A New Family of Transcriptional Regulators Activating Biosynthetic Gene Clusters for Secondary Metabolites
Abstract
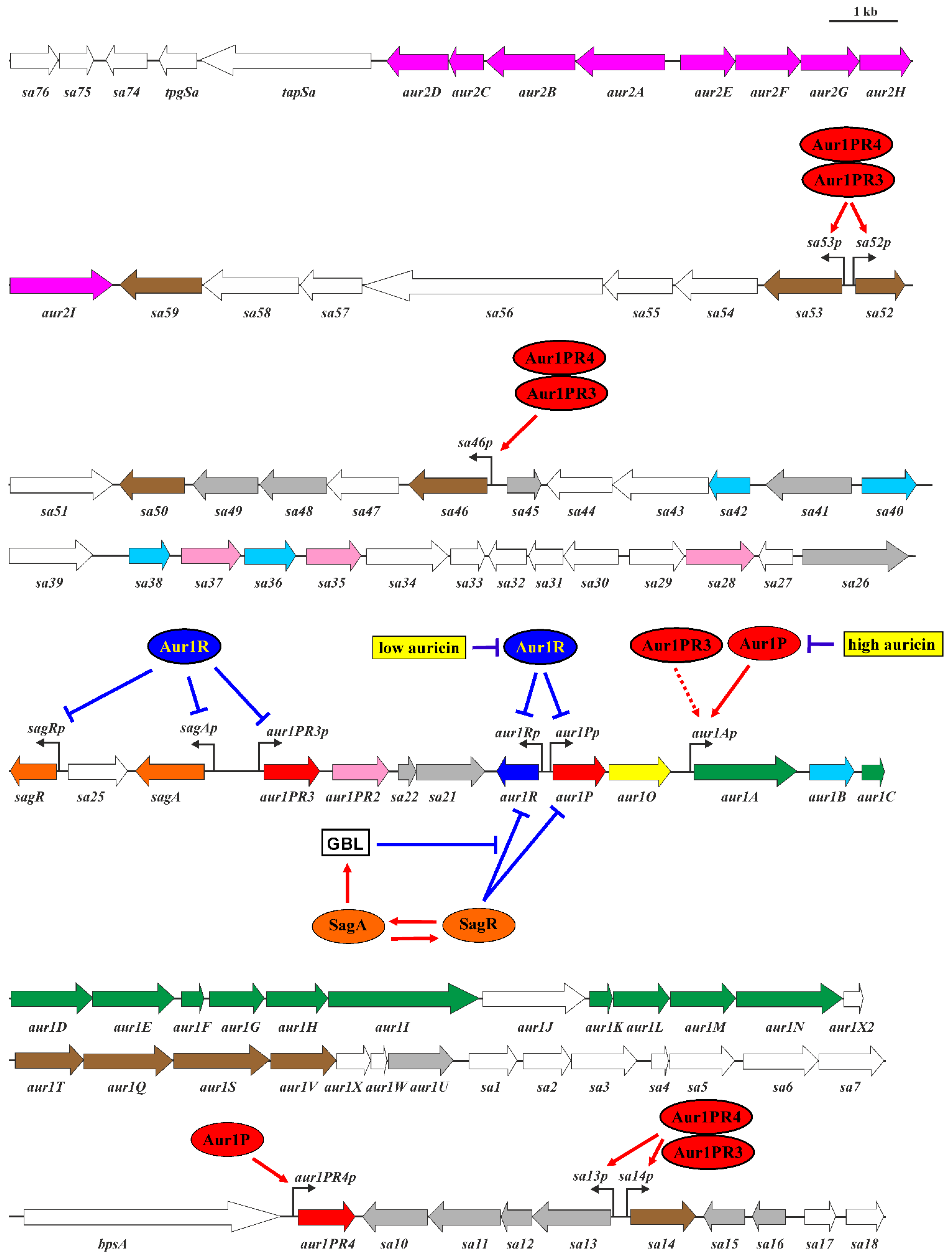
1. Introduction
2. Results and Discussion
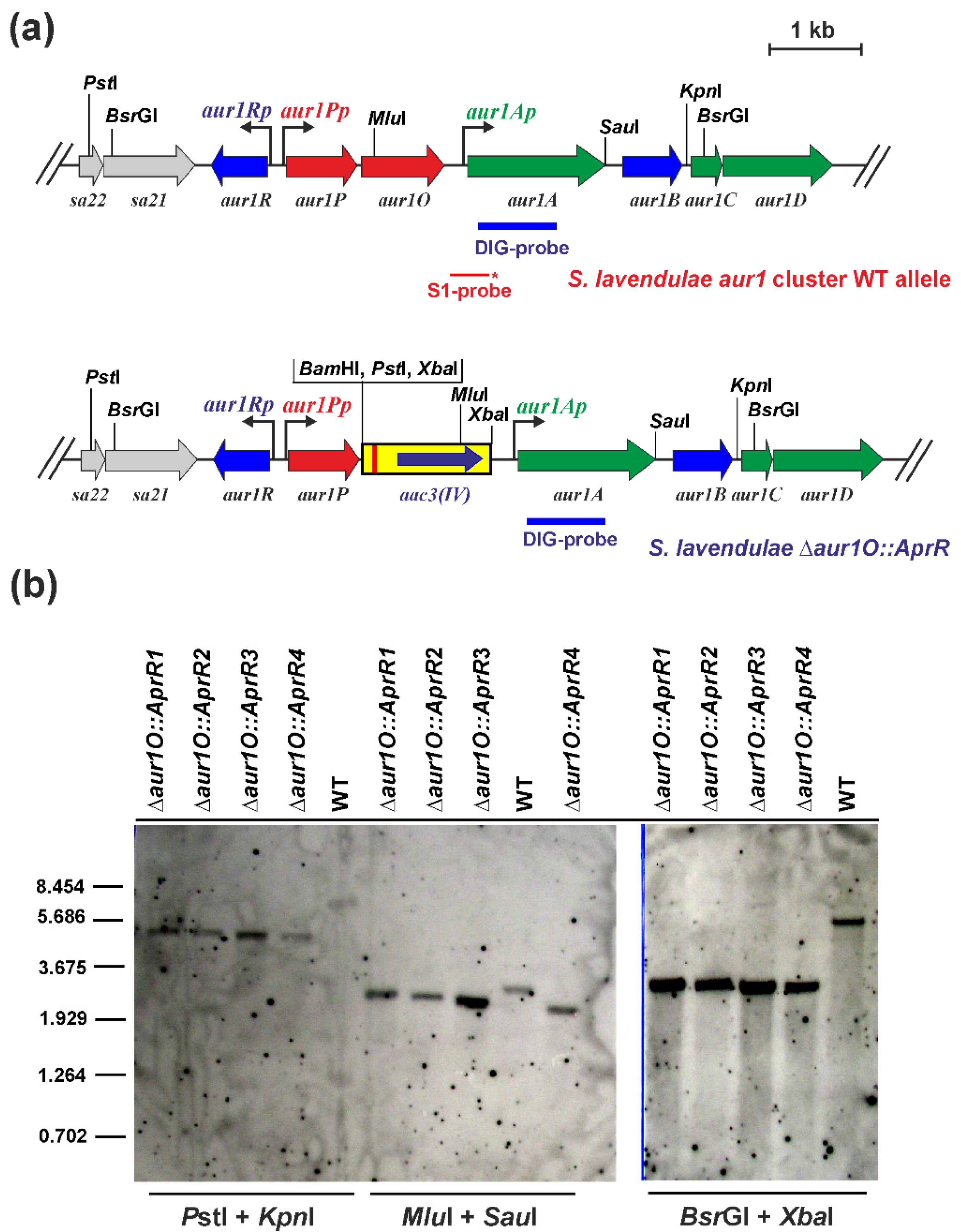
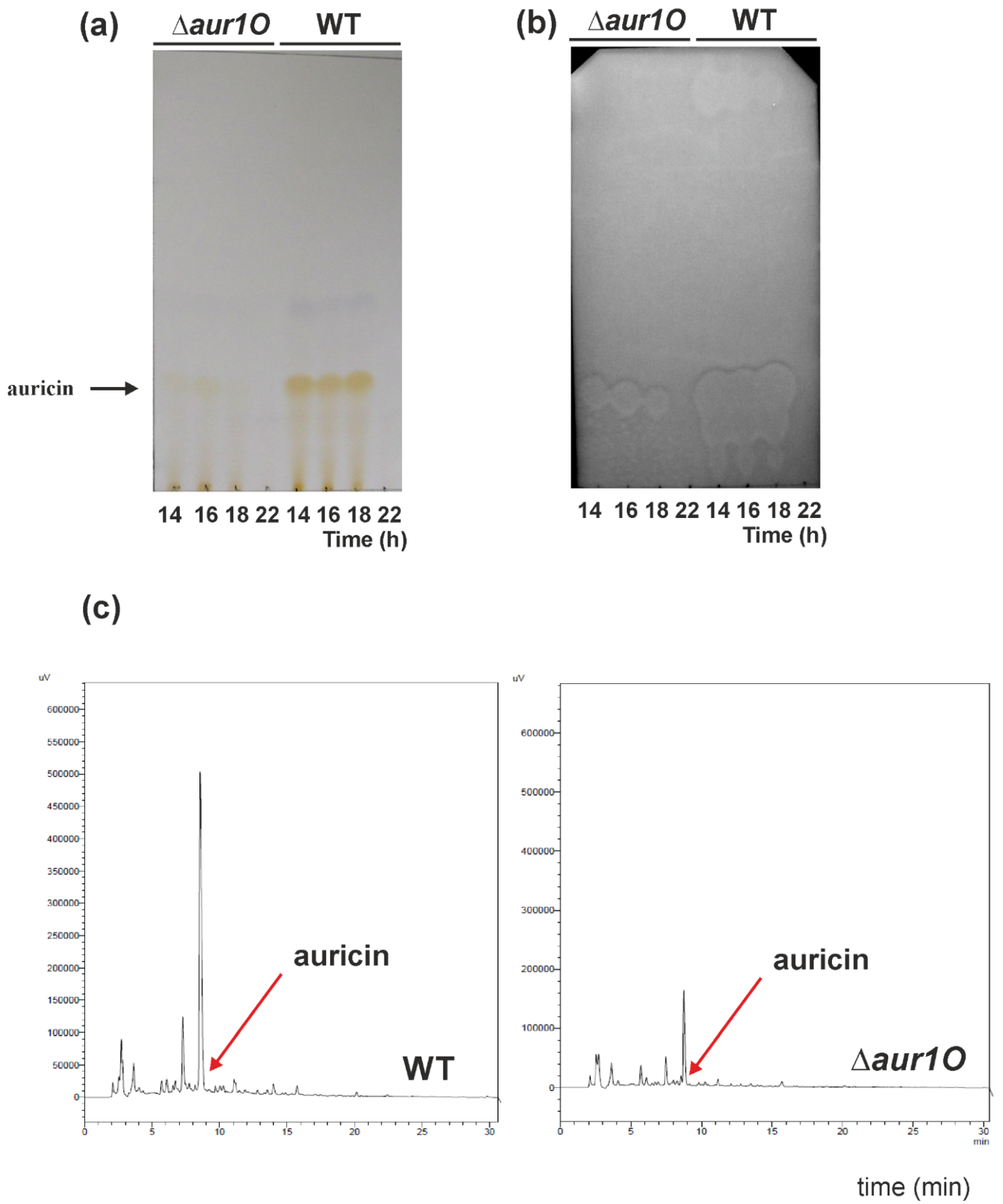
2.1. Characterization of the aur1O Gene in Auricin Biosynthesis
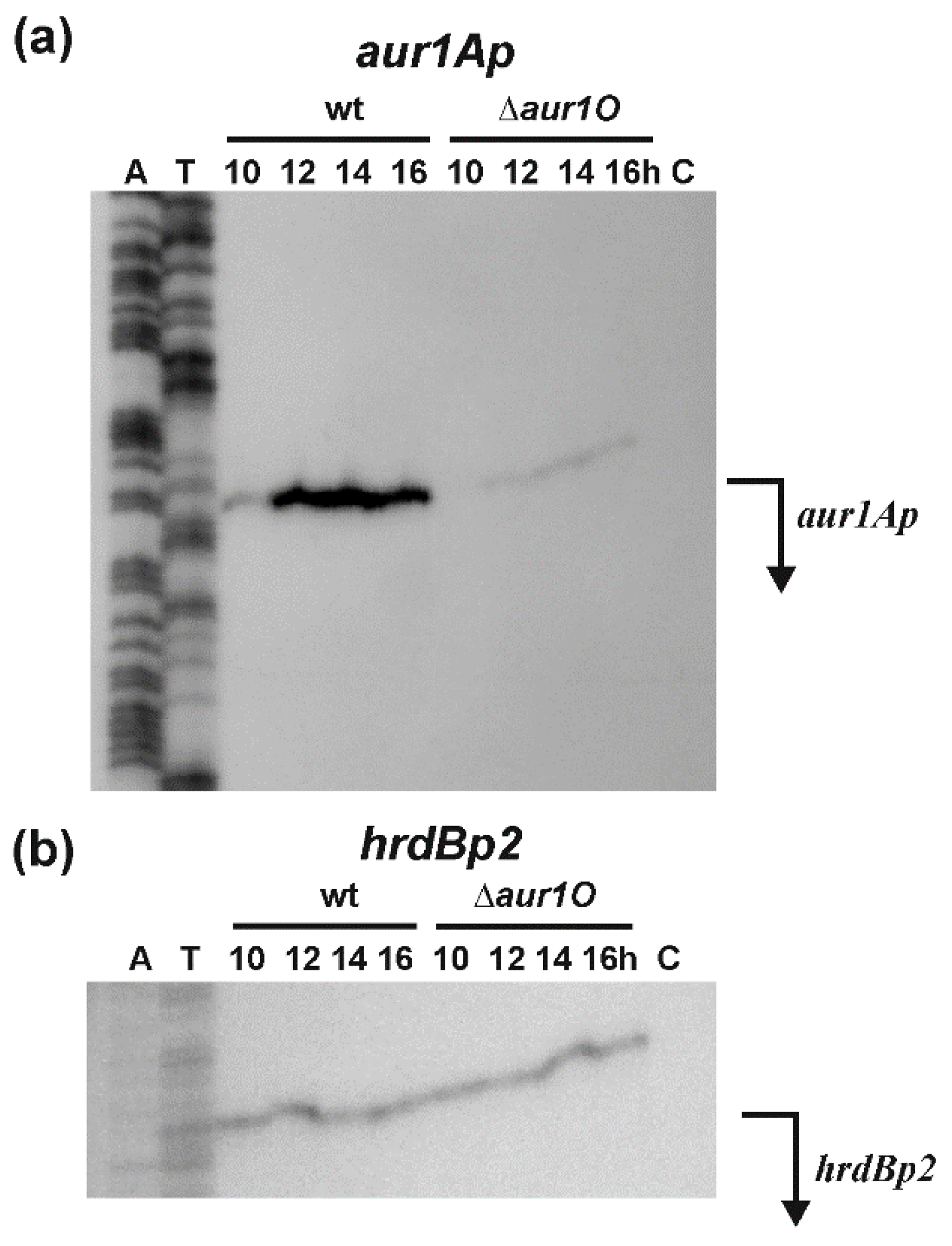
2.2. Transcriptional Analysis of the aur1Ap Promoter in the aur1O Mutant
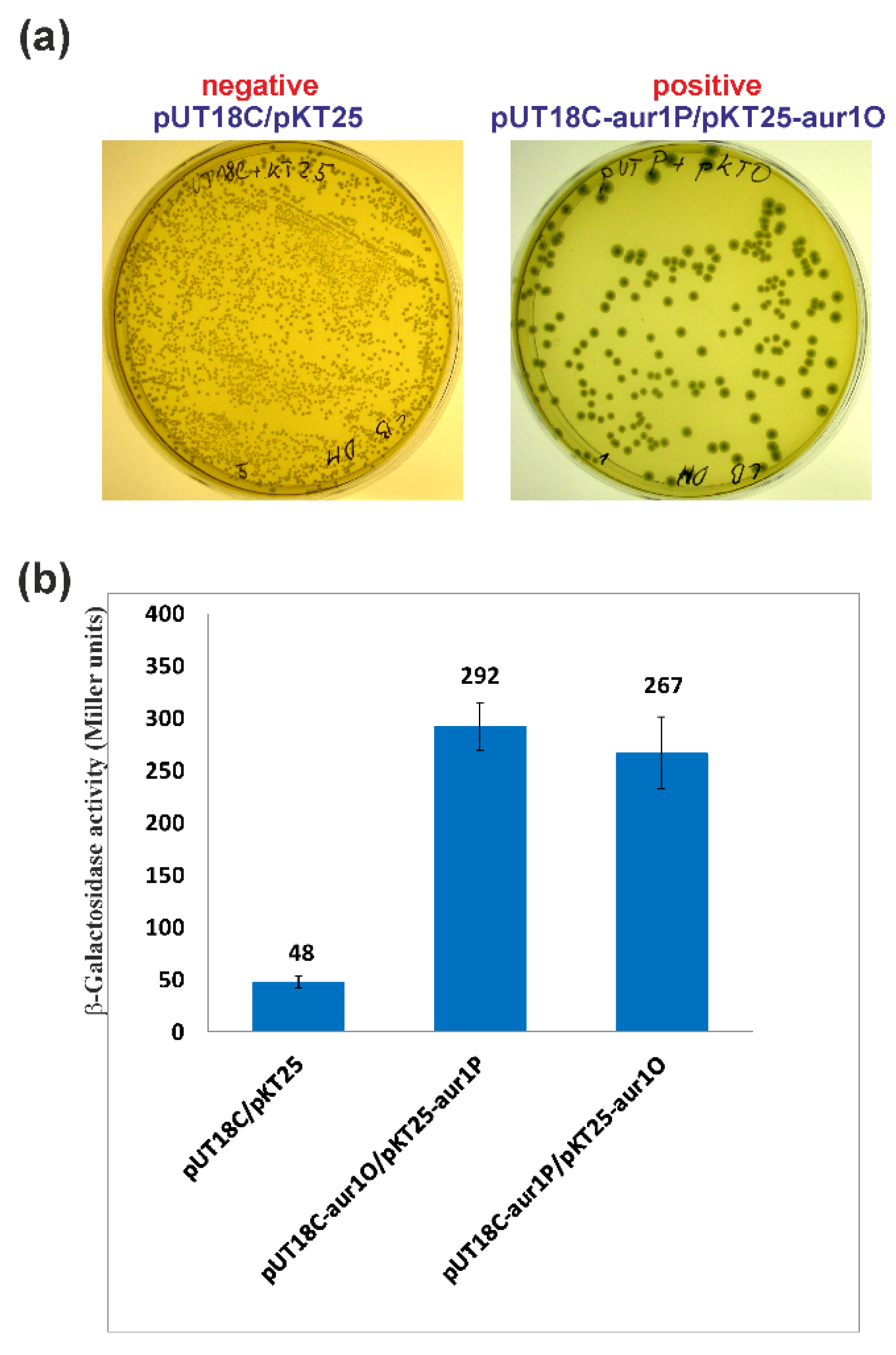
2.3. Activation of the aur1Ap Promoter by aur1P and aur1O in the Heterologous System
2.4. aur1O Interacts with aur1P
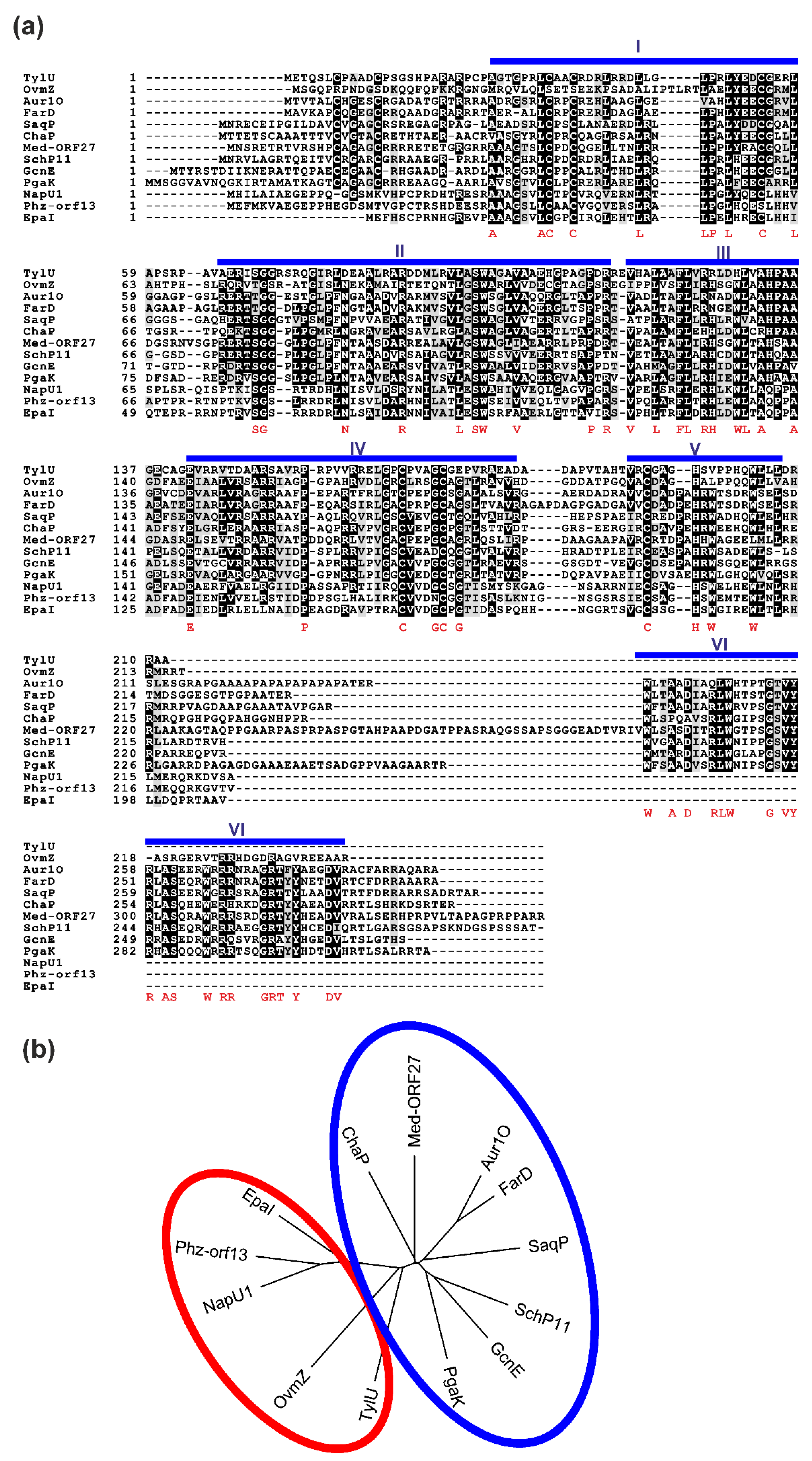
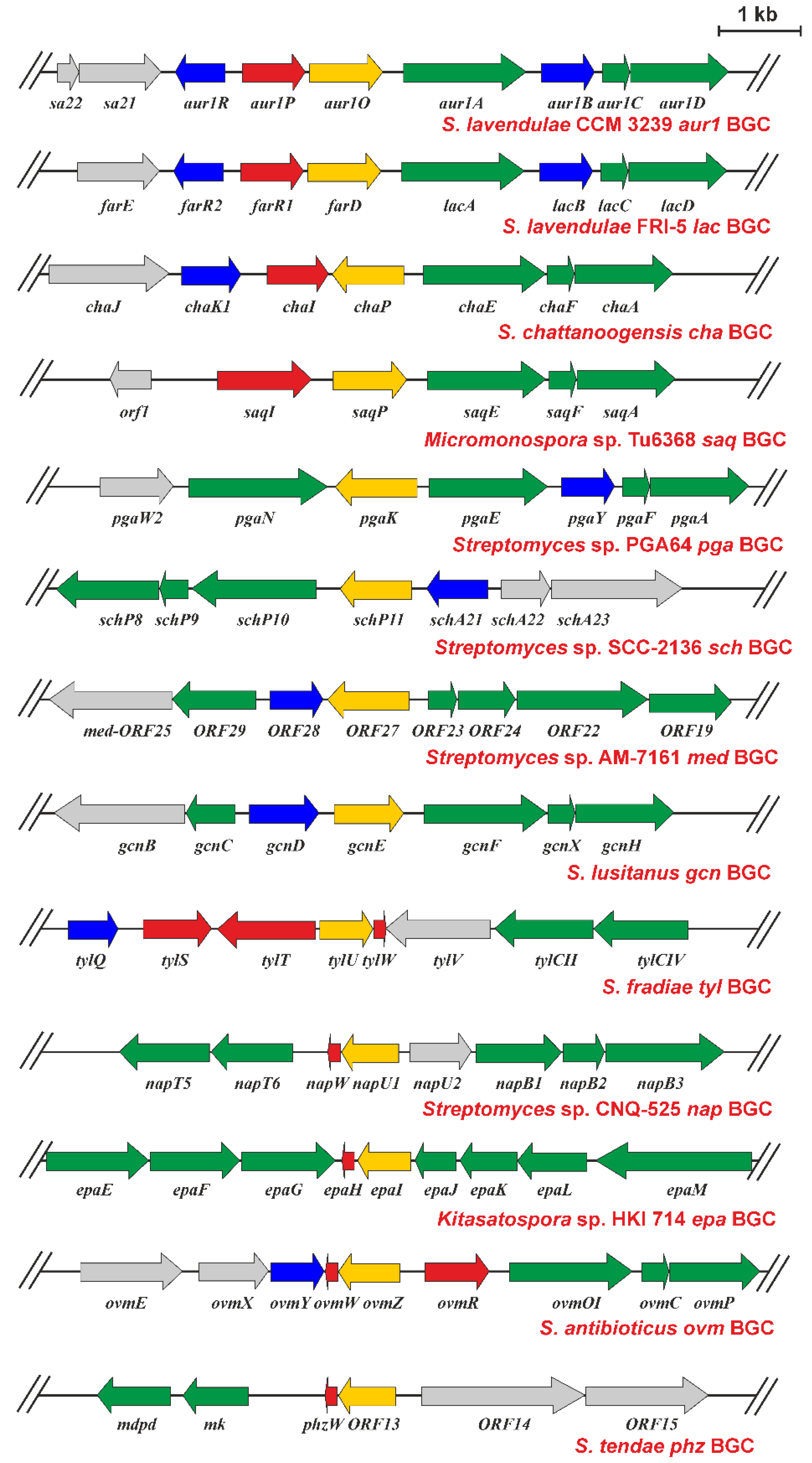
2.5. Presence of aur1O Homologues in Other BGCs
3. Material and Methods
3.1. Bacterial Strains, Plasmids, and Culture Conditions
3.2. Recombinant DNA Techniques
3.3. Disruption of the S. lavendulae subsp. lavendulae CCM 3239 aur1O Gene
3.4. RNA Isolation and S1-Nuclease Mapping
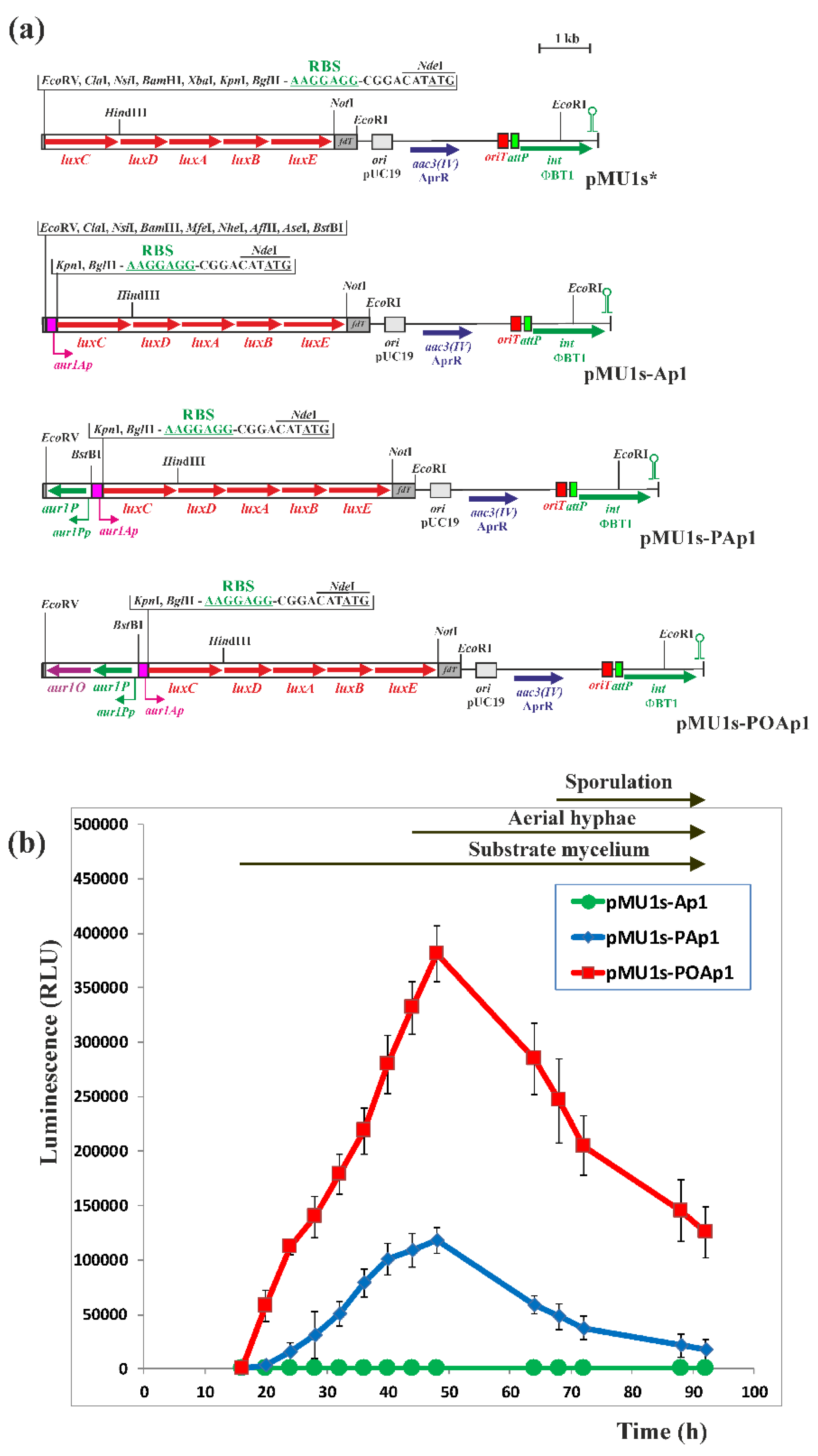
3.5. Construction of luxCDABE-Based Luciferase Reporter Plasmids and Bioluminescence Measurement
3.6. BACTH System to Investigate Protein-Protein Interactions
3.7. Analysis of Auricin Production
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Amp | ampicillin |
| Apr | apramycin |
| AprR | apramycin resistance |
| ARR | atypical response regulator |
| B | Bacillus |
| BACTH | bacterial two-hybrid |
| BGC | biosynthetic gene cluster |
| CCM | Czech Collection of Microorganisms |
| CFU | colony forming units |
| GBL | γ-butyrolactone |
| HPLC | high pressure liquid chromatography |
| HTH | helix-turn-helix |
| Kan | kanamycin |
| E | Escherichia |
| LB | Luria–Bertani (medium) |
| IPTG | isopropyl-β-d-thiogalactopyranoside |
| PCR | polymerase chain reaction |
| PKS | polyketide synthase |
| RLU | relative luminescence units |
| S | Streptomyces |
| SARP | Streptomyces antibiotic regulatory protein |
| TLC | thin layer chromatography |
