Identification and Expression Analysis of R2R3-MYB Family Genes Associated with Salt Tolerance in Cyclocarya paliurus
Abstract
:1. Introduction
2. Results
2.1. Identification of R2R3-MYB Genes and Prediction of Encoded Proteins
2.2. Phylogenetic Analysis of R2R3-MYB Family Genes
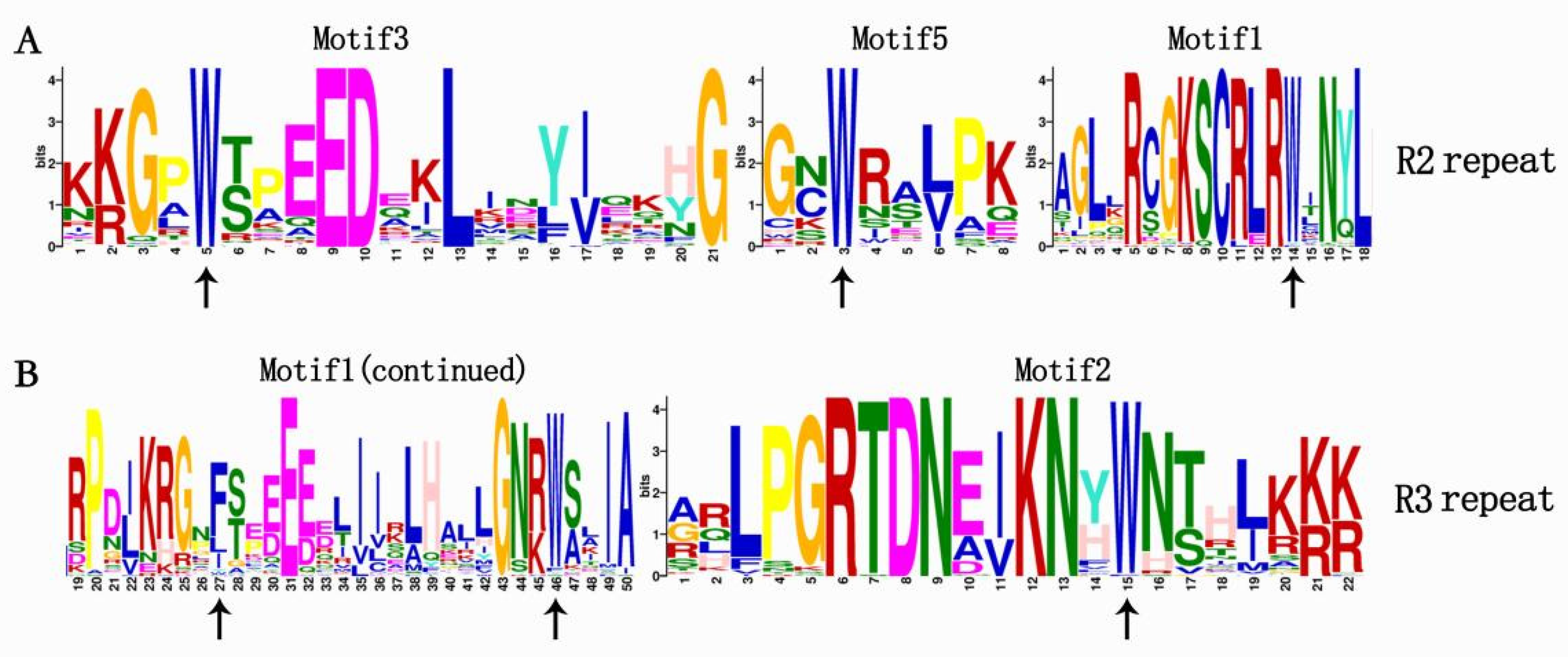
2.3. Gene Structure and Conserved Motifs Analysis of R2R3-MYB Proteins
2.4. Promoter Motif Analysis
2.5. Profiling of Expressed CpaMYB Genes and Verification of Selected Gene Expression
2.6. Profiling of Expressed CpaMYB Genes in Hydroponic Experiment
3. Discussion
3.1. Characterization of the R2R3-MYB Gene Family in C. paliurus
3.2. Responses of R2R3-MYB Genes to Salt Stress in C. paliurus
3.3. Phytohormone Signal Response to Salt Stress
4. Materials and Methods
4.1. Plant Materials and Experiment Treatments
4.1.1. Pot Experiment
4.1.2. Hydroponic Experiment
4.2. Gene Identification and Coding Protein Prediction
4.3. Multiple Sequence Alignment and Phylogenetic Analysis
4.4. Exon–Intron Structural and Conserved Motif Analysis
4.5. Promoter Cis-Acting Element Prediction
4.6. Expression Profile under Salt Stress and qRT-PCR Analysis
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Conflicts of Interest
References
- Zhu, J.-K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Han, G.; Sun, C.; Sui, N. Research advances of MYB transcription factors in plant stress resistance and breeding. Plant Signal. Behav. 2019, 14, 1613131. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, Z.; Fang, S.; Liu, Y.; Shang, X. Integrative analysis of metabolome and transcriptome reveals molecular reg-ulatory mechanism of flavonoid biosynthesis in Cyclocarya paliurus under salt stress. Ind. Crops Prod. 2021, 170, 113823. [Google Scholar] [CrossRef]
- Anumalla, M.; Roychowdhury, R.; Geda, C.; Bharathkumar, S.; Goutam, K.; Mohandev, T. Mechanism of stress signal transduction and involvement of stress inducible transcription factors and genes in response to abiotic stresses in plant. Int. J. Recent. Sci. Res. 2016, 7, 12754–12771. [Google Scholar]
- Aragüés, R.; Medina, E.T.; Zribi, W.; Clavería, I.; Álvaro-Fuentes, J.; Faci, J. Soil salinization as a threat to the sustainability of deficit irrigation under present and expected climate change scenarios. Irrig. Sci. 2014, 33, 67–79. [Google Scholar] [CrossRef] [Green Version]
- Colombani, N.; Osti, A.; Volta, G.; Mastrocicco, M. Impact of Climate Change on Salinization of Coastal Water Resources. Water Resour. Manag. 2016, 30, 2483–2496. [Google Scholar] [CrossRef]
- Khan, S.-A.; Li, M.-Z.; Wang, S.-M.; Yin, H.-J. Revisiting the Role of Plant Transcription Factors in the Battle against Abiotic Stress. Int. J. Mol. Sci. 2018, 19, 1634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Ambawat, S.; Sharma, P.; Yadav, N.R.; Yadav, R.C. MYB transcription factor genes as regulators for plant responses: An over-view. Physiol. Mol. Biol. Plants 2013, 19, 307–321. [Google Scholar] [CrossRef] [Green Version]
