Interactions between miRNAs and Double-Strand Breaks DNA Repair Genes, Pursuing a Fine-Tuning of Repair
Abstract
:1. Introduction
2. MiRNA-Mediated Regulation of NHEJ
3. Ku70/80
4. DNA-PKcs
5. DCLRE1C, DNA Pol M and Pol L
6. XRCC4: DNA Ligase IV Complex and XLF
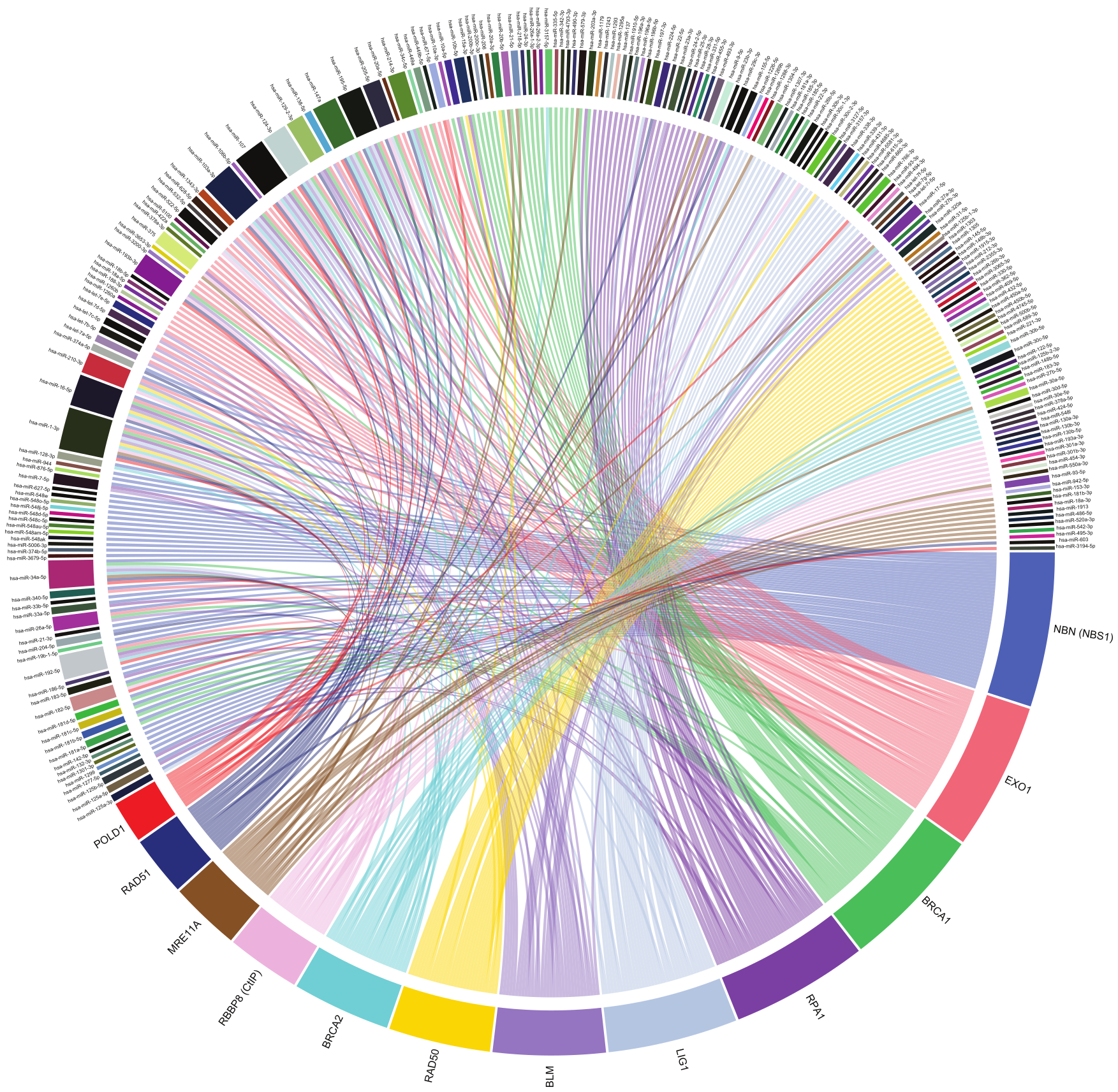
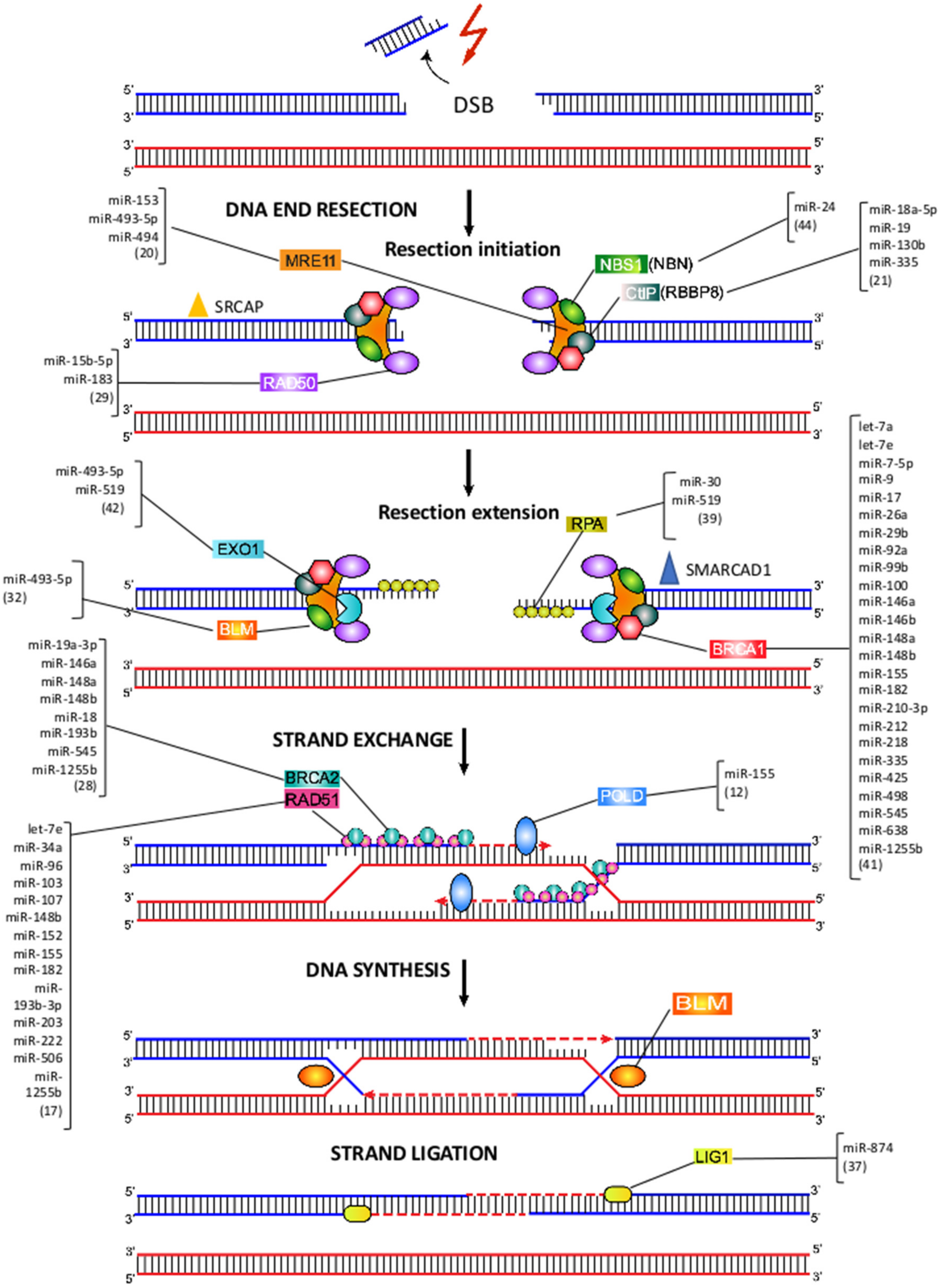
7. MiRNA-Mediated Regulation of HRR
8. MRE11-RAD50-NBS1 Complex
9. CtIP, EXO1 and RPA
10. BRCA1 and BRCA2
11. RAD51
12. Pol δ
13. BLM and LIG1
14. Other Validated MiRNA–MRNA Interactions
15. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Goldstein, M.; Kastan, M.B. The DNA damage response: Implications for tumor responses to radiation and chemotherapy. Annu. Rev. Med. 2015, 66, 129–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jackson, S.P.; Bartek, J. The DNA-damage response in human biology and disease. Nature 2009, 461, 1071–1078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lieber, M.R.; Wilson, T.E. Snap Shot: Nonhomologous DNA end joining (NHEJ). Cell 2010, 142, 496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lieber, M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010, 79, 181–211. [Google Scholar] [CrossRef] [Green Version]
- Chang, H.H.Y.; Pannunzio, N.R.; Adachi, N.; Lieber, M.R. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 2017, 18, 495–506. [Google Scholar] [CrossRef]
- Chiruvella, K.K.; Liang, Z.; Wilson, T.E. Repair of double-strand breaks by end joining. Cold Spring Harb. Perspect. Biol. 2013, 5, a012757. [Google Scholar] [CrossRef]
- Karanam, K.; Kafri, R.; Loewer, A.; Lahav, G. Quantitative live cell imaging reveals a gradual shift between DNA repair mechanisms and a maximal use of HR in mid S phase. Mol. Cell 2012, 47, 320–329. [Google Scholar] [CrossRef] [Green Version]
- Heyer, W.D.; Ehmsen, K.T.; Liu, J. Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 2010, 44, 113–139. [Google Scholar] [CrossRef] [Green Version]
- Harper, J.W.; Elledge, S.J. The DNA damage response: Ten years after. Mol. Cell 2007, 28, 739–745. [Google Scholar] [CrossRef]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef] [Green Version]
- Berezikov, E. Evolution of microRNA diversity and regulation in animals. Nat. Rev. Genet. 2011, 12, 846–860. [Google Scholar] [CrossRef] [PubMed]
- Lewis, B.P.; Shih, I.H.; Jones-Rhoades, M.W.; Bartel, D.P.; Burge, C.B. Prediction of mammalian microRNA targets. Cell 2003, 115, 787–798. [Google Scholar] [CrossRef] [Green Version]
- Bartel, D.P.; Chen, C.Z. Micromanagers of gene expression: The potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 2004, 5, 396–400. [Google Scholar] [CrossRef] [PubMed]
- Hornstein, E.; Shomron, N. Canalization of development by microRNAs. Nat. Genet. 2006, 38, S20–S24. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wan, G.; Berger, F.G.; He, X.; Lu, X. The ATM kinase induces microRNA biogenesis in the DNA damage response. Mol. Cell 2011, 41, 371–383. [Google Scholar] [CrossRef] [Green Version]
- Karagkouni, D.; Paraskevopoulou, M.D.; Chatzopoulos, S.; Vlachos, I.S.; Tastsoglou, S.; Kanellos, I.; Papadimitriou, D.; Kavakiotis, I.; Maniou, S.; Skoufos, G.; et al. DIANA-TarBase v8: A decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res 2018, 46, D239–D245. [Google Scholar] [CrossRef] [Green Version]
- Gu, Z.; Gu, L.; Eils, R.; Schlesner, M.; Brors, B. Circlize Implements and enhances circular visualization in R. Bioinformatics 2014, 30, 2811–2812. [Google Scholar] [CrossRef] [Green Version]
- Pannunzio, N.R.; Watanabe, G.; Lieber, M.R. Nonhomologous DNA end-joining for repair of DNA double-strand breaks. J. Biol. Chem. 2018, 293, 10512–10523. [Google Scholar] [CrossRef] [Green Version]
- Beucher, A.; Birraux, J.; Tchouandong, L.; Barton, O.; Shibata, A.; Conrad, S.; Goodarzi, A.A.; Krempler, A.; Jeggo, P.A.; Löbrich, M. ATM and Artemis promote homologous recombination of radiation-induced DNA double-strand breaks in G2. EMBO J. 2009, 28, 3413–3427. [Google Scholar] [CrossRef] [Green Version]
