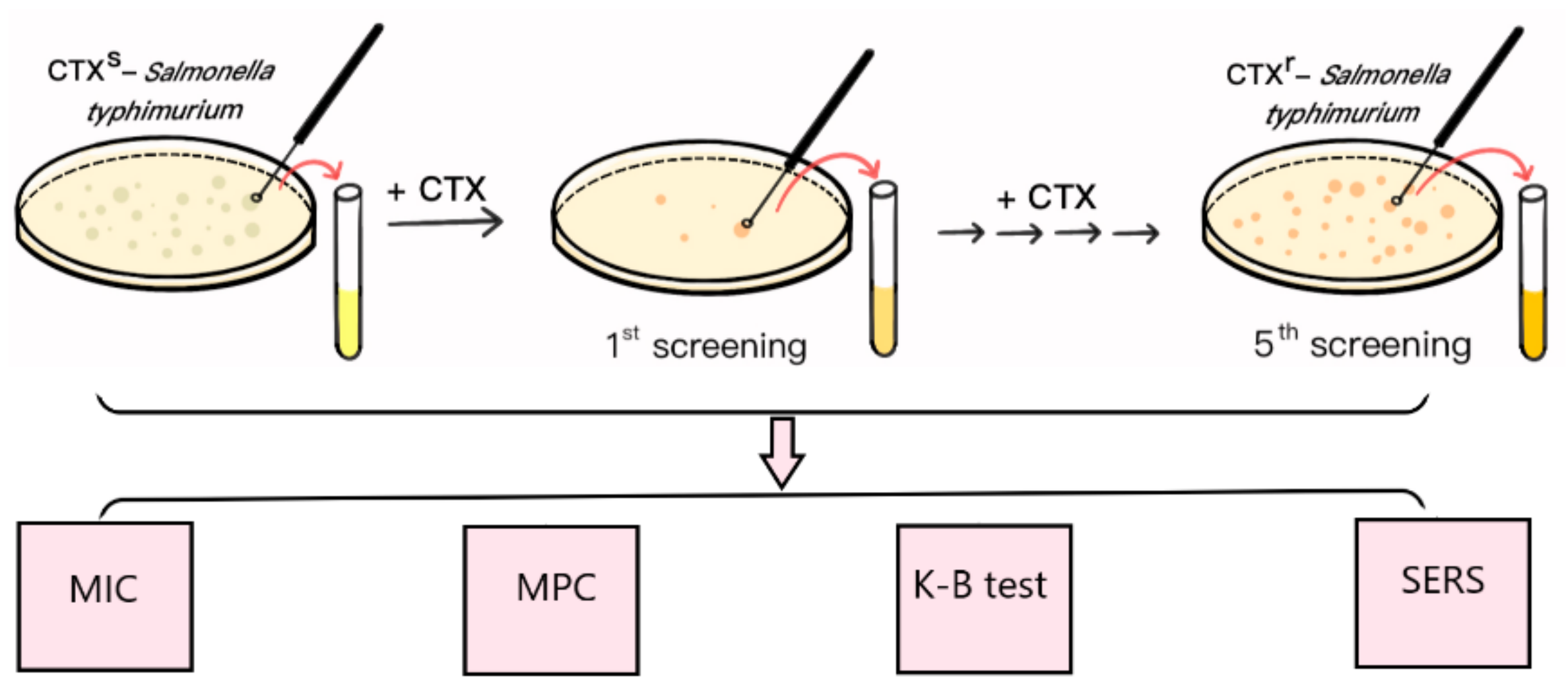
2.1. Mutation and Identification of Drug-Resistant Strains
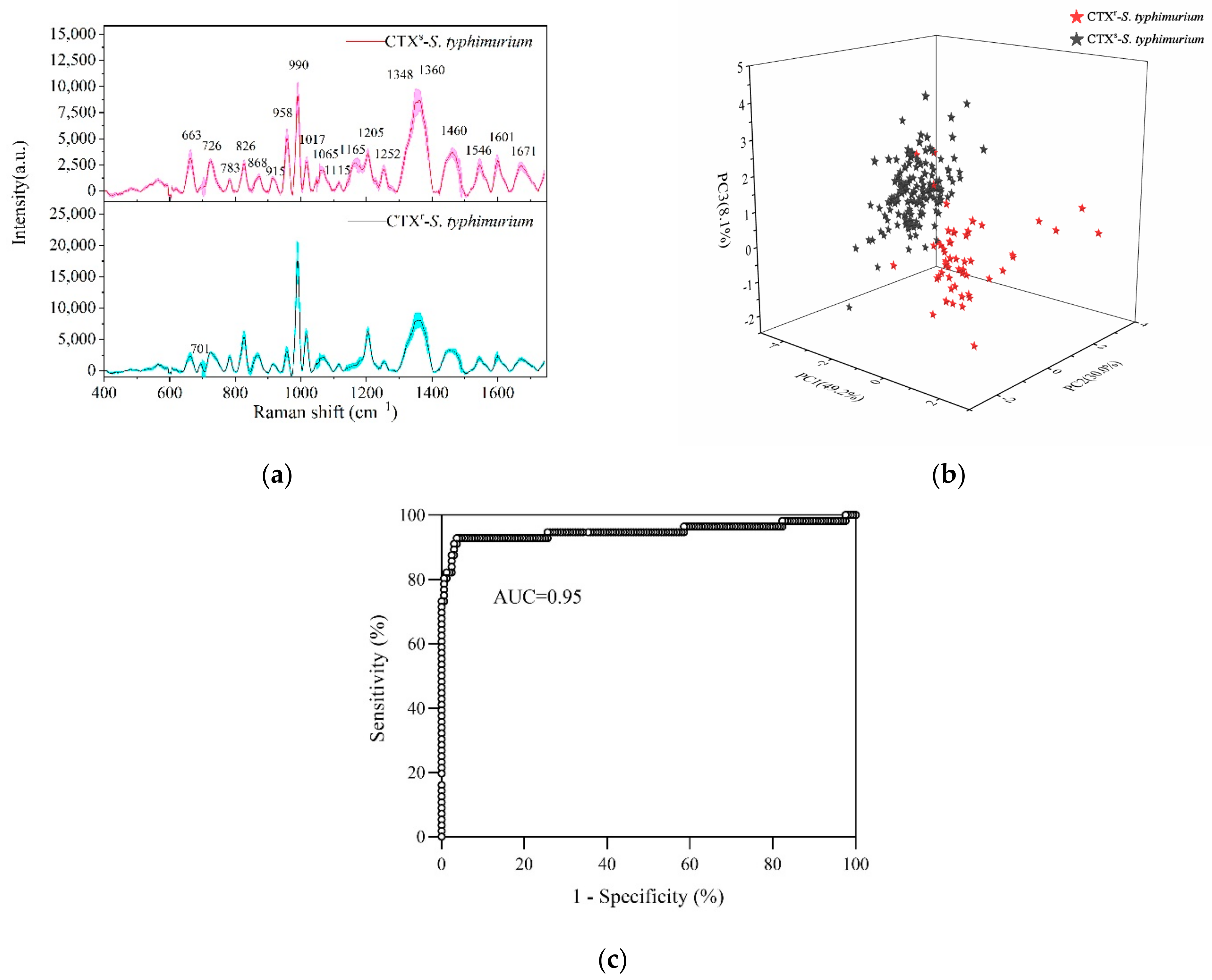
2.2. Rapid Detection and Discrimination of CTXs-S. typhimurium and CTXr-S. typhimurium by SERS
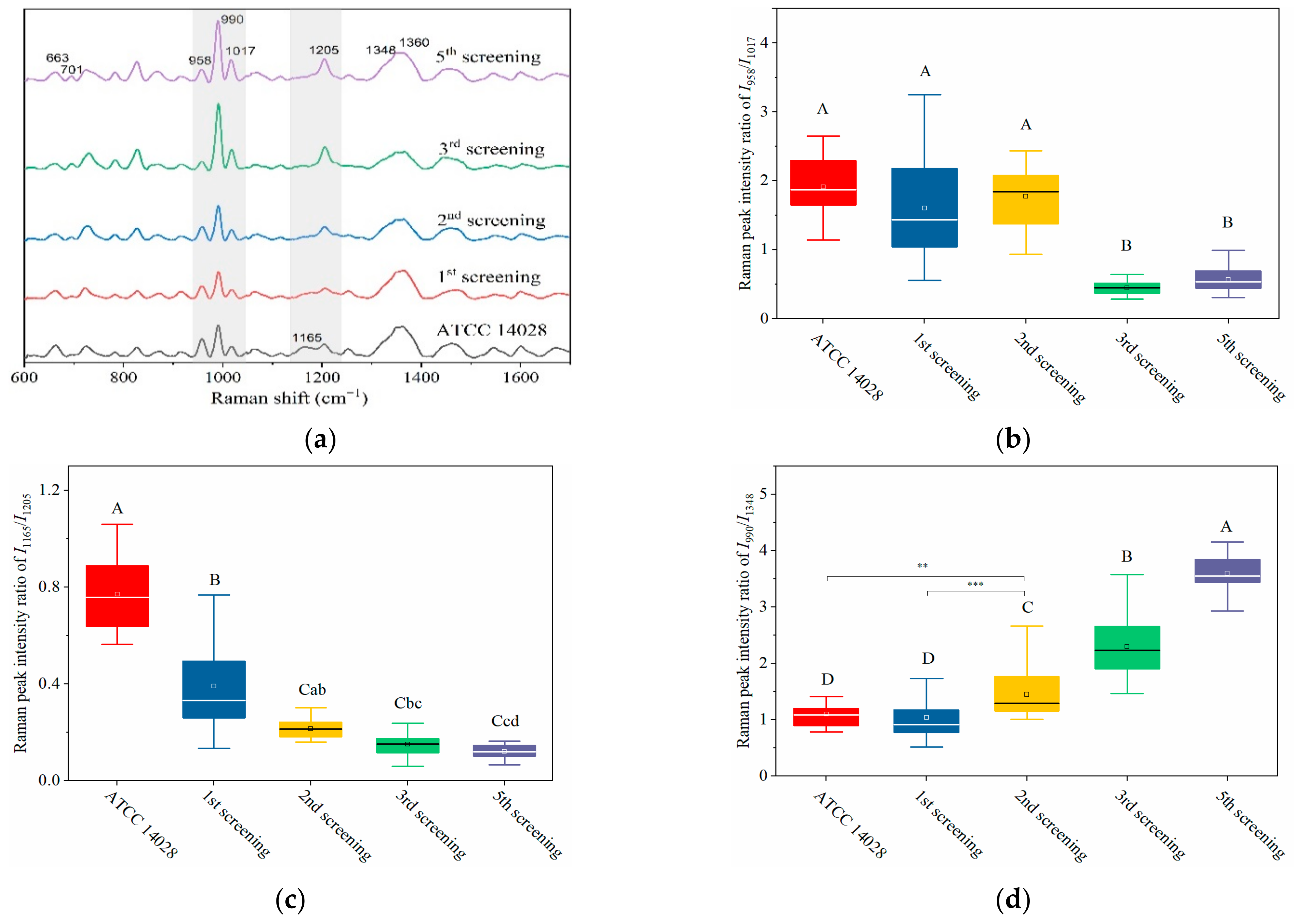
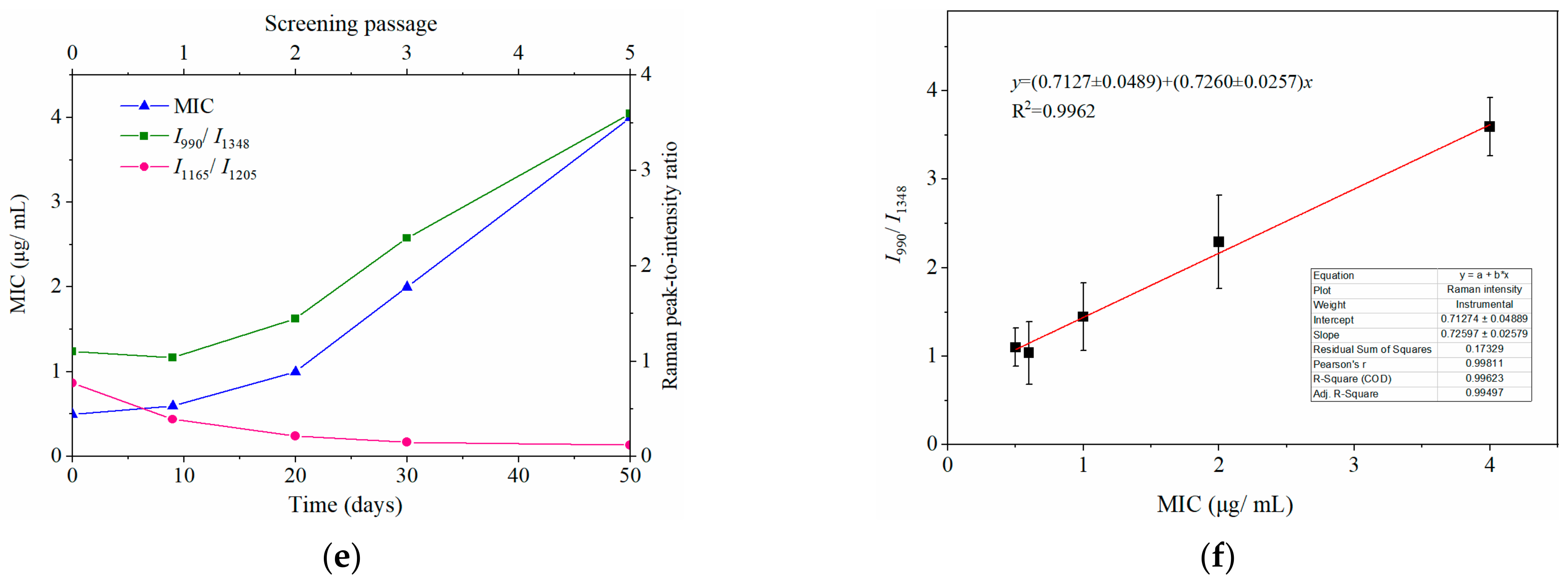
2.3. Dynamically Monitoring the Increasing Drug Resistance of S. typhimurium by SERS
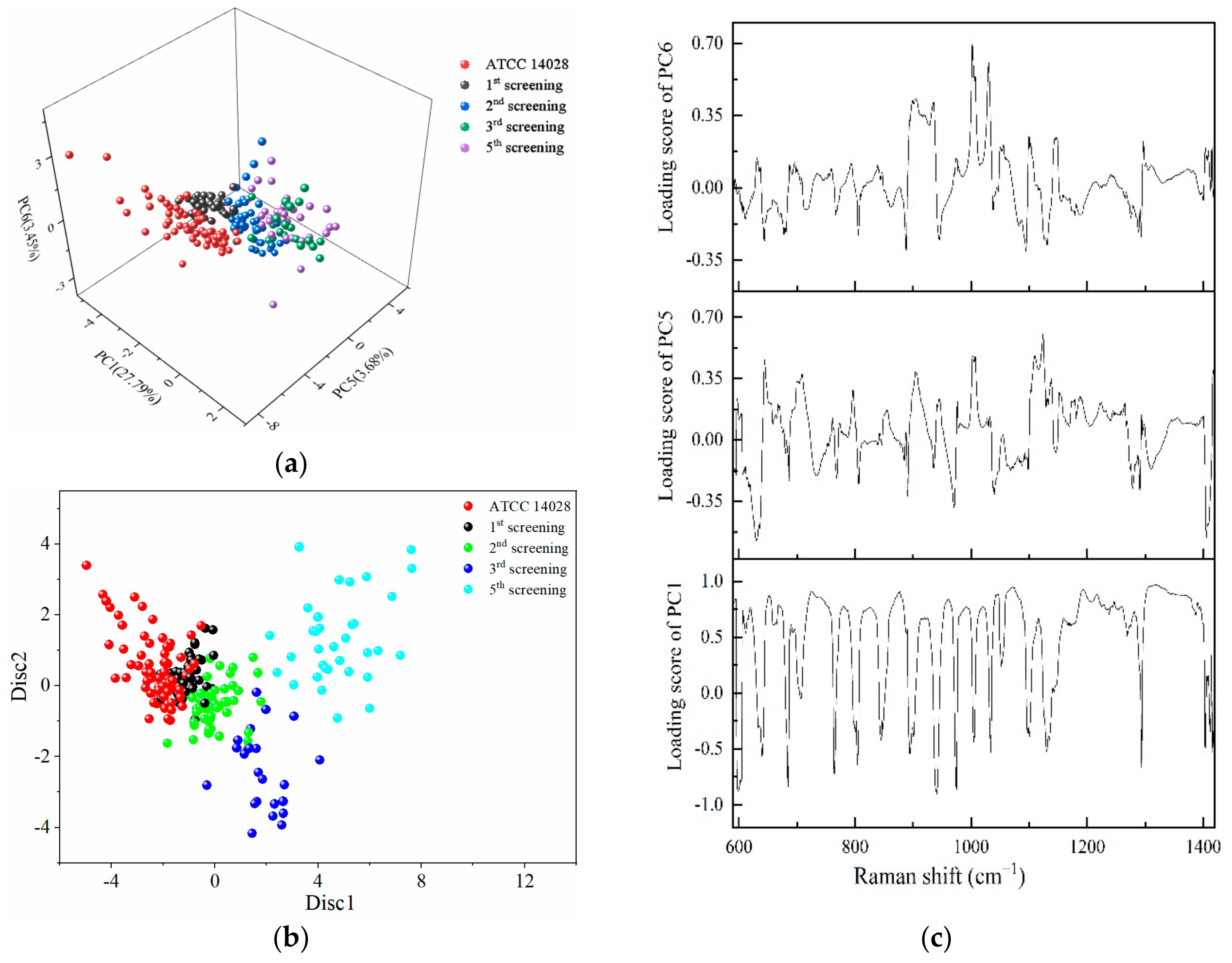
2.4. PCA-LDA for the Identification of Different Degrees of Drug-Resistant Strains
3. Materials and Methods
3.1. Bacteria and Antibiotics
3.2. Mutation and Screening of Drug-Resistant Strains
3.3. Identification of Drug-Resistant Strains
3.4. SERS Test and Data Processing
3.5. Transmission Electron Microscopy of the Combination of AuNPs and Bacterial Cells
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rossolini, G.M.; Arena, F.; Pecile, P.; Pollini, S. Update on the antibiotic resistance crisis. Curr. Opin. Pharmacol. 2014, 18, 56–60. [Google Scholar] [CrossRef] [PubMed]
- Andersson, D.I.; Hughes, D. Microbiological effects of sublethal levels of antibiotics. Nat. Rev. Microbiol. 2014, 12, 465–478. [Google Scholar] [CrossRef] [PubMed]
- Pehrsson, E.; Tsukayama, P.; Patel, S.; Mejía-Bautista, M.; Sosa-Soto, G.; Navarrete, K.M.; Calderon, M.; Cabrera, L.; Hoyos-Arango, W.; Bertoli, M.T.; et al. Interconnected microbiomes and resistomes in low-income human habitats. Nat. Cell Biol. 2016, 533, 212–216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiegand, I.; Hilpert, K.; Hancock, R.E.W. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef]
- Notingher, I.; Hench, L.L. Raman microspectroscopy: A noninvasive tool for studies of individual living cells in vitro. Expert Rev. Med. Devic. 2006, 3, 215–234. [Google Scholar] [CrossRef]
- Wang, Y.; Huang, W.E.; Cui, L.; Wagner, M. Single cell stable isotope probing in microbiology using Raman microspectroscopy. Curr. Opin. Biotechnol. 2016, 41, 34–42. [Google Scholar] [CrossRef] [Green Version]
- Lorenz, B.; Wichmann, C.; Stöckel, S.; Rösch, P.; Popp, J. Cultivation-Free Raman Spectroscopic Investigations of Bacteria. Trends Microbiol. 2017, 25, 413–424. [Google Scholar] [CrossRef]
- Germond, A.; Ichimura, T.; Horinouchi, T.; Fujita, H.; Furusawa, C.; Watanabe, T.M. Raman spectral signature reflects tran-scriptomic features of antibiotic resistance in Escherichia coli. Commun. Biol. 2018, 1, 85. [Google Scholar] [CrossRef]
