Genetic Manipulation in Mucorales and New Developments to Study Mucormycosis
Abstract
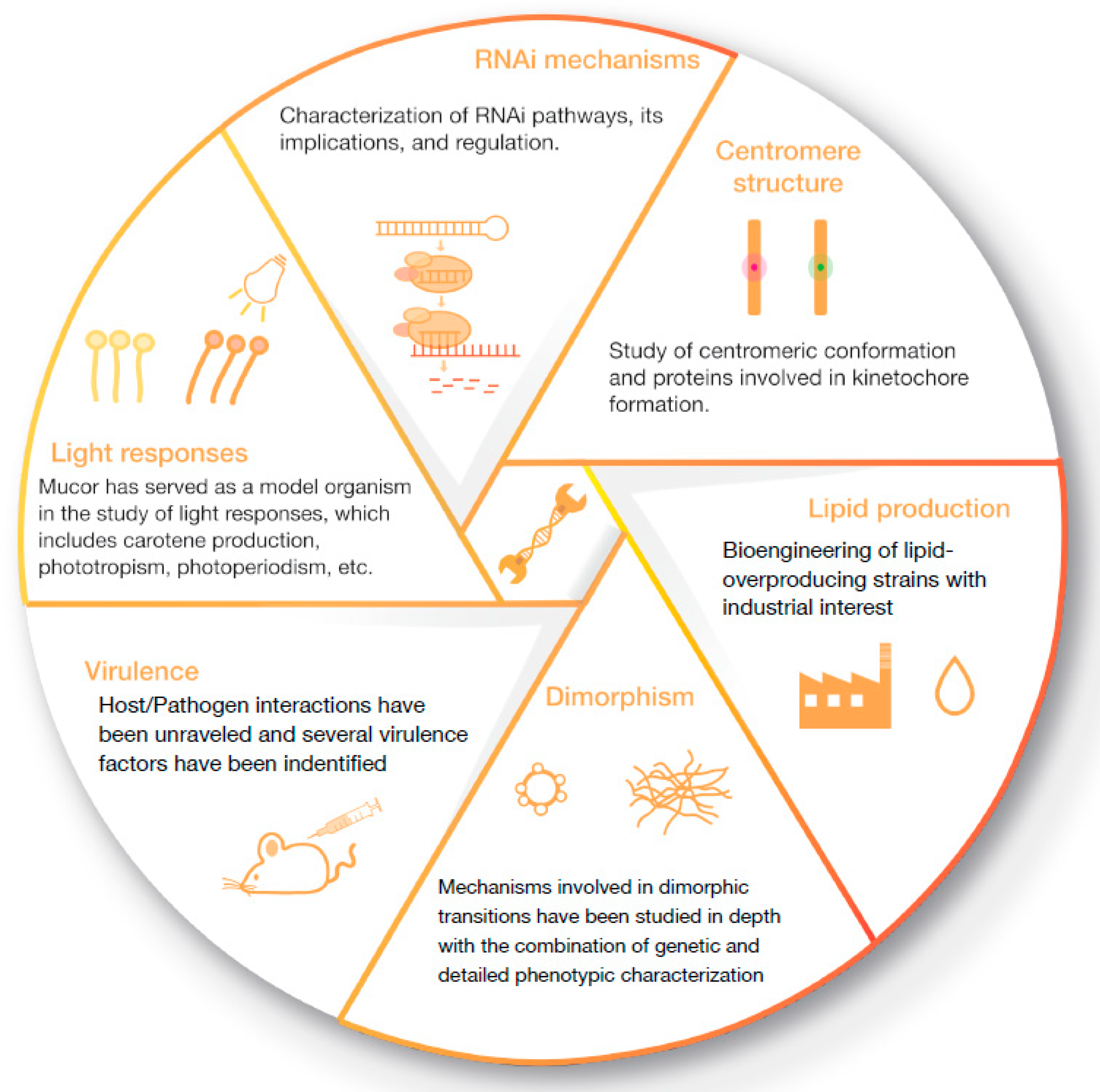
1. Introduction
2. Mucor lusitanicus, the Primary Genetic Model in Mucorales
2.1. Plasmid Transformation, RNAi, and Functional Genomics
2.2. Homologous Recombination and Its Derived Genetic Tools in M. lusitanicus
3. Rhizopus microsporus, the New Genetic Model to Study Mucormycosis
3.1. A New R. microsporus Strain for Genetic Transformation: Auxotrophic Isolation and Plasmid Transformation
3.2. Development of a Stable Homologous Recombination Strategy Based on the CRISPR-Cas9 Machinery in R. microsporus
3.3. Uracil Auxotrophy Is Directly Related to the Virulence of R. microsporus
4. Attempts to Transform Other Mucorales
4.1. Homologous Recombination in Rhizopus delemar
4.2. CRISPR-Cas9-Based Mutagenesis in the Fungus Lichtheimia corymbifera
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cerdá-Olmedo, E. Phycomyces and the biology of light and color. FEMS Microbiol. Rev. 2001, 25, 503–512. [Google Scholar] [CrossRef] [PubMed]
- Cerda-Olmedo, E.; Lipson, E. Phycomyces; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 1987. [Google Scholar]
- Obraztsova, I.N.; Prados, N.; Holzmann, K.; Avalos, J.; Cerdá-Olmedo, E. Genetic damage following introduction of DNA in Phycomyces. Fungal Genet. Biol. 2003, 41, 168–180. [Google Scholar] [CrossRef] [PubMed]
- Campbell, E.I.; Unkles, S.E.; Macro, J.A.; Hondel, C.V.D.; Contreras, R.; Kinghorn, J.R. Improved transformation efficiency of Aspergillus niger using the homologous niaD gene for nitrate reductase. Curr. Genet. 1989, 16, 53–56. [Google Scholar] [CrossRef] [PubMed]
- Kawai, S.; Hashimoto, W.; Murata, K. Transformation of Saccharomyces cerevisiae and other fungi: Methods and possible underlying mechanism. Bioeng. Bugs 2010, 1, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, E.; Sutton, D.A.; Cano, J.; Fothergill, A.W.; Stchigel, A.; Rinaldi, M.G.; Guarro, J. Spectrum of Zygomycete Species Identified in Clinically Significant Specimens in the United States. J. Clin. Microbiol. 2009, 47, 1650–1656. [Google Scholar] [CrossRef]
- Roden, M.M.; Zaoutis, T.E.; Buchanan, W.L.; Knudsen, T.A.; Sarkisova, T.A.; Schaufele, R.L.; Sein, M.; Sein, T.; Chiou, C.C.; Chu, J.H.; et al. Epidemiology and Outcome of Zygomycosis: A Review of 929 Reported Cases. Clin. Infect. Dis. 2005, 41, 634–653. [Google Scholar] [CrossRef] [PubMed]