- Lieber, M.R.; Ma, Y.; Pannicke, U.; Schwarz, K. Mechanism and regulation of human non-homologous DNA end-joining. Nat. Rev. Mol. Cell Biol. 2003, 4, 712–720. [Google Scholar] [CrossRef]
- Vannini, I.; Fanini, F.; Fabbri, M. Emerging roles of microRNAs in cancer. Curr. Opin. Genet. Dev. 2018, 48, 128–133. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.; Xiao, W.; Zhu, N.; Liu, Z.; Yang, J.; Wang, Y.; Hong, M. MicroR-545 enhanced radiosensitivity via suppressing Ku70 expression in Lewis lung carcinoma xenograft model. Cancer Cell Int. 2015, 15, 56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dang, Y.; Wang, X.; Hao, Y.; Zhang, X.; Zhao, S.; Ma, J.; Qin, Y.; Chen, Z.J. MicroRNA-379-5p is associate with biochemical premature ovarian insufficiency through PARP1 and XRCC6. Cell Death Dis. 2018, 9, 106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, F.; Liu, J.L.; Li, J.P.; Xiao, F.; Zhang, Z.X.; Zhang, L. MicroRNA-124 (miR-124) regulates Ku70 expression and is correlated with neuronal death induced by ischemia/reperfusion. J. Mol. Neurosci. 2014, 52, 148–155. [Google Scholar] [CrossRef]
- Hatano, K.; Kumar, B.; Zhang, Y.; Coulter, J.B.; Hedayati, M.; Mears, B.; Ni, X.; Kudrolli, T.A.; Chowdhury, W.H.; Rodriguez, R.; et al. A functional screen identifies miRNAs that inhibit DNA repair and sensitize prostate cancer cells to ionizing radiation. Nucleic Acids Res. 2015, 43, 4075–4086. [Google Scholar] [CrossRef] [Green Version]
- Wei, S.; Zhang, Z.Y.; Fu, S.L.; Xie, J.G.; Liu, X.S.; Xu, Y.J.; Zhao, J.P.; Xiong, W.N. Hsa-miR-623 suppresses tumor progression in human lung adenocarcinoma. Cell Death Dis. 2016, 7, e2388. [Google Scholar] [CrossRef] [Green Version]
- Someya, M.; Yamamoto, H.; Nojima, M.; Hori, M.; Tateoka, K.; Nakata, K.; Takagi, M.; Saito, M.; Hirokawa, N.; Tokino, T.; et al. Relation between Ku80 and microRNA-99a expression and late rectal bleeding after radiotherapy for prostate cancer. Radiother. Oncol. 2015, 115, 235–239. [Google Scholar] [CrossRef]
- Tseng, S.H.; Yang, C.C.; Yu, E.H.; Chang, C.; Lee, Y.S.; Liu, C.J.; Chang, K.W.; Lin, S.C. K14-EGFP-miR-31 transgenic mice have high susceptibility to chemical-induced squamous cell tumorigenesis that is associating with Ku80 repression. Int. J. Cancer 2015, 136, 1263–1275. [Google Scholar] [CrossRef]
- Zhang, Z.Y.; Fu, S.L.; Xu, S.Q.; Zhou, X.; Liu, X.S.; Xu, Y.J.; Zhao, J.P.; Wei, S. By downregulating Ku80, hsa-miR-526b suppresses non-small cell lung cancer. Oncotarget 2015, 6, 1462–1477. [Google Scholar] [CrossRef] [Green Version]
- Choi, Y.E.; Meghani, K.; Brault, M.E.; Leclerc, L.; He, Y.J.; Day, T.A.; Elias, K.M.; Drapkin, R.; Weinstock, D.M.; Dao, F.; et al. Platinum and PARP Inhibitor Resistance Due to Overexpression of MicroRNA-622 in BRCA1-Mutant Ovarian Cancer. Cell Rep. 2016, 14, 429–439. [Google Scholar] [CrossRef] [Green Version]
- Smolinska, A.; Swoboda, J.; Fendler, W.; Lerch, M.M.; Sendler, M.; Moskwa, P. MiR-502 is the first reported miRNA simultaneously targeting two components of the classical non-homologous end joining (C-NHEJ) in pancreatic cell lines. Heliyon 2020, 6, e03187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blackford, A.N.; Jackson, S.P. ATM, ATR, and DNA-PK: The Trinity at the Heart of the DNA Damage Response. Mol. Cell 2017, 66, 801–817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, D.; Ng, W.L.; Zhang, X.; Wang, P.; Zhang, Z.; Mo, Y.Y.; Mao, H.; Hao, C.; Olson, J.J.; Curran, W.J.; et al. Targeting DNA-PKcs and ATM with miR-101 sensitizes tumors to radiation. PLoS ONE 2010, 5, e11397. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Wang, H.; Ng, W.L.; Curran, W.J.; Wang, Y. Radiosensitizing effects of ectopic miR-101 on non-small-cell lung cancer cells depend on the endogenous miR-101 level. Int. J. Radiat. Oncol. Biol. Phys. 2011, 81, 1524–1529. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; He, Y.; Wang, Y.; Chen, W.; Hu, B.; Gu, Y. micorRNA-101 silences DNA-PKcs and sensitizes pancreatic cancer cells to gemcitabine. Biochem. Biophys. Res. Commun. 2017, 483, 725–731. [Google Scholar] [CrossRef] [PubMed]