- Nie, S.; Emory, S.R. Probing Single Molecules and Single Nanoparticles by Surface-Enhanced Raman Scattering. Science 1997, 275, 1102–1106. [Google Scholar] [CrossRef]
- Schlücker, S. Surface-Enhanced Raman Spectroscopy: Concepts and Chemical Applications. Angew. Chem. Int. Ed. 2014, 53, 4756–4795. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.S.; Jones, S.; Pramanik, A.; Ray, P.C. Nanoarchitecture Based SERS for Biomolecular Fingerprinting and Label-Free Disease Markers Diagnosis. Acc. Chem. Res. 2016, 49, 2725–2735. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zong, S.; Wu, L.; Zhu, D.; Cui, Y. SERS-Activated Platforms for Immunoassay: Probes, Encoding Methods, and Applications. Chem. Rev. 2017, 117, 7910–7963. [Google Scholar] [CrossRef] [PubMed]
- Hatzenpichler, R.; Krukenberg, V.; Spietz, R.; Jay, Z. Next-generation physiology approaches to study microbiome function at single cell level. Nat. Rev. Genet. 2020, 18, 241–256. [Google Scholar] [CrossRef]
- Zhou, X.; Hu, Z.; Yang, D.; Xie, S.; Jiang, Z.; Niessner, R.; Haisch, C.; Zhou, H.; Sun, P. Bacteria Detection: From Powerful SERS to Its Advanced Compatible Techniques. Adv. Sci. 2020, 7, 2001739. [Google Scholar] [CrossRef]
- Zhang, M.; Hong, W.; Abutaleb, N.S.; Li, J.; Dong, P.; Zong, C.; Wang, P.; Seleem, M.; Cheng, J. Rapid Determination of Antimicrobial Susceptibility by Stimulated Raman Scattering Imaging of D2O Metabolic Incorporation in a Single Bacterium. Adv. Sci. 2020, 7, 2001452. [Google Scholar] [CrossRef]
- Liu, T.Y.; Tsai, K.T.; Wang, H.H.; Chen, Y.; Chen, Y.H.; Chao, Y.C.; Chang, H.H.; Lin, C.H.; Wang, J.K.; Wang, Y.L. Functionalized arrays of Raman-enhancing nanoparticles for capture and culture-free analysis of bacteria in human blood. Nat. Commun. 2011, 2, 538. [Google Scholar] [CrossRef]
- Boardman, A.K.; Wong, W.S.; Premasiri, W.R.; Ziegler, L.D.; Lee, J.; Miljkovic, M.; Klapperich, C.M.; Sharon, A.; Sauer-Budge, A.F. Rapid Detection of Bacteria from Blood with Surface-Enhanced Raman Spectroscopy. Anal. Chem. 2016, 88, 8026–8035. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Wang, C.; Shi, L.; Shao, L.; Fu, P.; Wang, K.; Xiao, R.; Wang, S.; Gu, B. Rapid identification and antibiotic susceptibility test of pathogens in blood based on magnetic separation and surface-enhanced Raman scattering. Microchim. Acta 2019, 186, 475. [Google Scholar] [CrossRef]