- Ji, Q.; Wang, D.; Zhou, J.; Xu, Y.; Shen, B.; Zhou, F. Genome-wide characterization and expression analyses of the MYB super-family genes during developmental stages in chinese jujube. PeerJ 2019, 7, e6353. [Google Scholar]
- Yang, L.; Liu, H.; Hao, Z.; Zong, Y.; Xia, H.; Shen, Y.; Li, H. Genome-Wide Identification and Expression Analysis of R2R3-MYB Family Genes Associated with Petal Pigment Synthesis in Liriodendron. Int. J. Mol. Sci. 2021, 22, 11291. [Google Scholar] [CrossRef]
- Pradhan, S.; Shyamli, P.S.; Suranjika, S.; Parida, A. Genome Wide Identification and Analysis of the R2R3-MYB Transcription Factor Gene Family in the Mangrove Avicennia marina. Agronomy 2021, 11, 123. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Y.; Fan, C.; Wei, Y.; Meng, J.; Li, Z.; Zhong, C. Genome-wide analysis of MYB transcription factors and their responses to salt stress in Casuarina equisetifolia. BMC Plant Biol. 2021, 21, 328. [Google Scholar] [CrossRef]
- Zhang, C.; Ma, R.; Xu, J.; Yan, J.; Guo, L.; Song, J.; Feng, R.; Yu, M. Genome-wide identification and classification of MYB su-perfamily genes in peach. PLoS ONE 2018, 13, e0199192. [Google Scholar]
- Oppenheimer, D.G.; Herman, P.L.; Sivakumaran, S.; Esch, J.; Marks, M.D. A myb gene required for leaf trichome differenti-ation in Arabidopsis is expressed in stipules. Cell 1991, 67, 483–493. [Google Scholar] [CrossRef]
- Liu, H.; Ma, J.; Li, H. Transcriptomic and microstructural analyses in Liriodendron tulipifera Linn. reveal candidate genes in-volved in nectary development and nectar secretion. BMC Plant Biol. 2019, 19, 531. [Google Scholar] [CrossRef] [Green Version]
- Baumann, K.; Perez-Rodriguez, M.; Bradley, D.; Venail, J.; Bailey, P.; Jin, H.; Koes, R.; Roberts, K.; Martin, C. Control of cell and petal morphogenesis by R2R3 MYB transcription factors. Development 2007, 134, 1691–1701. [Google Scholar] [CrossRef] [Green Version]
- Grotewold, E.; Drummond, B.J.; Bowen, B.; Peterson, T. The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell 1994, 76, 543–553. [Google Scholar] [CrossRef]
- Urao, T.; Yamaguchi-Shinozaki, K.; Urao, S.; Shinozaki, K. An Arabidopsis myb Homolog Is Induced by Dehydration Stress and Its Gene Product Binds to the Conserved MYB Recognition Sequence. Plant Cell 1993, 5, 1529. [Google Scholar] [CrossRef] [Green Version]
- Yo, Y.; Klessig, D.F. Isolation and characterization of a tobacco mosaic virus-inducible MYB oncogene homolog from tobacco. Proc. Natl. Acad. Sci. USA 1997, 93, 14972–14977. [Google Scholar]