- Cornely, O.A.; Alastruey-Izquierdo, A.; Arenz, D.; Chen, S.C.A.; Dannaoui, E.; Hochhegger, B.; Hoenigl, M.; Jensen, H.E.; Lagrou, K.; Lewis, R.E.; et al. Global guideline for the diagnosis and management of mucormycosis: An initiative of the European Confederation of Medical Mycology in cooperation with the Mycoses Study Group Education and Research Consortium. Lancet Infect. Dis. 2019, 19, e405–e421. [Google Scholar] [CrossRef]
- Chayakulkeeree, M.; Ghannoum, M.A.; Perfect, J.R. Zygomycosis: The re-emerging fungal infection. Eur. J. Clin. Microbiol. 2006, 25, 215–229. [Google Scholar] [CrossRef]
- Kontoyiannis, D.P. Antifungal Resistance: An Emerging Reality and A Global Challenge. J. Infect. Dis. 2017, 216, S431–S435. [Google Scholar] [CrossRef]
- Veisi, A.; Bagheri, A.; Eshaghi, M.; Rikhtehgar, M.H.; Kanavi, M.R.; Farjad, R. Rhino-orbital mucormycosis during steroid therapy in COVID-19 patients: A case report. Eur. J. Ophthalmol. 2021. [Google Scholar] [CrossRef]
- John, T.; Jacob, C.; Kontoyiannis, D. When Uncontrolled Diabetes Mellitus and Severe COVID-19 Converge: The Perfect Storm for Mucormycosis. J. Fungi 2021, 7, 298. [Google Scholar] [CrossRef] [PubMed]
- Sridhara, S.R.; Paragache, G.; Panda, N.K.; Chakrabarti, A. Mucormycosis in Immunocompetent Individuals: An Increasing Trend. J. Otolaryngol. 2005, 34, 402–406. [Google Scholar] [CrossRef] [PubMed]
- Prakash, H.; Chakrabarti, A. Global Epidemiology of Mucormycosis. J. Fungi 2019, 5, 26. [Google Scholar] [CrossRef] [PubMed]
- Jeong, W.; Keighley, C.; Wolfe, R.; Lee, W.L.; Slavin, M.A.; Kong, D.C.M.; Chen, S.C.-A. The epidemiology and clinical manifestations of mucormycosis: A systematic review and meta-analysis of case reports. Clin. Microbiol. Infect. 2019, 25, 26–34. [Google Scholar] [CrossRef]
- Hassan, M.A.; Voigt, K. Pathogenicity patterns of mucormycosis: Epidemiology, interaction with immune cells and virulence factors. Med Mycol. 2019, 57, S245–S256. [Google Scholar] [CrossRef]
- Dannaoui, E. Antifungal resistance in mucorales. Int. J. Antimicrob. Agents 2017, 50, 617–621. [Google Scholar] [CrossRef]
- Caramalho, R.; Tyndall, J.D.A.; Monk, B.C.; Larentis, T.; Lass-Flörl, C.; Lackner, M. Intrinsic short-tailed azole resistance in mucormycetes is due to an evolutionary conserved aminoacid substitution of the lanosterol 14α-demethylase. Sci. Rep. 2017, 7, 15898. [Google Scholar] [CrossRef]
- Caetano, L.A.; Faria, T.; Springer, J.; Loeffler, J.; Viegas, C. Antifungal-resistant Mucorales in different indoor environments. Mycology 2019, 10, 75–83. [Google Scholar] [CrossRef]