- Premasiri, W.R.; Chen, Y.; Williamson, P.M.; Bandarage, D.C.; Pyles, C.; Ziegler, L.D. Rapid urinary tract infection diagnostics by surface-enhanced Raman spectroscopy (SERS): Identification and antibiotic susceptibilities. Anal. Bioanal. Chem. 2017, 409, 3043–3054. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Yang, D.; Ivleva, N.P.; Mircescu, N.E.; Niessner, R.; Haisch, C. SERS Detection of Bacteria in Water by in Situ Coating with Ag Nanoparticles. Anal. Chem. 2014, 86, 1525–1533. [Google Scholar] [CrossRef]
- Witkowska, E.; Korsak, D.; Kowalska, A.; Księżopolska-Gocalska, M.; Niedziółka-Jönsson, J.; Roźniecka, E.; Michałowicz, W.; Albrycht, P.; Podrażka, M.; Holyst, R.; et al. Surface-enhanced Raman spectroscopy introduced into the International Standard Organization (ISO) regulations as an alternative method for detection and identification of pathogens in the food industry. Anal. Bioanal. Chem. 2017, 409, 1555–1567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eichorst, S.A.; Strasser, F.; Woyke, T.; Schintlmeister, A.; Wagner, M.; Woebken, D. Advancements in the application of Na-noSIMS and Raman microspectroscopy to investigate the activity of microbial cells in soils. FEMS Microbiol. Ecol. 2015, 91, fiv106. [Google Scholar] [CrossRef] [Green Version]
- Bittel, M.; Cordella, C.B.Y.; Assaf, A.; Jouanneau, S.; Durand, M.J.; Thouand, G. Potential of Raman Spectroscopy To Monitor Arsenic Toxicity on Bacteria: Insights toward Multiparametric Bioassays. Environ. Sci. Technol. 2015, 49, 12324–12332. [Google Scholar] [CrossRef] [PubMed]
- Carey, P.R.; Heidari-Torkabadi, H. New techniques in antibiotic discovery and resistance: Raman spectroscopy. Ann. N. Y. Acad. Sci. 2015, 1354, 67–81. [Google Scholar] [CrossRef] [PubMed]
- Schröder, U.-C.; Kirchhoff, J.; Hübner, U.; Mayer, G.; Glaser, U.; Henkel, T.; Pfister, W.; Fritzsche, W.; Popp, J.; Neugebauer, U. On-chip spectroscopic assessment of microbial susceptibility to antibiotics within 3.5 hours. J. Biophotonics 2017, 10, 1547–1557. [Google Scholar] [CrossRef]
- Liu, C.-Y.; Han, Y.-Y.; Shih, P.-H.; Lian, W.-N.; Wang, H.-H.; Lin, C.-H.; Hsueh, P.-R.; Wang, J.-K.; Wang, Y.-L. Rapid bacterial antibiotic susceptibility test based on simple surface-enhanced Raman spectroscopic biomarkers. Sci. Rep. 2016, 6, 23375. [Google Scholar] [CrossRef] [Green Version]