- Zhao, K.; Cheng, Z.; Guo, Q.; Yao, W.; Liu, H.; Zhou, B.; Jiang, T. Characterization of the Poplar R2R3-MYB Gene Family and Over-Expression of PsnMYB108 Confers Salt Tolerance in Transgenic Tobacco. Front. Plant Sci. 2020, 11, 571881. [Google Scholar] [CrossRef] [PubMed]
- Rahaie, M.; Xue, G.-P.; Naghavi, M.R.; Alizadeh, H.; Schenk, P.M. A MYB gene from wheat (Triticum aestivum L.) is up-regulated during salt and drought stresses and differentially regulated between salt-tolerant and sensitive genotypes. Plant Cell Rep. 2010, 29, 835–844. [Google Scholar] [CrossRef] [PubMed]
- Yoo, J.H.; Park, C.Y.; Kim, J.C.; Heo, W.D.; Cheong, M.S.; Park, H.C.; Kim, M.C.; Moon, B.C.; Choi, M.S.; Kang, Y.H.; et al. Direct Interaction of a Divergent CaM Isoform and the Transcription Factor, MYB2, Enhances Salt Tolerance in Arabidopsis. J. Biol. Chem. 2005, 280, 3697–3706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, C.; Seo, J.S.; Han, S.; Koo, Y.; Kim, C.; Song, S.; Nahm, B.; Choi, Y.; Cheong, J.-J. Overexpression of AtMYB44 enhances stomatal closure to confer sbiotic stress tolerance in transgenic Arabidopsis. Plant Physiol. 2008, 146, 623–635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, R.; Wang, Y.; Zheng, H.; Lu, W.; Wu, C.A.; Huang, J.; Yan, K.; Yang, G.; Zheng, C. Salt-induced transcription factor MYB74 is regulated by the RNA-directed DNA methylation pathway in Arabidopsis. J. Exp. Bot. 2015, 66, 5997–6008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, Q.; Chen, R.; Wei, X.; Liu, Y.; Zhao, S.; Yin, X.; Xie, T. Genome-wide identification of R2R3-MYB family in wheat and functional characteristics of the abiotic stress responsive gene TaMYB344. BMC Genom. 2020, 21, 792. [Google Scholar] [CrossRef]
- Rhoads, A.; Au, K.F. PacBio Sequencing and Its Applications. Genom. Proteom. Bioinform. 2015, 13, 278–289. [Google Scholar] [CrossRef] [Green Version]
- Fang, S.; Wang, J.; Wei, Z.; Zhu, Z. Methods to break seed dormancy in Cyclocarya paliurus (Batal) Iljinskaja. Sci. Hortic. 2006, 110, 305–309. [Google Scholar] [CrossRef]
- Deng, B.; Fang, S.; Yang, W.; Tian, Y.; Shang, X. Provenance variation in growth and wood properties of juvenile Cyclocarya paliurus. New For. 2014, 45, 625–639. [Google Scholar] [CrossRef]
- Yao, X.; Lin, Z.; Jiang, C.; Gao, M.; Wang, Q.; Yao, N.; Ma, Y.; Li, Y.; Fang, S.; Shang, X.; et al. Cyclocarya paliurus prevents high fat diet induced hyperlipidemia and obesity in Sprague–Dawley rats. Can. J. Physiol. Pharmacol. 2015, 93, 677–686. [Google Scholar] [CrossRef]
- Fang, S.; Yang, W.; Chu, X.; Shang, X.; She, C.; Fu, X. Provenance and temporal variations in selected flavonoids in leaves of Cyclocarya paliurus. Food Chem. 2011, 124, 1382–1386. [Google Scholar] [CrossRef]
- Yao, R.L.; Fang, S.Z.; Nichols, D.J. A strategy of Ca2+ alleviating Na+ toxicity insalt-treated Cyclocarya paliurus seedlings: Pho-tosynthetic and nutritional responses. Plant Growth Regul. 2012, 68, 351–359. [Google Scholar] [CrossRef]
