Structural Insights into Human Adenovirus Type 4 Virus-Associated RNA I
Abstract
:1. Introduction
2. Results
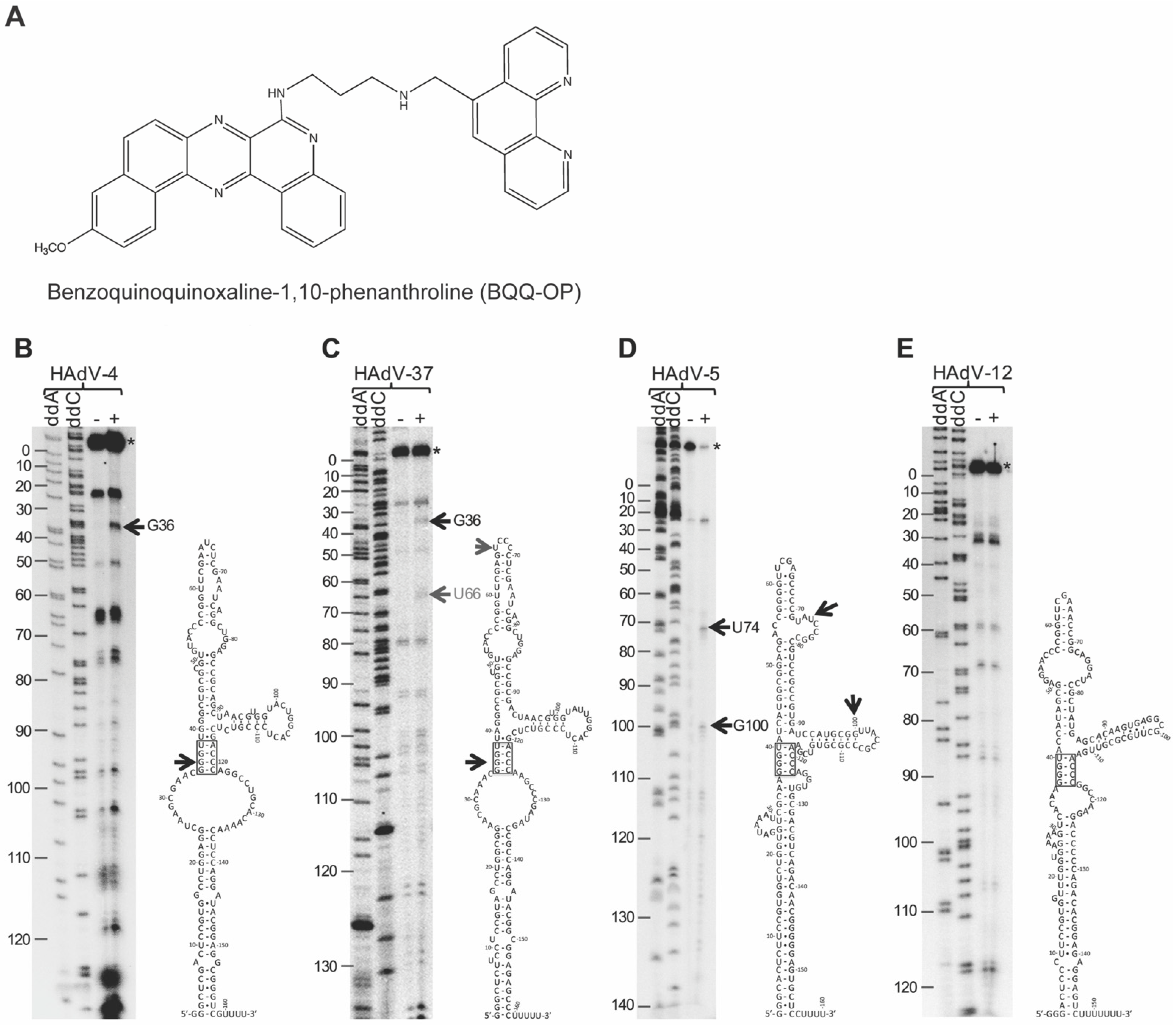
2.1. Differential Cleavage of HAdV-4, HAdV-5, HAdV-12, and HAdV-37 VA RNAI by BQQ–OP
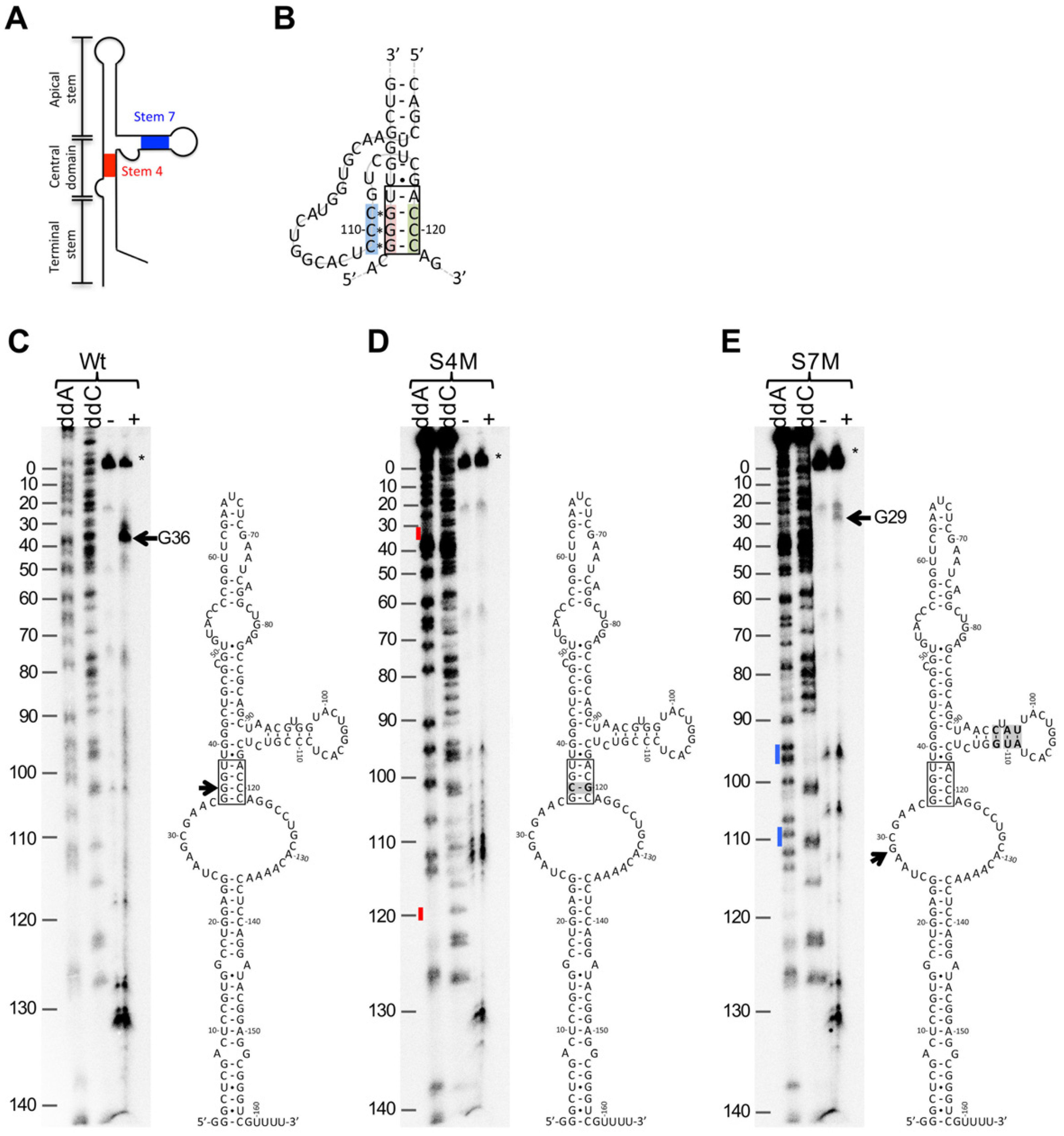
2.2. BQQ–OP Cleavage Specificity of the HAdV-4 VA-RNAI
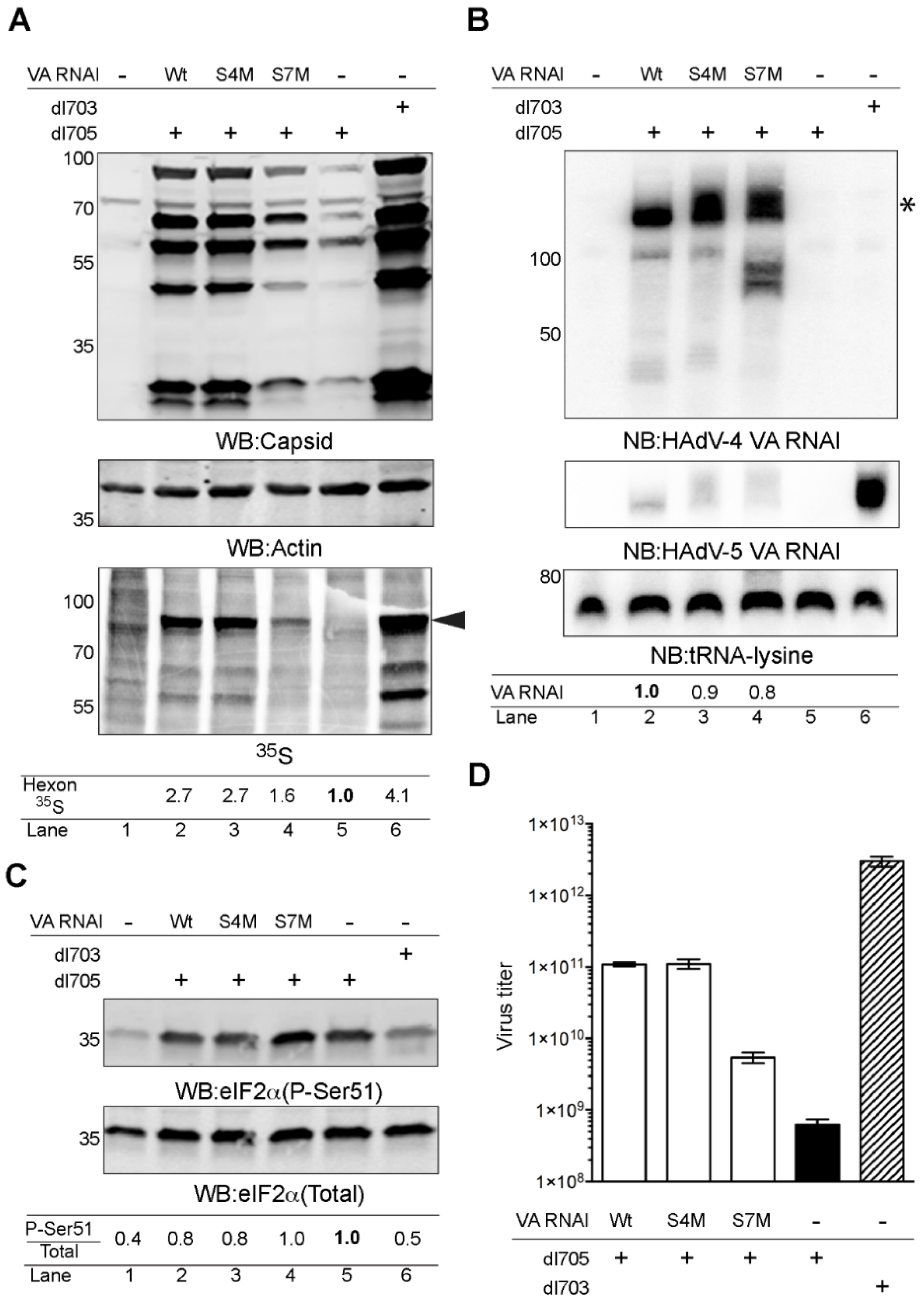
2.3. HAdV-4 VA RNAI (S7M) Is Partially Deficient in Complementing VA RNAI Lacking Virus Replication
3. Discussion
4. Materials and Methods
4.1. Plasmids and In Vitro Transcription Reaction
4.2. 3′-End Labelling of VA RNAI
4.3. Sanger Sequencing
4.4. BQQ–OP Triplex-Specific Cleavage
4.5. Cell Culture, Transient Transfection, Virus Infection, and Virus Titration
4.6. Metabolic Labelling of Proteins and Western Blot
4.7. Total RNA Isolation and Northern Blot
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Morris, K.V.; Mattick, J.S. The rise of regulatory RNA. Nat. Rev. Genet. 2014, 15, 423–437. [Google Scholar] [CrossRef] [Green Version]
- Batey, R.T.; Rambo, R.P.; Doudna, J.A. Tertiary Motifs in RNA Structure and Folding. Angew. Chem. Int. Ed. Engl. 1999, 38, 2326–2343. [Google Scholar] [CrossRef]
- Butcher, S.E.; Pyle, A.M. The molecular interactions that stabilize RNA tertiary structure: RNA motifs, patterns, and networks. Acc. Chem. Res. 2011, 44, 1302–1311. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.A. Unraveling the structure and biological functions of RNA triple helices. Wiley Interdiscip. Rev. RNA 2020, 11, e1598. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.A.; Kinzig, C.G.; DeGregorio, S.J.; Steitz, J.A. Hoogsteen-position pyrimidines promote the stability and function of the MALAT1 RNA triple helix. RNA 2016, 22, 743–749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fox, K.R.; Brown, T. Formation of stable DNA triplexes. Biochem. Soc. Trans. 2011, 39, 629–634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Escude, C.; Nguyen, C.H.; Kukreti, S.; Janin, Y.; Sun, J.S.; Bisagni, E.; Garestier, T.; Helene, C. Rational design of a triple helix-specific intercalating ligand. Proc. Natl. Acad. Sci. USA 1998, 95, 3591–3596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaid, A.; Sun, J.S.; Nguyen, C.H.; Bisagni, E.; Garestier, T.; Grierson, D.S.; Zain, R. Triple-helix directed cleavage of double-stranded DNA by benzoquinoquinoxaline-1,10-phenanthroline conjugates. Chembiochem Eur. J. Chem. Biol. 2004, 5, 1550–1557. [Google Scholar] [CrossRef]
- Zain, R.; Marchand, C.; Sun, J.; Nguyen, C.H.; Bisagni, E.; Garestier, T.; Helene, C. Design of a triple-helix-specific cleaving reagent. Chem. Biol. 1999, 6, 771–777. [Google Scholar] [CrossRef] [Green Version]
- Zain, R.; Polverari, D.; Nguyen, C.H.; Blouquit, Y.; Bisagni, E.; Garestier, T.; Grierson, D.S.; Sun, J.S. Optimization of triple-helix-directed DNA cleavage by benzoquinoquinoxaline-ethylenediaminetetraacetic acid conjugates. Chembiochem 2003, 4, 856–862. [Google Scholar] [CrossRef]
- Bergquist, H.; Nikravesh, A.; Fernandez, R.D.; Larsson, V.; Nguyen, C.H.; Good, L.; Zain, R. Structure-specific recognition of Friedreich’s ataxia (GAA)n repeats by benzoquinoquinoxaline derivatives. Chembiochem Eur. J. Chem. Biol. 2009, 10, 2629–2637. [Google Scholar] [CrossRef]