| TSS | transcription start site |
| X-Gal | 5-bromo-4-chloro-3-indolyl-β-d-galactopyranosid |
References
- Bibb, M.J. Regulation of secondary metabolism in streptomycetes. Curr. Opin. Microbiol. 2005, 8, 208–215. [Google Scholar] [CrossRef]
- Hopwood, D.A. Streptomyces in Nature and Medicine. The Antibiotic Makers; Oxford University Press: New York, NY, USA, 2007. [Google Scholar]
- Wei, J.; He, L.; Niu, G. Regulation of antibiotic biosynthesis in actinomycetes: Perspectives and challenges. Synth. Syst. Biotechnol. 2018, 3, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.; Li, X.; Zhan, X.; Mao, X.; Li, Y. The application of regulatory cascades in Streptomyces: Yield enhancement and metabolite mining. Front. Microbiol. 2020, 11, 406. [Google Scholar] [CrossRef] [PubMed]
- van der Heul, H.U.; Bilyk, B.L.; McDowall, K.J.; Seipke, R.F.; van Wezel, G.P. Regulation of antibiotic production in Actino-615 bacteria: New persepctives from the post-genomic era. Nat. Prod. Rep. 2018, 35, 575–604. [Google Scholar] [CrossRef]
- Liu, G.; Chater, K.F.; Chandra, G.; Niu, G.; Tan, H. Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol. Mol. Biol. Rev. 2013, 77, 112–143. [Google Scholar] [CrossRef] [PubMed]
- Kong, D.; Wang, X.; Nie, J.; Niu, G. Regulation of Antibiotic Production by Signaling Molecules in Streptomyces. Front. Microbiol. 2019, 10, 2927. [Google Scholar] [CrossRef] [PubMed]
- Kormanec, J.; Novakova, R.; Mingyar, E.; Feckova, L. Intriguing properties of the angucycline antibiotic auricin and complex regulation of its biosynthesis. Appl. Microbiol. Biotechnol. 2014, 98, 45–60. [Google Scholar] [CrossRef]
- Novakova, R.; Bistakova, J.; Homerova, D.; Rezuchova, B.; Kormanec, J. Cloning and characterization of a polyketide synthase gene cluster involved in biosynthesis of a proposed angucycline-like polyketide auricin in Streptomyces aureofaciens CCM3239. Gene 2002, 297, 197–208. [Google Scholar] [CrossRef]