- Wilkins, O.; Nahal, H.; Foong, J.; Provart, N.J.; Campbell, M.M. Expansion and Diversification of the Populus R2R3-MYB Family of Transcription Factors. Plant Physiol. 2008, 149, 981–993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, X.-J.; Li, S.-B.; Liu, S.-R.; Hu, C.-G.; Zhang, J.-Z. Genome-Wide Classification and Evolutionary and Expression Analyses of Citrus MYB Transcription Factor Families in Sweet Orange. PLoS ONE 2014, 9, e112375. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Lin-Wang, K.; Liu, Z.; Allan, A.C.; Qin, S.; Zhang, J.; Liu, Y. Genome-wide analysis and expression profiles of the StR2R3-MYB transcription factor superfamily in potato (Solanum tuberosum L.). Int. J. Biol. Macromol. 2020, 148, 817–832. [Google Scholar] [CrossRef]
- Wu, W.; Zhu, S.; Zhu, L.; Wang, D.; Liu, Y.; Liu, S.; Zhang, J.; Hao, Z.; Lu, Y.; Cheng, T.; et al. Characterization of the Liriodendron Chinense MYB Gene Family and Its Role in Abiotic Stress Response. Front. Plant Sci. 2021, 12, 641280. [Google Scholar] [CrossRef]
- Wang, J.; Liu, Y.; Bingqian, T.; Dai, X.; Xie, L.; Liu, F.; Zou, X. Genome-wide identification and capsaicinoid biosynthesis-related expression analysis of the R2R3-MYB gene family in Capsicum annuum L. Front. Genet. 2020, 11, 598183. [Google Scholar] [CrossRef]
- Katiyar, A.; Smita, S.; Lenka, S.K.; Rajwanshi, R.; Chinnusamy, V.; Bansal, K.C. Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genom. 2012, 13, 544. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Zhang, L.; Chen, P.; Liang, T.; Li, X.; Liu, H. UV-B photoreceptor UVR8 interacts with MYB73/MYB77 to regulate auxin responses and lateral root development. Embo J. 2020, 39, e101928. [Google Scholar] [CrossRef]
- Kim, J.H.; Nguyen, N.H.; Jeong, C.Y.; Nguyen, N.T.; Hong, S.W.; Lee, H. Loss of the R2R3 MYB, AtMyb73, causes hyper-induction of the SOS1 and SOS3 genes in response to high salinity in Arabidopsis. J. Plant Physiol. 2013, 170, 1461–1465. [Google Scholar] [CrossRef]
- Zou, C.; Li, L.; Miki, D.; Li, D.; Tang, Q.; Xiao, L.; Rajput, S.; Deng, P.; Peng, L.; Jia, W.; et al. The genome of broomcorn millet. Nat. Commun. 2019, 10, 436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, K.; Li, F.; Chen, J.; Li, Z.; Lin, W.; Cai, S.; Liu, J.; Lin, W. Asymmetric evolution and expansion of the NAC transcription factor in polyploidized Cotton. Front. Plant Sci. 2018, 9, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, H.; Zhang, L.; Liu, L.; Tang, X.F.; Yang, W.J.; Wu, Y.M.; Huang, Y.B.; Tang, Y.X. Biochemical and molecular characterization of plant MYB transcription factor family. Biochemistry 2009, 74, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Govindan, G.; Sankararamasubramanian, H.; Harikrishnan, M.; Ganpudi, A.; Parida, A. A MYB transcription factor from the grey mangrove is induced by stress and confers NaCl tolerance in tobacco. J. Exp. Bot. 2012, 63, 4549–4561. [Google Scholar]