- Zhen, Y.F.; Li, S.T.; Zhu, Y.R.; Wang, X.D.; Zhou, X.Z.; Zhu, L.Q. Identification of DNA-PKcs as a primary resistance factor of salinomycin in osteosarcoma cells. Oncotarget 2016, 7, 79417–79427. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.B.; Zhou, Z.T.; Yang, L.; Wei, M.X.; Tang, M.; Ruan, T.Y.; Xu, J.Y.; Zhou, X.Z.; Chen, G.; Lu, P.H. KU-0060648 inhibits hepatocellular carcinoma cells through DNA-PKcs-dependent and DNA-PKcs-independent mechanisms. Oncotarget 2016, 7, 17047–17059. [Google Scholar] [CrossRef]
- Li, Y.; Han, W.; Ni, T.T.; Lu, L.; Huang, M.; Zhang, Y.; Cao, H.; Zhang, H.Q.; Luo, W.; Li, H. Knockdown of microRNA-1323 restores sensitivity to radiation by suppression of PRKDC activity in radiation-resistant lung cancer cells. Oncol. Rep. 2015, 33, 2821–2828. [Google Scholar] [CrossRef] [Green Version]
- Li, N.; Ma, Y.; Ma, L.; Guan, Y.; Yang, D. MicroRNA-488-3p sensitizes malignant melanoma cells to cisplatin by targeting PRKDC. Cell Biol. Int. 2017, 41, 622–629. [Google Scholar] [CrossRef]
- Jeong, J.Y.; Kang, H.; Kim, T.H.; Kim, G.; Heo, J.H.; Kwon, A.Y.; Kim, S.; Jung, S.G.; An, H.J. MicroRNA-136 inhibits cancer stem cell activity and enhances the anti-tumor effect of paclitaxel against chemoresistant ovarian cancer cells by targeting Notch3. Cancer Lett. 2017, 386, 168–178. [Google Scholar] [CrossRef]
- Piotto, C.; Biscontin, A.; Millino, C.; Mognato, M. Functional validation of miRNAs targeting genes of DNA double-strand break repair to radiosensitize non-small lung cancer cells. Biochim. Biophys. Acta Gene Regul. Mech. 2018, 1861, 1102–1118. [Google Scholar] [CrossRef] [PubMed]
- Chu, Y.Y.; Ko, C.Y.; Wang, S.M.; Lin, P.I.; Wang, H.Y.; Lin, W.C.; Wu, D.Y.; Wang, L.H.; Wang, J.M. Bortezomib-induced miRNAs direct epigenetic silencing of locus genes and trigger apoptosis in leukemia. Cell Death Dis. 2017, 8, e3167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riballo, E.; Kühne, M.; Rief, N.; Doherty, A.; Smith, G.C.; Recio, M.J.; Reis, C.; Dahm, K.; Fricke, A.; Krempler, A.; et al. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol. Cell 2004, 16, 715–724. [Google Scholar] [CrossRef] [PubMed]
- Bebenek, K.; Pedersen, L.C.; Kunkel, T.A. Structure-function studies of DNA polymerase λ. Biochemistry 2014, 53, 2781–2792. [Google Scholar] [CrossRef]
- Moon, A.F.; Pryor, J.M.; Ramsden, D.A.; Kunkel, T.A.; Bebenek, K.; Pedersen, L.C. Sustained active site rigidity during synthesis by human DNA polymerase μ. Nat. Struct. Mol. Biol. 2014, 21, 253–260. [Google Scholar] [CrossRef] [Green Version]
- Bertocci, B.; De Smet, A.; Weill, J.C.; Reynaud, C.A. Nonoverlapping functions of DNA polymerases mu, lambda, and terminal deoxynucleotidyltransferase during immunoglobulin V(D)J recombination in vivo. Immunity 2006, 25, 31–41. [Google Scholar] [CrossRef]
- Gu, J.; Lu, H.; Tippin, B.; Shimazaki, N.; Goodman, M.F.; Lieber, M.R. XRCC4:DNA ligase IV can ligate incompatible DNA ends and can ligate across gaps. EMBO J. 2007, 26, 1010–1023. [Google Scholar] [CrossRef] [Green Version]
- Zeng, A.; Wei, Z.; Yan, W.; Yin, J.; Huang, X.; Zhou, X.; Li, R.; Shen, F.; Wu, W.; Wang, X.; et al. Exosomal transfer of miR-151a enhances chemosensitivity to temozolomide in drug-resistant glioblastoma. Cancer Lett. 2018, 436, 10–21. [Google Scholar] [CrossRef]
- Mo, L.J.; Song, M.; Huang, Q.H.; Guan, H.; Liu, X.D.; Xie, D.F.; Huang, B.; Huang, R.X.; Zhou, P.K. Exosome-packaged miR-1246 contributes to bystander DNA damage by targeting LIG4. Br. J. Cancer 2018, 119, 492–502. [Google Scholar] [CrossRef] [Green Version]
- Liu, R.; Shen, L.; Lin, C.; He, J.; Wang, Q.; Qi, Z.; Zhang, Q.; Zhou, M.; Wang, Z. MiR-1587 Regulates DNA Damage Repair and the Radiosensitivity of CRC Cells via Targeting LIG4. Dose Response 2020, 18, 1559325820936906. [Google Scholar] [CrossRef]