- Luo, G.; Gebremariam, T.; Lee, H.; French, S.W.; Wiederhold, N.; Patterson, T.F.; Filler, S.G.; Ibrahim, A.S. Efficacy of Liposomal Amphotericin B and Posaconazole in Intratracheal Models of Murine Mucormycosis. Antimicrob. Agents Chemother. 2013, 57, 3340–3347. [Google Scholar] [CrossRef]
- Maurer, E.; Binder, U.; Sparber, M.; Lackner, M.; Caramalho, R.; Lass-Flörl, C. Susceptibility profiles of amphotericin B and posaconazole against clinically relevant Mucorales species under hypoxic conditions. Antimicrob. Agents Chemother. 2015, 59, 1344–1346. [Google Scholar] [CrossRef][Green Version]
- Calo, S.; Shertz-Wall, C.; Lee, S.C.; Bastidas, R.J.; Nicolas, F.E.; Granek, J.A.; Mieczkowski, P.; Torres-Martínez, S.; Ruiz-Vázquez, R.M.; Cardenas, M.E.; et al. Antifungal drug resistance evoked via RNAi-dependent epimutations. Nature 2014, 513, 555–558. [Google Scholar] [CrossRef] [PubMed]
- Trieu, T.A.; Navarro-Mendoza, M.I.; Perez-Arques, C.; Sanchis, M.; Capilla, J.; Navarro-Rodríguez, P.; Lopez-Fernandez, L.; Torres-Martínez, S.; Garre, V.; Ruiz-Vázquez, R.M.; et al. RNAi-Based Functional Genomics Identifies New Virulence Determinants in Mucormycosis. PLoS Pathog. 2017, 13, e1006150. [Google Scholar] [CrossRef] [PubMed]
- Gebremariam, T.; Alkhazraji, S.; Soliman, S.S.M.; Gu, Y.; Jeon, H.H.; Zhang, L.; French, S.W.; Stevens, D.A.; Edwards, J.E.; Filler, S.G.; et al. Anti-CotH3 antibodies protect mice from mucormycosis by prevention of invasion and augmenting opsonophagocytosis. Sci. Adv. 2019, 5, eaaw1327. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Arques, C.; Navarro-Mendoza, M.I.; Murcia, L.; Lax, C.; Martínez-García, P.; Heitman, J.; Nicolás, F.E.; Garre, V. Mucor circinelloides Thrives inside the Phagosome through an Atf-Mediated Germination Pathway. mBio 2019, 10, 02765-18. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Arques, C.; Navarro-Mendoza, M.; Murcia, L.; Navarro, E.; Garre, V.; Nicolás, F. The RNAi Mechanism Regulates a New Exonuclease Gene Involved in the Virulence of Mucorales. Int. J. Mol. Sci. 2021, 22, 2282. [Google Scholar] [CrossRef]
- Pérez-Arques, C.; Navarro-Mendoza, M.; Murcia, L.; Lax, C.; Sanchis, M.; Capilla, J.; Navarro, E.; Garre, V.; Nicolás, F. A Mucoralean White Collar-1 Photoreceptor Controls Virulence by Regulating an Intricate Gene Network during Host Interactions. Microorganisms 2021, 9, 459. [Google Scholar] [CrossRef]
- López-Fernández, L.; Sanchis, M.; Navarro-Rodríguez, P.; Nicolás, F.E.; Silva-Franco, F.; Guarro, J.; Garre, V.; Navarro-Mendoza, M.I.; Pérez-Arques, C.; Capilla, J. Understanding Mucor circinelloides pathogenesis by comparative genomics and phenotypical studies. Virulence 2018, 9, 707–720. [Google Scholar] [CrossRef]