- Tao, Y.; Wang, Y.; Huang, S.; Zhu, P.; Huang, W.E.; Ling, J.; Xu, J.; Zhu, X.; Su, J.Q.; Ren, B.; et al. Metabolic-activity-based assessment of antimicrobial effects by D2O-labeled single-cell Raman microspectroscopy. Anal. Chem. 2017, 89, 4108–4115. [Google Scholar] [CrossRef]
- Yang, K.; Li, H.-Z.; Zhu, X.; Su, J.-Q.; Ren, B.; Zhu, Y.-G.; Cui, L. Rapid Antibiotic Susceptibility Testing of Pathogenic Bacteria Using Heavy-Water-Labeled Single-Cell Raman Spectroscopy in Clinical Samples. Anal. Chem. 2019, 91, 6296–6303. [Google Scholar] [CrossRef]
- Ayala, O.D.; Wakeman, C.A.; Pence, I.J.; Gaddy, J.A.; Slaughter, J.C.; Skaar, E.P.; Mahadevan-Jansen, A. Drug-resistant Staphylococcus aureus strains reveal distinct biochemical features with Raman microspectroscopy. ACS Infect. Dis. 2018, 4, 1197–1210. [Google Scholar] [CrossRef]
- Ho, C.-S.; Jean, N.; Hogan, C.A.; Blackmon, L.; Jeffrey, S.S.; Holodniy, M.; Banaei, N.; Saleh, A.A.E.; Ermon, S.; Dionne, J. Rapid identification of pathogenic bacteria using Raman spectroscopy and deep learning. Nat. Commun. 2019, 10, 1–8. [Google Scholar] [CrossRef]
- Han, Y.-Y.; Lin, Y.-C.; Cheng, W.-C.; Lin, Y.-T.; Teng, L.-J.; Wang, J.-K.; Wang, Y.-L. Rapid antibiotic susceptibility testing of bacteria from patients’ blood via assaying bacterial metabolic response with surface-enhanced Raman spectroscopy. Sci. Rep. 2020, 10, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Teng, L.; Wang, X.; Wang, X.; Gou, H.; Ren, L.; Wang, T.; Wang, Y.; Ji, Y.; Huang, W.E.; Xu, J. Label-free, rapid and quantitative phenotyping of stress response in E. coli via ramanome. Sci. Rep. 2016, 6, 34359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Y.; Wang, X.; Ma, B.; Xu, J. Ramanome technology platform for label-free screening and sorting of microbial cell factories at single-cell resolution. Biotechnol. Adv. 2019, 37, 107388. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, J.; Kong, L.; Li, B.; Li, H.; Huang, W.E.; Zheng, C. Raman-activated sorting of antibiotic-resistant bacteria in human gut microbiota. Environ. Microbiol. 2020, 22, 2613–2624. [Google Scholar] [CrossRef] [PubMed]