- Ceccaldi, R.; Rondinelli, B.; D’Andrea, A.D. Repair Pathway Choices and Consequences at the Double-Strand Break. Trends Cell Biol. 2016, 26, 52–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krajewska, M.; Fehrmann, R.S.; de Vries, E.G.; van Vugt, M.A. Regulators of homologous recombination repair as novel targets for cancer treatment. Front. Genet. 2015, 6, 96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Heyer, W.D. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 2008, 18, 99–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kowalczykowski, S.C. An Overview of the Molecular Mechanisms of Recombinational DNA Repair. Cold Spring Harb. Perspect. Biol. 2015, 7, a016410. [Google Scholar] [CrossRef] [Green Version]
- Jasin, M.; Rothstein, R. Repair of strand breaks by homologous recombination. Cold Spring Harb. Perspect. Biol. 2013, 5, a012740. [Google Scholar] [CrossRef]
- Xie, A.; Kwok, A.; Scully, R. Role of mammalian MRE11 in classical and alternative nonhomologous end joining. Nat. Struct. Mol. Biol. 2009, 16, 814–818. [Google Scholar] [CrossRef] [Green Version]
- Zha, S.; Boboila, C.; Alt, F.W. MRE11: Roles in DNA repair beyond homologous recombination. Nat. Struct. Mol. Biol. 2009, 16, 798–800. [Google Scholar] [CrossRef]
- Wilson, R.; Espinosa-Diez, C.; Kanner, N.; Chatterjee, N.; Ruhl, R.; Hipfinger, C.; Advani, S.J.; Li, J.; Khan, O.F.; Franovic, A.; et al. MicroRNA regulation of endothelial TREX1 reprograms the tumour microenvironment. Nat. Commun. 2016, 7, 13597. [Google Scholar] [CrossRef] [Green Version]
- Espinosa-Diez, C.; Wilson, R.; Chatterjee, N.; Hudson, C.; Ruhl, R.; Hipfinger, C.; Helms, E.; Khan, O.F.; Anderson, D.G.; Anand, S. MicroRNA regulation of the MRN complex impacts DNA damage, cellular senescence, and angiogenic signaling. Cell Death Dis. 2018, 9, 632. [Google Scholar] [CrossRef]
- Van den Bosch, M.; Bree, R.T.; Lowndes, N.F. The MRN complex: Coordinating and mediating the response to broken chromosomes. EMBO Rep. 2003, 4, 844–849. [Google Scholar] [CrossRef] [Green Version]
- Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meghani, K.; Fuchs, W.; Detappe, A.; Drané, P.; Gogola, E.; Rottenberg, S.; Jonkers, J.; Matulonis, U.; Swisher, E.M.; Konstantinopoulos, P.A.; et al. Multifaceted impact of microRNA 493-5p on genome-stabilizing pathways induces platinum and PARP inhibitor resistance in BRCA2-mutated carcinomas. Cell Rep. 2018, 23, 100–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.H.; Lee, B.R.; Choi, E.S.; Lee, K.M.; Choi, S.K.; Cho, J.H.; Jeon, W.B.; Kim, E. Reverse Expression of Aging-Associated Molecules through Transfection of miRNAs to Aged Mice. Mol. Ther. Nucleic Acids 2017, 6, 106–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naccarati, A.; Rosa, F.; Vymetalkova, V.; Barone, E.; Jiraskova, K.; Di Gaetano, C.; Novotny, J.; Levy, M.; Vodickova, L.; Gemignani, F.; et al. Double-strand break repair and colorectal cancer: Gene variants within 3’ UTRs and microRNAs binding as modulators of cancer risk and clinical outcome. Oncotarget 2016, 7, 23156–23169. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.; Wang, P.; Song, C.; Wang, K.; Yan, R.; Li, J.; Dai, L. Evaluation of miRNA-binding-site SNPs of MRE11A, NBS1, RAD51 and RAD52 involved in HRR pathway genes and risk of breast cancer in China. Mol. Genet. Genom. 2015, 290, 1141–1153. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.M.; Kerr, M.; Teo, M.T.; Jevons, S.J.; Koritzinsky, M.; Wouters, B.G.; Bhattarai, S.; Kiltie, A.E. Post-transcriptional regulation of MRE11 expression in muscle-invasive bladder tumours. Oncotarget 2014, 5, 993–1003. [Google Scholar] [CrossRef] [Green Version]
- Lammens, K.; Bemeleit, D.J.; Möckel, C.; Clausing, E.; Schele, A.; Hartung, S.; Schiller, C.B.; Lucas, M.; Angermüller, C.; Söding, J.; et al. The MRE11:RAD50 structure shows an ATP-dependent molecular clamp in DNA double-strand break repair. Cell 2011, 145, 54–66. [Google Scholar] [CrossRef] [Green Version]