- Lax, C.; Pérez-Arques, C.; Navarro-Mendoza, M.I.; Cánovas-Márquez, J.T.; Tahiri, G.; Pérez-Ruiz, J.A.; Osorio-Concepción, M.; Murcia-Flores, L.; Navarro, E.; Garre, V.; et al. Genes, pathways, and mechanisms involved in the virulence of mucorales. Genes 2020, 11, 317. [Google Scholar] [CrossRef]
- Binder, U.; Navarro-Mendoza, M.I.; Naschberger, V.; Bauer, I.; Nicolas, F.E.; Pallua, J.D.; Lass-Flörl, C.; Garre, V. Generation of a mucor circinelloides reporter strain—A promising new tool to study antifungal drug efficacy and mucormycosis. Genes 2018, 9, 613. [Google Scholar] [CrossRef]
- López-Muñoz, A.; Nicolás, F.E.; García-Moreno, D.; Pérez-Oliva, A.B.; Navarro-Mendoza, M.I.; Oñate, M.; Herrera-Estrella, A.; Torres-Martínez, S.; Ruiz-Vázquez, R.M.; Garre, V.; et al. An Adult Zebrafish Model Reveals that Mucormycosis Induces Apoptosis of Infected Macrophages. Sci. Rep. 2018, 8, 12. [Google Scholar] [CrossRef]
- Navarro-Mendoza, M.I.; Pérez-Arques, C.; Murcia, L.; Martínez-García, P.; Lax, C.; Sanchis, M.; Capilla, J.; Nicolás, F.E.; Garre, V. Components of a new gene family of ferroxidases involved in virulence are functionally specialized in fungal dimorphism. Sci. Rep. 2018, 8, 76. [Google Scholar] [CrossRef] [PubMed]
- Wagner, L.; Stielow, J.; De Hoog, G.; Bensch, K.; Schwartze, V.U.; Voigt, K.; Alastruey-Izquierdo, A.; Kurzai, O.; Walther, G. A new species concept for the clinically relevant Mucor circinelloides complex. Persoonia Mol. Phylogeny Evol. Fungi 2020, 44, 67–97. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, F.E.; Torres-Martínez, S.; Ruiz-Vázquez, R.M. Two classes of small antisense RNAs in fungal RNA silencing triggered by non-integrative transgenes. EMBO J. 2003, 22, 3983–3991. [Google Scholar] [CrossRef] [PubMed]
- Trieu, T.A.; Calo, S.; Nicolas, F.E.; Vila, A.; Moxon, S.; Dalmay, T.; Torres-Martínez, S.; Garre, V.; Ruiz-Vázquez, R.M. A Non-canonical RNA Silencing Pathway Promotes mRNA Degradation in Basal Fungi. PLoS Genet. 2015, 11, e1005168. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Mendoza, M.I.; Pérez-Arques, C.; Panchal, S.; Nicolás, F.E.; Mondo, S.J.; Ganguly, P.; Pangilinan, J.; Grigoriev, I.V.; Heitman, J.; Sanyal, K.; et al. Early Diverging Fungus Mucor circinelloides Lacks Centromeric Histone CENP-A and Displays a Mosaic of Point and Regional Centromeres. Curr. Biol. 2019, 29, 3791–3802.e6. [Google Scholar] [CrossRef]