- Novakova, R.; Knirchova, R.; Farkasovsky, M.; Feckova, L.; Rehakova, A.; Mingyar, E.; Kormanec, J. The gene cluster aur1 for the angucycline antibiotic auricin is located on a large linear plasmid pSA3239 in Streptomyces aureofaciens CCM 3239. FEMS Microbiol. Lett. 2013, 342, 130–137. [Google Scholar] [CrossRef][Green Version]
- Busche, T.; Novakova, R.; Al’Dilaimi, A.; Homerova, D.; Feckova, L.; Rezuchova, B.; Mingyar, E.; Csolleiova, D.; Bekeova, C.; Winkler, A.; et al. Complete genome sequence of Streptomyces lavendulae subsp. lavendulae CCM 3239 (formerly “Streptomyces aureofaciens CCM 3239”), a producer of the angucycline-type antibiotic auricin. Genome Announc. 2018, 6, e00103-18. [Google Scholar]
- Bekeova, C.; Rehakova, A.; Feckova, L.; Vlckova, S.; Novakova, R.; Mingyar, E.; Kormanec, J. Characterisation of the genes involved in the biosynthesis and attachment of the aminodeoxysugar D-forosamine in the auricin gene cluster of Streptomyces aureofaciens CCM3239. Appl. Microbiol. Biotechnol. 2016, 100, 3177–3195. [Google Scholar] [CrossRef] [PubMed]
- Mingyar, E.; Novakova, R.; Knirschova, R.; Feckova, L.; Bekeova, C.; Kormanec, J. Unusual features of the large linear plasmid pSA3239 from Streptomyces aureofaciens CCM 3239. Gene 2018, 642, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Kutas, P.; Feckova, L.; Rehakova, A.; Novakova, R.; Homerova, D.; Mingyar, E.; Rezuchova, B.; Sevcikova, B. Strict control of auricin production in Streptomyces aureofaciens CCM 3239 involves a feedback mechanism. Appl. Microbiol. Biotechnol. 2013, 97, 2413–2421. [Google Scholar] [CrossRef] [PubMed]
- Matulova, M.; Feckova, L.; Novakova, R.; Mingyar, E.; Csolleiova, D.; Zduriencikova, M.; Sedlak, J.; Patoprsty, V.; Sasinkova, V.; Uhliarikova, I.; et al. A structural analysis of the angucycline-like antibiotic auricin from Streptomyces lavendulae subsp. lavendulae CCM 3239 revealed its high similarity to griseusins. Antibiotics 2019, 8, 102. [Google Scholar] [CrossRef]