- Amiri, H.; Nekhotiaeva, N.; Sun, J.S.; Nguyen, C.H.; Grierson, D.S.; Good, L.; Zain, R. Benzoquinoquinoxaline derivatives stabilize and cleave H-DNA and repress transcription downstream of a triplex-forming sequence. J. Mol. Biol. 2005, 351, 776–783. [Google Scholar] [CrossRef] [PubMed]
- Bergquist, H.; Rocha, C.S.; Alvarez-Asencio, R.; Nguyen, C.H.; Rutland, M.W.; Smith, C.I.; Good, L.; Nielsen, P.E.; Zain, R. Disruption of Higher Order DNA Structures in Friedreich’s Ataxia (GAA)n Repeats by PNA or LNA Targeting. PLoS ONE 2016, 11, e0165788. [Google Scholar] [CrossRef] [PubMed]
- Umek, T.; Sollander, K.; Bergquist, H.; Wengel, J.; Lundin, K.E.; Smith, C.I.E.; Zain, R. Oligonucleotide Binding to Non-B-DNA in MYC. Molecules 2019, 24, 1000. [Google Scholar] [CrossRef] [Green Version]
- Lion, T. Adenovirus infections in immunocompetent and immunocompromised patients. Clin. Microbiol. Rev. 2014, 27, 441–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Assadian, F.; Sandstrom, K.; Bondeson, K.; Laurell, G.; Lidian, A.; Svensson, C.; Akusjarvi, G.; Bergqvist, A.; Punga, T. Distribution and Molecular Characterization of Human Adenovirus and Epstein-Barr Virus Infections in Tonsillar Lymphocytes Isolated from Patients Diagnosed with Tonsillar Diseases. PLoS ONE 2016, 11, e0154814. [Google Scholar]
- Fu, Y.; Tang, Z.; Ye, Z.; Mo, S.; Tian, X.; Ni, K.; Ren, L.; Liu, E.; Zang, N. Human adenovirus type 7 infection causes a more severe disease than type 3. BMC Infect. Dis. 2019, 19, 36. [Google Scholar] [CrossRef]
- Rogers, A.E.; Lu, X.; Killerby, M.; Campbell, E.; Gallus, L.; Kamau, E.; Froh, I.B.; Nowak, G.; Erdman, D.D.; Sakthivel, S.K.; et al. Outbreak of Acute Respiratory Illness Associated with Adenovirus Type 4 at the U.S. Naval Academy, 2016. MSMR 2019, 26, 21–27. [Google Scholar] [PubMed]