- Li, C.H.; Cervantes, M.; Springer, D.J.; Boekhout, T.; Ruiz-Vazquez, R.M.; Torres-Martinez, S.R.; Heitman, J.; Lee, S.C. Sporangiospore Size Dimorphism Is Linked to Virulence of Mucor circinelloides. PLoS Pathog. 2011, 7, e1002086. [Google Scholar] [CrossRef]
- Valle-Maldonado, M.I.; Patiño-Medina, J.A.; Pérez-Arques, C.; Reyes-Mares, N.Y.; Jácome-Galarza, I.E.; Ortíz-Alvarado, R.; Vellanki, S.; Ramírez-Díaz, M.I.; Lee, S.C.; Garre, V.; et al. The heterotrimeric G-protein beta subunit Gpb1 controls hyphal growth under low oxygen conditions through the protein kinase A pathway and is essential for virulence in the fungus Mucor circinelloides. Cell. Microbiol. 2020, 22, e13236. [Google Scholar] [CrossRef]
- Lax, C.; Navarro-Mendoza, M.I.; Pérez-Arques, C.; Navarro, E.; Nicolás, F.E.; Garre, V. Stable and reproducible homologous recombination enables CRISPR-based engineering in the fungus Rhizopus microsporus. Cell Rep. Methods 2021, 100124. [Google Scholar] [CrossRef]
- Hodgkin, J. The model organism diaspora. Heredity 2019, 123, 14–17. [Google Scholar] [CrossRef]
- van Heeswijck, R.; Roncero, M.I.G. High frequency transformation of Mucor with recombinant plasmid DNA. Carlsberg Res. Commun. 1984, 49, 691–702. [Google Scholar] [CrossRef]
- Vellanki, S.; Navarro-Mendoza, M.I.; Garcia, A.; Murcia, L.; Pérez-Arques, C.; Garre, V.; Nicolas, F.E.; Lee, S.C. Mucor circinelloides: Growth, Maintenance, and Genetic Manipulation. Curr. Protoc. Microbiol. 2018, 49, e53. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, A.; López-García, S.; Garre, V. High reliability transformation of the basal fungus Mucor circinelloides by electroporation. J. Microbiol. Methods 2011, 84, 442–446. [Google Scholar] [CrossRef] [PubMed]
- Silva, F.; Torres-Martinez, S.; Garre, V. Distinct white collar-1 genes control specific light responses in Mucor circinelloides. Mol. Microbiol. 2006, 61, 1023–1037. [Google Scholar] [CrossRef] [PubMed]
- Silva, F.; Navarro, E.; Peñaranda, A.; Murcia-Flores, L.; Torres-Martínez, S.; Garre, V. A RING-finger protein regulates carotenogenesis via proteolysis-independent ubiquitylation of a White Collar-1-like activator. Mol. Microbiol. 2008, 70, 1026–1036. [Google Scholar] [CrossRef]
- Torres-Martínez, S.; Ruiz-Vázquez, R.M. RNAi pathways in Mucor: A tale of proteins, small RNAs and functional diversity. Fungal. Genet. Biol. 2016, 90, 44–52. [Google Scholar] [CrossRef]
- Zhao, L.; Cánovas-Márquez, J.T.; Tang, X.; Chen, H.; Chen, Y.Q.; Chen, W.; Garre, V.; Song, Y.; Ratledge, C. Role of malate transporter in lipid accumulation of oleaginous fungus Mucor circinelloides. Appl. Microbiol. Biotechnol. 2016, 100, 1297–1305. [Google Scholar] [CrossRef]