- Novakova, R.; Homerova, D.; Feckova, L.; Kormanec, J. Characterization of a regulatory gene essential for the production of the angucycline-like polyketide antibiotic auricin in Streptomyces aureofaciens CCM 3239. Microbiology 2005, 151, 2693–2706. [Google Scholar] [CrossRef] [PubMed]
- Novakova, R.; Kutas, P.; Feckova, L.; Kormanec, J. The role of the TetR-family transcriptional regulator Aur1R in negative regulation of the auricin gene cluster in Streptomyces aureofaciens CCM 3239. Microbiology 2010, 156, 2374–2383. [Google Scholar] [CrossRef] [PubMed]
- Novakova, R.; Rehakova, A.; Kutas, P.; Feckova, L.; Kormanec, J. The role of two SARP-family transcriptional regulators in regulation of the auricin gene cluster in Streptomyces aureofaciens CCM 3239. Microbiology 2011, 157, 1629–1639. [Google Scholar] [CrossRef]
- Rehakova, A.; Novakova, R.; Feckova, L.; Mingyar, E.; Kormanec, J. A gene determining a new member of the SARP family contributes to transcription of genes for the synthesis of the angucycline polyketide auricin in Streptomyces aureofaciens CCM3239. FEMS Microbiol. Lett. 2013, 346, 45–55. [Google Scholar] [CrossRef]
- Mingyar, E.; Feckova, L.; Novakova, R.; Bekeova, C.; Kormanec, J. A γ-butyrolactone autoregulator-receptor system involved in the regulation of auricin production in Streptomyces aureofaciens CCM 3239. Appl. Microbiol. Biotechnol. 2015, 99, 309–325. [Google Scholar] [CrossRef] [PubMed]
- Gust, B.; Challis, G.L.; Fowler, K.; Kieser, T.; Chater, K.F. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. USA 2003, 18, 1541–1548. [Google Scholar] [CrossRef] [PubMed]
- Ausubel, F.M.; Brent, R.; Kingstone, R.E.; Moore, D.O.; Seidman, J.S.; Smith, J.A.; Struh, K. Current Protocols in Molecular Biology; Wiley: New York, NY, USA, 1995. [Google Scholar]