- Senger, R.S.; Scherr, D. Resolving complex phenotypes with Raman spectroscopy and chemometrics. Curr. Opin. Biotechnol. 2020, 66, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Heidari Baladehi, M.; Hekmatara, M.; He, Y.; Bhaskar, Y.; Wang, Z.; Liu, L.; Ji, Y.; Xu, J. Culture-free identification and met-abolic profiling of microalgal single cells via ensemble learning of Ramanomes. Anal. Chem. 2021, 93, 8872–8880. [Google Scholar] [CrossRef]
- Wei, X.D.; Zheng, D.W.; Zhang, P.; Lin, T.F.; Wang, H.Q.; Zhu, Y.W. Surface-enhanced Raman scattering investigation of bovine serum albumin by Au nanoparticles with different sizes. J. Appl. Biomater. Funct. Mater. 2018, 16, 157–162. [Google Scholar]
- Sharma, B.; Frontiera, R.R.; Henry, A.-I.; Ringe, E.; Van Duyne, R.P. SERS: Materials, applications, and the future. Mater. Today 2012, 15, 16–25. [Google Scholar] [CrossRef]
- Wang, H.; Levin, C.S.; Halas, N.J. Nanosphere arrays with controlled sub-10-nm gaps as surface-enhanced Raman spectros-copy substrates. J. Am. Chem. Soc. 2005, 127, 14992–14993. [Google Scholar] [CrossRef]
- Premasiri, W.R.; Lee, J.C.; Sauer-Budge, A.; Théberge, R.; Costello, C.E.; Ziegler, L.D. The biochemical origins of the surface-enhanced Raman spectra of bacteria: A metabolomics profiling by SERS. Anal. Bioanal. Chem. 2016, 408, 4631–4647. [Google Scholar] [CrossRef] [Green Version]
- Mosier-Boss, P.A. Review on SERS of Bacteria. Biosensors 2017, 7, 51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mosier-Boss, P.A.; Sorensen, K.C.; George, R.D.; Sims, P.C.; Obraztsova, A. Surface enhanced Raman scattering of bacteria using capped and uncapped silver nanoparticles. Spectrochim. Acta A 2020, 242, 118742. [Google Scholar] [CrossRef]
- Tadesse, L.F.; Ho, C.S.; Chen, D.H.; Arami, H.; Banaei, N.; Gambhir, S.S.; Jeffrey, S.S.; Saleh, A.A.E.; Dionne, J. Plasmonic and electrostatic interactions enable uniformly enhanced liquid bacterial surface-enhanced Raman scattering (SERS). Nano Lett. 2020, 20, 7655–7661. [Google Scholar] [CrossRef]
- Xie, Y.; Xu, L.; Wang, Y.; Shao, J.; Wang, L.; Wang, H.; Qian, H.; Yao, W. Label-free detection of the foodborne pathogens of Enterobacteriaceae by surface-enhanced Raman spectroscopy. Anal. Methods 2013, 5, 946–952. [Google Scholar] [CrossRef]
- Chen, L.; Mungroo, N.; Daikuara, L.; Neethirajan, S. Label-free NIR-SERS discrimination and detection of foodborne bacteria by in situ synthesis of Ag colloids. J. Nanobiotechnology 2015, 13, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.-J.; Lei, Z.-C.; Li, J.; Zong, C.; Yang, C.J.; Ren, B. Label-Free Surface-Enhanced Raman Spectroscopy Detection of DNA with Single-Base Sensitivity. J. Am. Chem. Soc. 2015, 137, 5149–5154. [Google Scholar] [CrossRef]