- Lee, S.C.; Li, A.; Calo, S.; Heitman, J. Calcineurin Plays Key Roles in the Dimorphic Transition and Virulence of the Human Pathogenic Zygomycete Mucor circinelloides. PLoS Pathog. 2013, 9, e1003625. [Google Scholar] [CrossRef]
- Navarro, E.; Lorca-Pascual, J.; Quiles-Rosillo, M.; Nicolas, F.; Garre, V.; Torres-Martínez, S.; Ruiz-Vázquez, R. A negative regulator of light-inducible carotenogenesis in Mucor circinelloides. Mol. Genet. Genom. 2001, 266, 463–470. [Google Scholar] [CrossRef]
- Zhang, Y.; Navarro, E.; Cánovas-Márquez, J.T.; Almagro, L.; Chen, H.; Chen, Y.Q.; Zhang, H.; Torres-Martínez, S.; Chen, W.; Garre, V. A new regulatory mechanism controlling carotenogenesis in the fungus Mucor circinelloides as a target to generate β-carotene over-producing strains by genetic engineering. Microb. Cell Factories 2016, 15, 1–14. [Google Scholar] [CrossRef]
- Roncero, M.I.G. Enrichment method for the isolation of auxotrophic mutants of Mucor using the polyene antibiotic N-glycosyl-polifungin. Carlsberg Res. Commun. 1984, 49, 685–690. [Google Scholar] [CrossRef]
- Anaya, N.; Roncero, M.I.G. Transformation of a methionine auxotrophic mutant of Mucor circinelloides by direct cloning of the corresponding wild type gene. MGG Mol. Gen. Genet. 1991, 230, 449–455. [Google Scholar] [CrossRef] [PubMed]
- Nicolás, F.E.; Ruiz-Vázquez, R.M. Functional Diversity of RNAi-Associated sRNAs in Fungi. Int. J. Mol. Sci. 2013, 14, 15348–15360. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Vázquez, R.M.; Nicolás, F.E.; Torres-Martínez, S.; Garre, V. Distinct RNAi Pathways in the Regulation of Physiology and Development in the Fungus Mucor circinelloides. Adv. Genet. 2015, 91, 55–102. [Google Scholar] [CrossRef] [PubMed]
- Cánovas-Márquez, J.; Navarro-Mendoza, M.; Pérez-Arques, C.; Lax, C.; Tahiri, G.; Pérez-Ruiz, J.; Lorenzo-Gutiérrez, D.; Calo, S.; López-García, S.; Navarro, E.; et al. Role of the Non-Canonical RNAi Pathway in the Antifungal Resistance and Virulence of Mucorales. Genes 2021, 12, 586. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Arques, C.; Navarro-Mendoza, M.I.; Murcia, L.; Navarro, E.; Garre, V.; Nicolás, F.E. A non-canonical RNAi pathway controls virulence and genome stability in Mucorales. PLoS Genet. 2020, 16, e1008611. [Google Scholar] [CrossRef]