- Shirode, A.B.; Kovvuru, P.; Chittur, S.V.; Henning, S.M.; Heber, D.; Reliene, R. Antiproliferative effects of pomegranate extract in MCF-7 breast cancer cells are associated with reduced DNA repair gene expression and induction of double-strand breaks. Mol. Carcinog. 2014, 53, 458–470. [Google Scholar] [CrossRef]
- Ramirez, H.A.; Pastar, I.; Jozic, I.; Stojadinovic, O.; Stone, R.C.; Ojeh, N.; Gil, J.; Davis, S.C.; Kirsner, R.S.; Tomic-Canic, M. Staphylococcus aureus Triggers Induction of miR-15B-5P to Diminish DNA Repair and Deregulate Inflammatory Response in Diabetic Foot Ulcers. J. Investig. Dermatol. 2018, 138, 1187–1196. [Google Scholar] [CrossRef] [Green Version]
- Lovric, J.; Mano, M.; Zentilin, L.; Eulalio, A.; Zacchigna, S.; Giacca, M. Terminal differentiation of cardiac and skeletal myocytes induces permissivity to AAV transduction by relieving inhibition imposed by DNA damage response proteins. Mol. Ther. 2012, 20, 2087–2097. [Google Scholar] [CrossRef] [Green Version]
- Kakarougkas, A.; Jeggo, P.A. DNA DSB repair pathway choice: An orchestrated handover mechanism. Br. J. Radiol. 2014, 87, 20130685. [Google Scholar] [CrossRef] [PubMed]
- You, Z.; Bailis, J.M. DNA damage and decisions: CtIP coordinates DNA repair and cell cycle checkpoints. Trends Cell Biol. 2010, 20, 402–409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hühn, D.; Kousholt, A.N.; Sørensen, C.S.; Sartori, A.A. miR-19, a component of the oncogenic miR-17∼92 cluster, targets the DNA-end resection factor CtIP. Oncogene 2015, 34, 3977–3984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, N.T.; Nakamura, K.; Davies, R.; Nahas, S.A.; Brown, C.; Tunuguntla, R.; Gatti, R.A.; Hu, H. ATM-dependent MiR-335 targets CtIP and modulates the DNA damage response. PLoS Genet. 2013, 9, e1003505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miao, W.J.; Yuan, D.J.; Zhang, G.Z.; Liu, Q.; Ma, H.M.; Jin, Q.Q. lncRNA CASC2/miR-18a-5p axis regulates the malignant potential of nasopharyngeal carcinoma by targeting RBBP8. Oncol. Rep. 2019, 41, 1797–1806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, L.; Yang, B.; Wang, Y.; Liu, T.; He, Z.; Zhao, H.; Xie, L.; Mu, H. The CTIP-mediated repair of TNF-α-induced DNA double-strand break was impaired by miR-130b in cervical cancer cell. Cell Biochem. Funct. 2020, 37, 534–544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maréchal, A.; Zou, L. RPA-coated single-stranded DNA as a platform for post-translational modifications in the DNA damage response. Cell Res. 2015, 25, 9–23. [Google Scholar] [CrossRef] [Green Version]
- Zou, Y.; Liu, Y.; Wu, X.; Shell, S.M. Functions of human replication protein A (RPA): From DNA replication to DNA damage and stress responses. J. Cell. Physiol. 2006, 208, 267–273. [Google Scholar] [CrossRef] [Green Version]
- Abdelmohsen, K.; Srikantan, S.; Tominaga, K.; Kang, M.J.; Yaniv, Y.; Martindale, J.L.; Yang, X.; Park, S.S.; Becker, K.G.; Subramanian, M.; et al. Growth inhibition by miR-519 via multiple p21-inducing pathways. Mol. Cell. Biol. 2012, 32, 2530–2548. [Google Scholar] [CrossRef] [Green Version]
- Zou, Z.; Ni, M.; Zhang, J.; Chen, Y.; Ma, H.; Qian, S.; Tang, L.; Tang, J.; Yao, H.; Zhao, C.; et al. miR-30a can inhibit DNA replication by targeting RPA1 thus slowing cancer cell proliferation. Biochem. J. 2016, 473, 2131–2139. [Google Scholar] [CrossRef]
- Choi, Y.E.; Pan, Y.; Park, E.; Konstantinopoulos, P.; De, S.; D’Andrea, A.; Chowdhury, D. MicroRNAs down-regulate homologous recombination in the G1 phase of cycling cells to maintain genomic stability. Elife 2014, 3, e02445. [Google Scholar] [CrossRef]