- Sundaram, J.; Park, B.; Hinton, A.; Lawrence, K.C.; Kwon, Y. Detection and differentiation of Salmonella serotypes using surface enhanced Raman scattering (SERS) technique. J. Food Meas. Charact. 2013, 7, 1–12. [Google Scholar] [CrossRef]
- Krafft, C.; Neudert, L.; Simat, T.; Salzer, R. Near infrared Raman spectra of human brain lipids. Spectrochim. Acta. A Mol. Biomol. Spectrosc. 2005, 61, 1529–1535. [Google Scholar] [CrossRef] [PubMed]
- Stone, N.; Kendall, C.; Smith, J.; Crow, P.; Barr, H. Raman spectroscopy for identification of epithelial cancers. Faraday Discuss. 2004, 126, 141–157. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Samuelson, D.; Rasco, B.A.; Konkel, M.E. Antimicrobial effect of diallyl sulphide on Campylobacter jejuni biofilms. J. Antimicrob. Chemother. 2012, 67, 1915–1926. [Google Scholar] [CrossRef]
- Notingher, I.; Green, C.; Dyer, C.; Perkins, E.; Hopkins, N.; Lindsay, C.; Hench, L.L. Discrimination between ricin and sulphur mustard toxicity in vitro using Raman spectroscopy. J. R. Soc. Interface. 2004, 1, 79–90. [Google Scholar] [CrossRef] [Green Version]
- Neupane, G.P.; Kim, D.M.; Kim, S.H.; Lee, B.K. In vitro synergism of ciprofloxacin and cefotaxime against nalidixic acid-resistant Salmonella enterica serotypes Paratyphi A and Paratyphi, B. Antimicrob. Agents Chemother. 2010, 54, 3696–3701. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Ding, S.; Wang, G.; Xu, X.; Zhou, G. In situ characterization and analysis of Salmonella biofilm formation under meat processing environments using a combined microscopic and spectroscopic approach. Int. J. Food Microbiol. 2013, 167, 293–302. [Google Scholar] [CrossRef]
- Movasaghi, Z.; Rehman, S.; Rehman, I.U. Raman spectroscopy of biological tissues. Appl. Spectrosc. Rev. 2007, 42, 493–541. [Google Scholar] [CrossRef]
- Lv, X.; Ge, W.; Li, Q.; Wu, Y.; Jiang, H.; Wang, X. Rapid and Ultrasensitive Electrochemical Detection of Multidrug-Resistant Bacteria Based on Nanostructured Gold Coated ITO Electrode. ACS Appl. Mater. Interfaces 2014, 6, 11025–11031. [Google Scholar] [CrossRef] [PubMed]
- Schulz, H.; Baranska, M. Identification and quantification of valuable plant substances by IR and Raman spectroscopy. Vib. Spectrosc. 2007, 43, 13–25. [Google Scholar] [CrossRef]