- Bibb, M.J.; White, J.; Ward, J.M.; Janssen, G.R. The mRNA for the 23S rRNA methylase encoded by the ermE gene of Saccharopolyspora erythraea is translated in the absence of a conventional ribosome-binding site. Mol. Microbiol. 1994, 14, 533–545. [Google Scholar] [CrossRef] [PubMed]
- Kormanec, J.; Farkasovsky, M. Differential expression of principal sigma factor homologues of Streptomyces aureofaciens correlates with the developmental stage. Nucleic Acids Res. 1993, 21, 3647–3652. [Google Scholar] [CrossRef]
- Maxam, A.M.; Gilbert, W. Sequencing end-labelled DNA with base specific chemical cleavages. Methods Enzymol. 1980, 65, 449–560. [Google Scholar]
- Craney, A.; Hohenauer, T.; Xu, Y.; Navani, N.K.; Li, Y.; Nodwell, J. A synthetic luxCDABE gene cluster optimized for expression in high-GC bacteria. Nucleic Acids Res. 2007, 35, e46. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Escribano, J.P.; Bibb, M.J. Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Microb. Biotechnol. 2011, 4, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Karimova, G.; Pidoux, J.; Ullmann, A.; Ladant, D.A. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. USA 1998, 95, 5752–5756. [Google Scholar] [CrossRef]
- Zhou, Z.; Xu, Q.; Bu, Q.; Guo, Y.; Liu, S.; Liu, Y.; Du, Y.; Li, Y. Genome mining-directed activation of a silent angucycline biosynthetic gene cluster in Streptomyces chattanoogensis. Chembiochem 2015, 16, 496–502. [Google Scholar] [CrossRef]
- Erb, A.; Luzhetskyy, A.; Hardter, U.; Bechthold, A. Cloning and sequencing of the biosynthetic gene cluster for saquayamycin Z and galtamycin B and the elucidation of the assembly of their saccharide chains. ChemBioChem 2009, 10, 1392–1401. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Xiang, S.; Li, L.; Wang, B.; Rajasarkka, J.; Grondahl-Yli-Hannuksela, K.; Ai, G.; Metsa-Ketela, M.; Yang, K. Targeted activation of silent natural product biosynthesis pathway by reporter-gided mutant selection. Metab. Engineer. 2015, 29, 134–142. [Google Scholar] [CrossRef]