- Roy, R.; Chun, J.; Powell, S.N. BRCA1 and BRCA2: Different roles in a common pathway of genome protection. Nat. Rev. Cancer 2011, 12, 68–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paul, A.; Paul, S. The breast cancer susceptibility genes (BRCA) in breast and ovarian cancers. Front. Biosci. 2014, 19, 605–618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, X.; Ayub, B.; Liu, Z.; Serna, V.A.; Qiang, W.; Liu, Y.; Hernando, E.; Zabludoff, S.; Kurita, T.; Kong, B.; et al. Anti-miR182 reduces ovarian cancer burden, invasion, and metastasis: An in vivo study in orthotopic xenografts of nude mice. Mol. Cancer Ther. 2014, 13, 1729–1739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, P.T.; Pongkulapa, T.; Cho, H.Y.; Han, J.; Pasquale, N.J.; Rabie, H.; Kim, J.H.; Choi, J.W.; Lee, K.B. Overcoming Chemoresistance in Cancer via Combined MicroRNA Therapeutics with Anticancer Drugs Using Multifunctional Magnetic Core-Shell Nanoparticles. ACS Appl. Mater. Interfaces 2018, 10, 26954–26963. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Zhang, M.; Peng, F.; Fang, L.; Zhang, Y.; Liang, H.; Zhou, W.; Ao, L.; Guo, Z. The BRCA1/2-directed miRNA signature predicts a good prognosis in ovarian cancer patients with wild-type BRCA1/2. Oncotarget 2015, 6, 2397–2406. [Google Scholar] [CrossRef] [Green Version]
- Slattery, M.L.; Herrick, J.S.; Mullany, L.E.; Samowitz, W.S.; Sevens, J.R.; Sakoda, L.; Wolff, R.K. The co-regulatory networks of tumor suppressor genes, oncogenes, and miRNAs in colorectal cancer. Genes Chromosomes Cancer 2017, 56, 769–787. [Google Scholar] [CrossRef] [Green Version]
- Liu, T.; Huang, J. DNA End Resection: Facts and Mechanisms. Genom. Proteom. Bioinform. 2016, 14, 126–130. [Google Scholar] [CrossRef] [Green Version]
- Schlegel, B.P.; Jodelka, F.M.; Nunez, R. BRCA1 promotes induction of ssDNA by ionizing radiation. Cancer Res. 2006, 66, 5181–5189. [Google Scholar] [CrossRef] [Green Version]
- McMillen, B.D.; Aponte, M.M.; Liu, Z.; Helenowski, I.B.; Scholtens, D.M.; Buttin, B.M.; Wei, J.J. Expression analysis of MIR182 and its associated target genes in advanced ovarian carcinoma. Mod. Pathol. 2012, 25, 1644–1653. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Liu, J.; Segura, M.F.; Shao, C.; Lee, P.; Gong, Y.; Hernando, E.; Wei, J.J. MiR-182 overexpression in tumourigenesis of high-grade serous ovarian carcinoma. J. Pathol. 2012, 228, 204–215. [Google Scholar] [CrossRef] [PubMed]
- Moskwa, P.; Buffa, F.M.; Pan, Y.; Panchakshari, R.; Gottipati, P.; Muschel, R.J.; Beech, J.; Kulshrestha, R.; Abdelmohsen, K.; Weinstock, D.M.; et al. miR-182-mediated downregulation of BRCA1 impacts DNA repair and sensitivity to PARP inhibitors. Mol. Cell 2011, 41, 210–220. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Martinez-Ruiz, H.; Murnane, J.; Powell, S.N.; Barcellos-Hoff, M.H. Abstract 831: TGFβ controls the DNA damage response via miR-182 regulation of BRCA1 and ATM. Cancer Res. 2017, 77, 831. [Google Scholar]
- Garcia, A.I.; Buisson, M.; Bertrand, P.; Rimokh, R.; Rouleau, E.; Lopez, B.S.; Lidereau, R.; Mikaélian, I.; Mazoyer, S. Down-regulation of BRCA1 expression by miR-146a and miR-146b-5p in triple negative sporadic breast cancers. EMBO Mol. Med. 2011, 3, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Hua, J.; Jia, Z.; Ding, J.; Han, Z.; Dong, Y.; Lin, Q.; Yao, Y. Expression of miR-146a-5p in breast cancer and its role in proliferation of breast cancer cells. Oncol. Lett. 2018, 15, 9884–9888. [Google Scholar] [PubMed] [Green Version]
- Mueller, A.C.; Sun, D.; Dutta, A. The miR-99 family regulates the DNA damage response through its target SNF2H. Oncogene 2013, 32, 1164–1172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, Y.; Lin, X.; Xu, W.; Huang, Q.; Shi, L.; Pan, Y.; Zhang, Y.; Zhu, Y.; Li, C.; Liu, L.; et al. MiR-210-3p protects endometriotic cells from oxidative stress-induced cell cycle arrest by targeting BARD1. Cell Death Dis. 2019, 10, 144. [Google Scholar] [CrossRef] [Green Version]
- Luo, H.; Liang, H.; Chen, Y.; Chen, S.; Xu, Y.; Xu, L.; Liu, J.; Zhou, K.; Peng, J.; Guo, G.; et al. miR-7-5p overexpression suppresses cell proliferation and promotes apoptosis through inhibiting the ability of DNA damage repair of PARP-1 and BRCA1 in TK6 cells exposed to hydroquinone. Chem. Biol. Interact. 2018, 283, 84–90. [Google Scholar] [CrossRef]
- He, X.; Fan, S. hsa-miR-212 modulates the radiosensitivity of glioma cells by targeting BRCA1. Oncol. Rep. 2018, 39, 977–984. [Google Scholar] [CrossRef] [Green Version]