- Chen, Y.-J.; Chen, Y.-Y.; Wang, K.-H.; Wang, C.-H.; Yang, C.-Y.; Chau, L.-K.; Wang, S.-C. Integration of a Thermoelectric Heating Unit with Ionic Wind-Induced Droplet Centrifugation Chip to Develop Miniaturized Concentration Device for Rapid Determination of Salmonella on Food Samples Using Antibody-Functionalized SERS Tags. Sensors 2020, 20, 7177. [Google Scholar] [CrossRef]
- Ruiz-Chica, A.J.; Medina, M.A.; Sanchez-Jimenez, F.; Ramirez, F.J. Characterization by Raman spectroscopy of conformational changes on guanine–cytosine and adenine–thymine oligonucleotides induced by aminooxy analogues of spermidine. J. Raman Spectrosc. 2004, 35, 93–100. [Google Scholar] [CrossRef]
- Nanda, S.S.; Kim, B.J.; Kim, K.W.; Nasir, T.; Park, J.; Yun, K.; Hembram, K.; Papaefthymiou, G.C.; Choi, J.Y.; Yi, D.K. A new device concept for bacterial sensing by Raman spectroscopy and voltage-gated monolayer graphene. Nanoscale 2019, 11, 8528–8537. [Google Scholar] [CrossRef] [PubMed]
- Hanlon, E.B.; Manoharan, R.; Koo, T.W.; Shafer, K.E.; Motz, J.T.; Fitzmaurice, M.; Kramer, J.R.; Itzkan, I.; Dasari, R.R.; Feld, M.S. Prospects for in vivo Raman spectroscopy. Phys. Med. Biol. 2000, 45, R1–59. [Google Scholar] [CrossRef] [Green Version]

| Raman Shift (cm−1) | Peak Assignments |
|---|---|
| 663 | δ(G) [5,18,44,45,46], υ(C-S) in Cys [5,8,44,45,47] |
| 701 | τ(C-C) in Tyr [45], Cholesterol, cholesterol ester [48] |
| 726 | A [5,8,18,44,46], N-acetyl-d-glucosamine (cell wall) [47], CoA/acetyl-CoA [18], C-S (protein), ρ(CH2) [49] |
| 783 | υ(PO2−) [5,45,50], C, A ring breathing [5,8,18,45,46,50,51] |
| 826 | υ(PO2-) [5,50], nucleic acids [8,50,52], Tyr [8], υ(C-C) in 1,4 glycosidic link [45] |
| 868 | υ(CN), υs(CON), δ(CCH) aliphatic [44], υ(CC) [47,53], υs(lipid) [5,8,44], υ(COC) [45,53], ribose [45] |
| 915 | υ(C-C) of Pro [50], Glucose, ribose vibration [54], Deoxyribose [18], C-O, C-OH, υ(C-COO−) (carbohydrates) [53] |
| 958 | δ(C=C) [44], υ(C-N) [18,45], υ(C-O) [18], |
| 990 | Phe [44,47], β-sheet [8] |
| 1017 | Phe [44] |
| 1048 | Carbohydrates [18,47], C-O [18,47], δ(C-OH) [18], polysaccharide [45] |
| 1065 | Phospholipids [18], Phe [55], fatty acid [48] |
| 1115 | Tyr, δ(NH3+) [44], δ(CH2,6) and C1-Cα-Hα bend [56] |
| 1165 | υ(C-C)/υ(C-N) of protein [5,8] |
| 1205 | Tyr [8,18], Phe, Try [5,8,18] (protein), υ(CN) [57], δ(H-C-C) [57] |