- Basnet, D.B.; Oh, T.-J.; Vu, T.T.H.; Sthapit, B.; Liou, K.; Lee, H.C.; Yoo, J.-C.; Sohng, J.K. Angucyclines Sch 47554 and Sch 47555 from Streptomyces sp. SCC-2136: Cloning, sequencing, and characterization. Mol. Cells 2006, 22, 154–162. [Google Scholar]
- Zhang, Y.; Huang, H.; Chen, Q.; Luo, M.; Sun, A.; Song, Y.; Ma, J.; Ju, J. Identification of the Grincamycin gene cluster unveils divergent roles for GcnQ in different hosts, tailoring the L-rhodinose moiety. Org. Lett. 2013, 15, 3254–3257. [Google Scholar] [CrossRef] [PubMed]
- Lombo, F.; Brana, A.F.; Salas, J.A.; Mendez, C. Genetic Organization of the Biosynthetic Gene Cluster for the Antitumor Angucycline Oviedomycin in Streptomyces antibioticus ATCC 11891l. ChemBioChem 2004, 5, 1181–1187. [Google Scholar] [CrossRef]
- Pait, I.G.; Kitani, S.; Kurniawan, Y.N.; Asa, M.; Iwai, T.; Ikeda, J.; Nihira, T. Identification and characterization of lbpA, an indigoidine biosynthetic gene in the γ-butyrolactone signaling system of Streptomyces lavendulae FRI-5. J. Biosci. Bioeng. 2017, 124, 369–375. [Google Scholar] [CrossRef]
- Ichinose, K.; Ozawa, M.; Itou, K.; Kunieda, K.; Ebizuka, Y. Cloning, sequencing and heterologous expression of the medermycin biosynthetic gene cluster of Streptomyces sp. AM-7161: Towards comparative analysis of the benzoisochromanequinone gene clusters. Microbiology 2003, 149, 1633–1645. [Google Scholar] [CrossRef]
- Bate, N.; Bignell, D.R.D.; Cundliffe, E. Regulation of tylosin biosynthesis involving ‘SARP-helper’ activity. Mol. Microbiol. 2006, 62, 148–156. [Google Scholar] [CrossRef]
- Heine, D.; Martin, K.; Hertweck, C. Genomics-guided discovery of endophenazines from Kitasatospora sp. HKI 714. J. Nat. Prod. 2014, 77, 1083–1097. [Google Scholar] [CrossRef]
- Saleh, O.; Bonitz, T.; Flinspach, K.; Kulik, A.; Barkard, N.; Muhlenweg, A.; Vente, A.; Polnick, S.; Lammerhofer, M.; Gust, B.; et al. Activation of a silent phenazine biosynthetic gene cluster reveals a novel natural product and a new resistance mechanism against phenazines. Med. Chem. Commun. 2012, 3, 1009. [Google Scholar] [CrossRef]