- Ullah, A.; Qamar, M.T.U.; Nisar, M.; Hazrat, A.; Rahim, G.; Khan, A.H.; Hayat, K.; Ahmed, S.; Ali, W.; Khan, A.; et al. Characterization of a novel cotton MYB gene, GhMYB108-like responsive to abiotic stresses. Mol. Biol. Rep. 2020, 47, 1573–1581. [Google Scholar] [CrossRef]
- Zhang, C.-Y.; Liu, H.-C.; Zhang, X.-S.; Guo, Q.-X.; Bian, S.-M.; Wang, J.-Y.; Zhai, L.-L. VcMYB4a, an R2R3-MYB transcription factor from Vaccinium corymbosum, negatively regulates salt, drought, and temperature stress. Gene 2020, 757, 144935. [Google Scholar] [CrossRef]
- Gao, F.; Zhao, H.X.; Yao, H.P.; Li, C.L.; Chen, H.; Wang, A.H.; Park, S.U.; Wu, Q. Identification, isolation and expression analysis of eight stress-related R2R3-MYB genes in tartary buckwheat (Fagopyrum tataricum). Plant Cell Rep. 2016, 35, 1385–1396. [Google Scholar] [CrossRef]
- Lippold, F.; Sanchez, D.H.; Musialak, M.; Schlereth, A.; Scheible, W.-R.; Hincha, D.K.; Udvardi, M.K. AtMyb41 Regulates Transcriptional and Metabolic Responses to Osmotic Stress in Arabidopsis. Plant Physiol. 2009, 149, 1761–1772. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Liu, X.; Wang, X.; Zhou, M.; Zhou, X.; Ye, X.; Wei, X. An R2R3 MYB transcription factor in wheat, TaPIMP1, mediates host resistance to Bipolaris sorokiniana and drought stresses through regulation of defense- and stress-related genes. New Phytol. 2012, 196, 1155–1170. [Google Scholar] [CrossRef]
- Denekamp, M.; Smeekens, S.C. Integration of Wounding and Osmotic Stress Signals Determines the Expression of the AtMYB102 Transcription Factor Gene. Plant Physiol. 2003, 132, 1415–1423. [Google Scholar] [CrossRef] [Green Version]
- Xiong, L.; Schumaker, K.S.; Zhu, J.-K. Cell Signaling during Cold, Drought, and Salt Stress. Plant Cell 2002, 14, S165–S183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 2006, 57, 781–803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vannini, C.; Locatelli, F.; Bracale, M.; Magnani, E.; Marsoni, M.; Osnato, M.; Mattana, M.; Baldoni, E.; Coraggio, I. Overexpression of the rice Osmyb4 gene increases chilling and freezing tolerance of Arabidopsis thaliana plants. Plant J. 2004, 37, 115–127. [Google Scholar] [CrossRef]
- Ascencio-Ibañez, J.; Sozzani, R.; Lee, T.J.; Chu, T.M.; Wolfinger, R.; Cella, R.; Hanley-Bowdoin, L. Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 2008, 148, 436–454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, D.; Cheng, H.; Wu, W.; Soo, H.M.; Peng, J. Gibberellin Mobilizes Distinct DELLA-Dependent Transcriptomes to Regulate Seed Germination and Floral Development in Arabidopsis. Plant Physiol. 2006, 142, 509–525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, Z.; Li, S.; An, X.; Liu, X.; Qin, H.; Wang, D. Transgenic expression of MYB15 confers enhanced sensitivity to abscisic acid and improved drought tolerance in Arabidopsis thaliana. J. Genet. Genom. 2009, 36, 17–29. [Google Scholar] [CrossRef]