- Ma, Y.; Mathews, M.B. Comparative analysis of the structure and function of adenovirus virus-associated RNAs. J. Virol. 1993, 67, 6605–6617. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Mathews, M.B. Secondary and tertiary structure in the central domain of adenovirus type 2 VA RNA I. RNA 1996, 2, 937–951. [Google Scholar] [PubMed]
- Vachon, V.K.; Conn, G.L. Adenovirus VA RNA: An essential pro-viral non-coding RNA. Virus Res. 2016, 212, 39–52. [Google Scholar] [CrossRef]
- Punga, T.; Darweesh, M.; Akusjarvi, G. Synthesis, Structure, and Function of Human Adenovirus Small Non-Coding RNAs. Viruses 2020, 12, 1182. [Google Scholar] [CrossRef]
- Bhat, R.A.; Thimmappaya, B. Adenovirus mutants with DNA sequence perturbations in the intragenic promoter of VAI RNA gene allow the enhanced transcription of VAII RNA gene in HeLa cells. Nucleic Acids Res. 1984, 12, 7377–7388. [Google Scholar] [CrossRef] [Green Version]
- Thimmappaya, B.; Weinberger, C.; Schneider, R.J.; Shenk, T. Adenovirus VAI RNA is required for efficient translation of viral mRNAs at late times after infection. Cell 1982, 31, 543–551. [Google Scholar] [CrossRef]
- Inturi, R.; Kamel, W.; Akusjarvi, G.; Punga, T. Complementation of the human adenovirus type 5 VA RNAI defect by the Vaccinia virus E3L protein and serotype-specific VA RNAIs. Virology 2015, 485, 25–35. [Google Scholar] [CrossRef] [Green Version]
- Katze, M.G.; DeCorato, D.; Safer, B.; Galabru, J.; Hovanessian, A.G. Adenovirus VAI RNA complexes with the 68 000 Mr protein kinase to regulate its autophosphorylation and activity. EMBO J. 1987, 6, 689–697. [Google Scholar] [CrossRef]