- Winter, J.M.; Moffitt, M.C.; Zazopoulos, E.; McAlpine, J.B.; Dorrestein, P.C.; Moore, B.S. Molecular basis for chloronium-mediated meroterpene cyclization. J. Biol. Chem. 2007, 282, 16362–16368. [Google Scholar] [CrossRef]
- Wang, L.; Tian, X.; Wang, J.; Yang, H.; Fan, K.; Xu, G.; Yang, K.; Tan, H. Autoregulation of antibiotic biosynthesis by binding of the end product to an atypical response regulator. Proc. Nat. Acad. Sci. USA 2009, 106, 8617–8622. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zhang, J.; Zhuo, J.; Li, Y.; Tian, Y.; Tan, H. Activation and molecular mechanism of a cryptic oviedomycin biosynthetic gene cluster via the disruption of a global regulatory gene, adpA, in Streptomyces ansochromogenes. J. Biol. Chem. 2017, 292, 19708–19720. [Google Scholar] [CrossRef]
- Kitani, S.; Iida, A.; Izumi, T.; Maeda, A.; Yamada, Y.; Nihira, T. Identification of genes involved in the butyrolactone autoregulator cascade that modulates secondary metabolism in Streptomyces lavendulae FRI-5. Gene 2008, 425, 9–16. [Google Scholar] [CrossRef]
- Novakova, R.; Rehakova, A.; Feckova, L.; Kutas, P.; Knirschova, R.; Kormanec, J. Genetic manipulation of pathway regulation for overproduction of angucycline-like antibiotic auricin in Streptomyces aureofaciens CCM 3239. Folia Microbiol. 2011, 56, 278–282. [Google Scholar] [CrossRef]
- Kormanec, J.; Rezuchova, B.; Novakova, R. Screening systems for stable markerless genomic deletions/integrations in Streptomyces species. In Antimicrobial Therapies. Methods in Molecular Biology; Barreiro, C., Barredo, J.L., Eds.; Humana: New York, NY, USA, 2021; Volume 2296, pp. 91–141. [Google Scholar]
- Kormanec, J. Analyzing the developmental expression of sigma factors with S1-nuclease mapping. In Nuclease Methods and Protocols. Methods in Molecular Biology; Chein, C.H., Ed.; Humana Press: Totowa, NJ, USA, 2001; Volume 160, pp. 481–494. [Google Scholar]