| 1252 | Amide II [5,8,18,44,48,50], υas(PO2−) [44,46], lipids [18], G, C (NH2) [58] |
| 1348 | A [5,8,46], G [5,8], Try [18,44]; δ(CH) [5,8,44,53], Amide III [48] |
| 1360 | δ(CH2) [44], Try [18,44] |
| 1460 | Amide II, δ(CH) (protein, DNA/RNA, lipid, carbohydrate) [5,8,18,44,47,53] |
| 1546 | υas(NO2) [8,44], υ(CH2), exopolysaccharide [44], υ(C=C) [53], Amide II [45,53] |
| 1601 | Amide I [18,50], Tyr [5,8,44,45] |
| 1671 | Amide I [5,8,44,45,53,55,59], ν(C=C) [60] |
| Screening Algebra | Prediction Group | Total | Sensitivity (%) | Specificity (%) | ||||
|---|---|---|---|---|---|---|---|---|
| ATCC 14028 | 1st Screening | 2nd Screening | 3rd Screening | 5th Screening | ||||
| ATCC 14028 | 66 | 3 | 6 | 0 | 0 | 75 | 88 | 96.1 |
| 1st screening | 6 | 40 | 6 | 0 | 0 | 52 | 77 | 98.3 |
| 2nd screening | 0 | 0 | 40 | 3 | 1 | 44 | 91 | 92.4 |
| 3rd screening | 0 | 0 | 2 | 19 | 0 | 21 | 90.5 | 97.1 |
| 5th screening | 0 | 0 | 0 | 3 | 33 | 36 | 91.7 | 99.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, P.; Wu, X.-H.; Su, L.; Wang, H.-Q.; Lin, T.-F.; Fang, Y.-P.; Zhao, H.-M.; Lu, W.-J.; Liu, M.-J.; Liu, W.-B.; et al. Rapid, Label-Free Prediction of Antibiotic Resistance in Salmonella typhimurium by Surface-Enhanced Raman Spectroscopy. Int. J. Mol. Sci. 2022, 23, 1356. https://doi.org/10.3390/ijms23031356
Zhang P, Wu X-H, Su L, Wang H-Q, Lin T-F, Fang Y-P, Zhao H-M, Lu W-J, Liu M-J, Liu W-B, et al. Rapid, Label-Free Prediction of Antibiotic Resistance in Salmonella typhimurium by Surface-Enhanced Raman Spectroscopy. International Journal of Molecular Sciences. 2022; 23(3):1356. https://doi.org/10.3390/ijms23031356
Chicago/Turabian StyleZhang, Ping, Xi-Hao Wu, Lan Su, Hui-Qin Wang, Tai-Feng Lin, Ya-Ping Fang, Hui-Min Zhao, Wen-Jing Lu, Meng-Jia Liu, Wen-Bo Liu, and et al. 2022. "Rapid, Label-Free Prediction of Antibiotic Resistance in Salmonella typhimurium by Surface-Enhanced Raman Spectroscopy" International Journal of Molecular Sciences 23, no. 3: 1356. https://doi.org/10.3390/ijms23031356
APA StyleZhang, P., Wu, X.-H., Su, L., Wang, H.-Q., Lin, T.-F., Fang, Y.-P., Zhao, H.-M., Lu, W.-J., Liu, M.-J., Liu, W.-B., & Zheng, D.-W. (2022). Rapid, Label-Free Prediction of Antibiotic Resistance in Salmonella typhimurium by Surface-Enhanced Raman Spectroscopy. International Journal of Molecular Sciences, 23(3), 1356. https://doi.org/10.3390/ijms23031356