- Zhang, L.; Zhao, G.; Jia, J.; Liu, X.; Kong, X. Molecular characterization of 60 isolated wheat MYB genes and analysis of their expression during abiotic stress. J. Exp. Bot. 2011, 63, 203–214. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Zhang, H.; Xin, D.W.; Wang, L.L.; Zhang, L.W.; Liu, C.Y.; Chen, Q.S.; Hu, G.H. Domain analysis and function prediction of TCP transcription factors family in soybean. Soybean Sci. 2012, 31, 707–717. [Google Scholar]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chinnusamy, V.; Ohta, M.; Kanrar, S.; Lee, B.H.; Hong, X.; Agarwal, M.; Zhu, J.K. ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003, 17, 1043–1054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hannah, M.A.; Heyer, A.G.; Hincha, D.K. A Global Survey of Gene Regulation during Cold Acclimation in Arabidopsis thaliana. PLoS Genet. 2005, 1, e26. [Google Scholar] [CrossRef]
- Chen, X.; Mao, X.; Huang, P.; Fang, S. Morphological characterization of flower buds development and related gene expression profiling at bud break stage in heterodichogamous Cyclocarya paliurus (Batal.) lljinskaja. Genes 2019, 10, 818. [Google Scholar] [CrossRef] [Green Version]
- Penfield, S. MYB61 is required for mucilage deposition and extrusion in the Arabidopsis seed coat. Plant Cell Online 2001, 13, 2777–2791. [Google Scholar] [CrossRef] [Green Version]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; Zhang, L.; Liu, Y.; Shang, X.; Fang, S. Identification and Expression Analysis of R2R3-MYB Family Genes Associated with Salt Tolerance in Cyclocarya paliurus. Int. J. Mol. Sci. 2022, 23, 3429. https://doi.org/10.3390/ijms23073429
Zhang Z, Zhang L, Liu Y, Shang X, Fang S. Identification and Expression Analysis of R2R3-MYB Family Genes Associated with Salt Tolerance in Cyclocarya paliurus. International Journal of Molecular Sciences. 2022; 23(7):3429. https://doi.org/10.3390/ijms23073429
Chicago/Turabian StyleZhang, Zijie, Lei Zhang, Yang Liu, Xulan Shang, and Shengzuo Fang. 2022. "Identification and Expression Analysis of R2R3-MYB Family Genes Associated with Salt Tolerance in Cyclocarya paliurus" International Journal of Molecular Sciences 23, no. 7: 3429. https://doi.org/10.3390/ijms23073429
APA StyleZhang, Z., Zhang, L., Liu, Y., Shang, X., & Fang, S. (2022). Identification and Expression Analysis of R2R3-MYB Family Genes Associated with Salt Tolerance in Cyclocarya paliurus. International Journal of Molecular Sciences, 23(7), 3429. https://doi.org/10.3390/ijms23073429