- Miller, J.H. Experiments in Molecular Genetics; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 1972. [Google Scholar]
- Kieser, T.; Bibb, M.J.; Buttner, M.J.; Chater, K.F.; Hopwood, D.A. Practical Streptomyces Genetics; The John Innes Foundation: Norwich, UK, 2000. [Google Scholar]
- Martinez-Hackert, E.; Stock, A.M. Structural relationships in the OmpR family of winged-helix transcription factors. J. Mol. Biol. 1997, 269, 301–312. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Novakova, R.; Mingyar, E.; Feckova, L.; Homerova, D.; Csolleiova, D.; Rezuchova, B.; Sevcikova, B.; Javorova, R.; Kormanec, J. A New Family of Transcriptional Regulators Activating Biosynthetic Gene Clusters for Secondary Metabolites. Int. J. Mol. Sci. 2022, 23, 2455. https://doi.org/10.3390/ijms23052455
Novakova R, Mingyar E, Feckova L, Homerova D, Csolleiova D, Rezuchova B, Sevcikova B, Javorova R, Kormanec J. A New Family of Transcriptional Regulators Activating Biosynthetic Gene Clusters for Secondary Metabolites. International Journal of Molecular Sciences. 2022; 23(5):2455. https://doi.org/10.3390/ijms23052455
Chicago/Turabian StyleNovakova, Renata, Erik Mingyar, Lubomira Feckova, Dagmar Homerova, Dominika Csolleiova, Bronislava Rezuchova, Beatrica Sevcikova, Rachel Javorova, and Jan Kormanec. 2022. "A New Family of Transcriptional Regulators Activating Biosynthetic Gene Clusters for Secondary Metabolites" International Journal of Molecular Sciences 23, no. 5: 2455. https://doi.org/10.3390/ijms23052455
APA StyleNovakova, R., Mingyar, E., Feckova, L., Homerova, D., Csolleiova, D., Rezuchova, B., Sevcikova, B., Javorova, R., & Kormanec, J. (2022). A New Family of Transcriptional Regulators Activating Biosynthetic Gene Clusters for Secondary Metabolites. International Journal of Molecular Sciences, 23(5), 2455. https://doi.org/10.3390/ijms23052455