- Sun, C.; Li, N.; Yang, Z.; Zhou, B.; He, Y.; Weng, D.; Fang, Y.; Wu, P.; Chen, P.; Yang, X.; et al. miR-9 regulation of BRCA1 and ovarian cancer sensitivity to cisplatin and PARP inhibition. J. Natl. Cancer Inst. 2013, 105, 1750–1758. [Google Scholar] [CrossRef] [Green Version]
- He, X.; Xiao, X.; Dong, L.; Wan, N.; Zhou, Z.; Deng, H.; Zhang, X. MiR-218 regulates cisplatin chemosensitivity in breast cancer by targeting BRCA1. Tumour Biol. 2015, 36, 2065–2075. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Peng, J.; Fu, Y.; An, S.; Rezaei, K.; Tabbara, S.; Teal, C.B.; Man, Y.G.; Brem, R.F.; Fu, S.W. miR-638 mediated regulation of BRCA1 affects DNA repair and sensitivity to UV and cisplatin in triple-negative breast cancer. Breast Cancer Res. 2014, 16, 435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heyn, H.; Engelmann, M.; Schreek, S.; Ahrens, P.; Lehmann, U.; Kreipe, H.; Schlegelberger, B.; Beger, C. MicroRNA miR-335 is crucial for the BRCA1 regulatory cascade in breast cancer development. Int. J. Cancer 2011, 129, 2797–2806. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, A.; Tsunoda, Y.; Tsuji, M.; Udaka, Y.; Oyamada, H.; Tsuchiya, H.; Oguchi, K. Decreased miR-206 expression in BRCA1 wild-type triple-negative breast cancer cells after concomitant treatment with gemcitabine and a Poly(ADP-ribose) polymerase-1 inhibitor. Anticancer Res. 2014, 34, 4893–4897. [Google Scholar] [PubMed]
- Matamala, N.; Vargas, M.T.; González-Cámpora, R.; Arias, J.I.; Menéndez, P.; Andrés-León, E.; Yanowsky, K.; Llaneza-Folgueras, A.; Miñambres, R.; Martínez-Delgado, B.; et al. MicroRNA deregulation in triple negative breast cancer reveals a role of miR-498 in regulating BRCA1 expression. Oncotarget 2016, 7, 20068–20079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.L.; Meng, F.M.; Li, H.J. High expression of lncRNA MEG3 participates in non-small cell lung cancer by regulating microRNA-7-5p. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 5938–5945. [Google Scholar]
- Gao, S.; Wang, Y.; Wang, M.; Li, Z.; Zhao, Z.; Wang, R.X.; Wu, R.; Yuan, Z.; Cui, R.; Jiao, K.; et al. MicroRNA-155, induced by FOXP3 through transcriptional repression of BRCA1, is associated with tumor initiation in human breast cancer. Oncotarget 2017, 8, 41451–41464. [Google Scholar] [CrossRef]
- Srinivasan, G.; Williamson, E.A.; Kong, K.; Jaiswal, A.S.; Huang, G.; Kim, H.S.; Schärer, O.; Zhao, W.; Burma, S.; Sung, P.; et al. MiR223-3p promotes synthetic lethality in BRCA1-deficient cancers. Proc. Natl. Acad. Sci. USA 2019, 116, 17438–17443. [Google Scholar] [CrossRef] [Green Version]
- O’Connor, M.J. Targeting the DNA Damage Response in Cancer. Mol. Cell 2015, 60, 547–560. [Google Scholar] [CrossRef] [Green Version]
- Xiao, M.; Cai, J.; Cai, L.; Jia, J.; Xie, L.; Zhu, Y.; Huang, B.; Jin, D.; Wang, Z. Let-7e sensitizes epithelial ovarian cancer to cisplatin through repressing DNA double-strand break repair. J. Ovarian Res. 2017, 10, 24. [Google Scholar] [CrossRef] [Green Version]
- Lai, T.H.; Ewald, B.; Zecevic, A.; Liu, C.; Sulda, M.; Papaioannou, D.; Garzon, R.; Blachly, J.S.; Plunkett, W.; Sampath, D. HDAC Inhibition Induces MicroRNA-182, which Targets RAD51 and Impairs HR Repair to Sensitize Cells to Sapacitabine in Acute Myelogenous Leukemia. Clin. Cancer Res. 2016, 22, 3537–3549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gasparini, P.; Lovat, F.; Fassan, M.; Casadei, L.; Cascione, L.; Jacob, N.K.; Carasi, S.; Palmieri, D.; Costinean, S.; Shapiro, C.L.; et al. Protective role of miR-155 in breast cancer through RAD51 targeting impairs homologous recombination after irradiation. Proc. Natl. Acad. Sci. USA 2014, 111, 4536–4541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Czochor, J.R.; Sulkowski, P.; Glazer, P.M. miR-155 Overexpression Promotes Genomic Instability by Reducing High-fidelity Polymerase Delta Expression and Activating Error-Prone DSB Repair. Mol. Cancer Res. 2016, 14, 363–373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, J.H.; Hwang, Y.H.; Lee, D.J.; Kim, D.H.; Park, J.M.; Wu, H.-G.; Kim, I.A. MicroRNA-203 Modulates the Radiation Sensitivity of Human Malignant Glioma Cells. Int. J. Radiat. Oncol. 2016, 94, 412–420. [Google Scholar] [CrossRef]