- Kitajewski, J.; Schneider, R.J.; Safer, B.; Munemitsu, S.M.; Samuel, C.E.; Thimmappaya, B.; Shenk, T. Adenovirus VAI RNA antagonizes the antiviral action of interferon by preventing activation of the interferon-induced eIF-2 alpha kinase. Cell 1986, 45, 195–200. [Google Scholar] [CrossRef]
- Ma, Y.; Mathews, M.B. Structure, function, and evolution of adenovirus-associated RNA: A phylogenetic approach. J. Virol. 1996, 70, 5083–5099. [Google Scholar] [CrossRef] [Green Version]
- Kamel, W.; Segerman, B.; Punga, T.; Akusjarvi, G. Small RNA sequence analysis of adenovirus VA RNA-derived miRNAs reveals an unexpected serotype-specific difference in structure and abundance. PLoS ONE 2014, 9, e105746. [Google Scholar] [CrossRef]
- Launer-Felty, K.; Wong, C.J.; Cole, J.L. Structural analysis of adenovirus VAI RNA defines the mechanism of inhibition of PKR. Biophys. J. 2015, 108, 748–757. [Google Scholar] [CrossRef] [Green Version]
- Wilson, J.L.; Vachon, V.K.; Sunita, S.; Schwartz, S.L.; Conn, G.L. Dissection of the adenoviral VA RNAI central domain structure reveals minimum requirements for RNA-mediated inhibition of PKR. J. Biol. Chem. 2014, 289, 23233–23245. [Google Scholar] [CrossRef] [Green Version]
- Hood, I.V.; Gordon, J.M.; Bou-Nader, C.; Henderson, F.E.; Bahmanjah, S.; Zhang, J. Crystal structure of an adenovirus virus-associated RNA. Nat. Commun. 2019, 10, 2871. [Google Scholar] [CrossRef]
- Machitani, M.; Yamaguchi, T.; Shimizu, K.; Sakurai, F.; Katayama, K.; Kawabata, K.; Mizuguchi, H. Adenovirus Vector-Derived VA-RNA-Mediated Innate Immune Responses. Pharmaceutics 2011, 3, 338–353. [Google Scholar] [CrossRef] [Green Version]