- Cortez, M.A.; Valdecanas, D.; Niknam, S.; Peltier, H.J.; Diao, L.; Giri, U.; Komaki, R.; Calin, G.A.; Gomez, D.R.; Chang, J.Y.; et al. In Vivo Delivery of miR-34a Sensitizes Lung Tumors to Radiation Through RAD51 Regulation. Mol. Ther. Nucleic Acids 2015, 4, e270. [Google Scholar] [CrossRef]
- Wiemer, E.A.; Berns, E.M. MicroRNA regulation of RAD51 in serous ovarian cancer modulates chemotherapy response. J. Natl. Cancer Inst. 2015, 107, djv161. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.; Yang, D.; Rupaimoole, R.; Pecot, C.V.; Sun, Y.; Mangala, L.S.; Li, X.; Ji, P.; Cogdell, D.; Hu, L.; et al. Augmentation of response to chemotherapy by microRNA-506 through regulation of RAD51 in serous ovarian cancers. J. Natl. Cancer Inst. 2015, 107, djv108. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.W.; Wang, Y.; Dhillon, K.K.; Calses, P.; Villegas, E.; Mitchell, P.S.; Tewari, M.; Kemp, C.J.; Taniguchi, T. Systematic screen identifies miRNAs that target RAD51 and RAD51D to enhance chemosensitivity. Mol. Cancer Res. 2013, 11, 1564–1573. [Google Scholar] [CrossRef] [Green Version]
- Neijenhuis, S.; Bajrami, I.; Miller, R.; Lord, C.J.; Ashworth, A. Identification of miRNA modulators to PARP inhibitor response. DNA Repair. 2013, 12, 394–402. [Google Scholar] [CrossRef]
- Wang, Y.; Huang, J.W.; Calses, P.; Kemp, C.J.; Taniguchi, T. MiR-96 downregulates REV1 and RAD51 to promote cellular sensitivity to cisplatin and PARP inhibition. Cancer Res. 2012, 72, 4037–4046. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Bao, W.; Liu, Y.; Wang, S.; Xu, S.; Li, X.; Li, Y.; Wu, S. miR-98-5p contributes to cisplatin resistance in epithelial ovarian cancer by suppressing miR-152 biogenesis via targeting Dicer1. Cell Death Dis. 2018, 9, 447. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Wang, Q.; Liu, R.; Chen, Z.; Zhang, X.; Zhou, P.; Wang, Z. LncRNA lnc-RI regulates homologous recombination repair of DNA double-strand breaks by stabilizing RAD51 mRNA as a competitive endogenous RNA. Nucleic Acids Res. 2018, 46, 717–729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McVey, M.; Khodaverdian, V.Y.; Meyer, D.; Cerqueira, P.G.; Heyer, W.D. Eukaryotic DNA Polymerases in Homologous Recombination. Annu. Rev. Genet. 2016, 50, 393–421. [Google Scholar] [CrossRef] [Green Version]
- Kaur, E.; Agrawal, R.; Sengupta, S. Functions of BLM Helicase in Cells: Is It Acting Like a Double-Edged Sword? Front. Genet. 2021, 12, 277. [Google Scholar] [CrossRef]
- Marin, J.J.G.; Macias, R.I.R.; Monte, M.J.; Romero, M.R.; Asensio, M.; Sanchez-Martin, A.; Cives-Losada, C.; Temprano, A.G.; Espinosa-Escudero, R.; Reviejo, M.; et al. Molecular Bases of Drug Resistance in Hepatocellular Carcinoma. Cancers 2020, 12, 1663. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peraza-Vega, R.I.; Valverde, M.; Rojas, E. Interactions between miRNAs and Double-Strand Breaks DNA Repair Genes, Pursuing a Fine-Tuning of Repair. Int. J. Mol. Sci. 2022, 23, 3231. https://doi.org/10.3390/ijms23063231
Peraza-Vega RI, Valverde M, Rojas E. Interactions between miRNAs and Double-Strand Breaks DNA Repair Genes, Pursuing a Fine-Tuning of Repair. International Journal of Molecular Sciences. 2022; 23(6):3231. https://doi.org/10.3390/ijms23063231
Chicago/Turabian StylePeraza-Vega, Ricardo I., Mahara Valverde, and Emilio Rojas. 2022. "Interactions between miRNAs and Double-Strand Breaks DNA Repair Genes, Pursuing a Fine-Tuning of Repair" International Journal of Molecular Sciences 23, no. 6: 3231. https://doi.org/10.3390/ijms23063231
APA StylePeraza-Vega, R. I., Valverde, M., & Rojas, E. (2022). Interactions between miRNAs and Double-Strand Breaks DNA Repair Genes, Pursuing a Fine-Tuning of Repair. International Journal of Molecular Sciences, 23(6), 3231. https://doi.org/10.3390/ijms23063231