- Conrad, N.K. The emerging role of triple helices in RNA biology. Wiley Interdiscip. Rev. RNA 2014, 5, 15–29. [Google Scholar] [CrossRef] [Green Version]
- Mun, K.; Punga, T. Cellular Zinc Finger Protein 622 Hinders Human Adenovirus Lytic Growth and Limits Binding of the Viral pVII Protein to Virus DNA. J. Virol. 2019, 93, e01628-18. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.; Bai, L.; Li, Z.; Samuel, C.E.; Akusjarvi, G.; Svensson, C. Poor growth of human adenovirus-12 compared to adenovirus-2 correlates with a failure to impair PKR activation during the late phase of infection. Virology 2015, 475, 120–128. [Google Scholar] [CrossRef] [Green Version]
- Inturi, R.; Thaduri, S.; Punga, T. Adenovirus precursor pVII protein stability is regulated by its propeptide sequence. PLoS ONE 2013, 8, e80617. [Google Scholar]
- Kamel, W.; Segerman, B.; Oberg, D.; Punga, T.; Akusjarvi, G. The adenovirus VA RNA-derived miRNAs are not essential for lytic virus growth in tissue culture cells. Nucleic Acids Res. 2013, 41, 4802–4812. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bergquist, H.; Inturi, R.; Zain, R.; Punga, T. Structural Insights into Human Adenovirus Type 4 Virus-Associated RNA I. Int. J. Mol. Sci. 2022, 23, 3103. https://doi.org/10.3390/ijms23063103
Bergquist H, Inturi R, Zain R, Punga T. Structural Insights into Human Adenovirus Type 4 Virus-Associated RNA I. International Journal of Molecular Sciences. 2022; 23(6):3103. https://doi.org/10.3390/ijms23063103
Chicago/Turabian StyleBergquist, Helen, Raviteja Inturi, Rula Zain, and Tanel Punga. 2022. "Structural Insights into Human Adenovirus Type 4 Virus-Associated RNA I" International Journal of Molecular Sciences 23, no. 6: 3103. https://doi.org/10.3390/ijms23063103