- Calo, S.; Nicolás, F.E.; Vila, A.; Torres-Martínez, S.; Ruiz-Vázquez, R.M. Two distinct RNA-dependent RNA polymerases are required for initiation and amplification of RNA silencing in the basal fungus Mucor circinelloides. Mol. Microbiol. 2011, 83, 379–394. [Google Scholar] [CrossRef]
- Papp, T.; Velayos, A.; Bartók, T.; Eslava, A.P.; Vágvölgyi, C.; Iturriaga, E.A. Heterologous expression of astaxanthin biosynthesis genes in Mucor circinelloides. Appl. Microbiol. Biotechnol. 2006, 69, 526–531. [Google Scholar] [CrossRef]
- Naz, T.; Nosheen, S.; Li, S.; Nazir, Y.; Mustafa, K.; Liu, Q.; Garre, V.; Song, Y. Comparative analysis of β-carotene production by mucor circinelloides strains CBS 277.49 and WJ11 under light and dark conditions. Metabolites 2020, 10, 38. [Google Scholar] [CrossRef]
- de Haro, J.P.; Calo, S.; Cervantes, M.; Nicolas, F.E.; Torres-Martínez, S.; Ruiz-Vázquez, R.M. A Single dicer Gene Is Required for Efficient Gene Silencing Associated with Two Classes of Small Antisense RNAs in Mucor circinelloides. Eukaryot. Cell 2009, 8, 1486–1497. [Google Scholar] [CrossRef]
- Cervantes, M.; Vila, A.; Nicolas, F.E.; Moxon, S.; De Haro, J.P.; Dalmay, T.; Torres-Martínez, S.; Ruiz-Vázquez, R.M. A Single Argonaute Gene Participates in Exogenous and Endogenous RNAi and Controls Cellular Functions in the Basal Fungus Mucor circinelloides. PLoS ONE 2013, 8, e69283. [Google Scholar] [CrossRef]
- Larsen, G.G.; Appel, K.F.; Wolff, A.-M.; Nielsen, J. Characterisation of the Mucor circinelloides regulated promoter gpd1P. Curr. Genet. 2004, 45, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Li, S.; Khan, A.K.; Garre, V.; Vongsangnak, W.; Song, Y. Increased Lipid Accumulation in Mucor circinelloides by Overexpression of Mitochondrial Citrate Transporter Genes. Ind. Eng. Chem. Res. 2019, 58, 2125–2134. [Google Scholar] [CrossRef]
- Arnau, J.; Jepsen, L.P.; Strømani, P. Integrative transformation by homologous recombination in the zygomycete Mucor circinelloides. Mol. Gen. Genet. MGG 1991, 225, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Thompson, L.H.; Schild, D. Homologous recombinational repair of DNA ensures mammalian chromosome stability. Mutat. Res. Mol. Mech. Mutagen. 2001, 477, 131–153. [Google Scholar] [CrossRef]
- Arnau, J.; Strøman, P. Gene replacement and ectopic integration in the zygomycete Mucor circinelloides. Curr. Genet. 1993, 23, 542–546. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, F.E.; De Haro, J.P.; Torres-Martínez, S.; Ruiz-Vázquez, R.M. Mutants defective in a Mucor circinelloides dicer-like gene are not compromised in siRNA silencing but display developmental defects. Fungal Genet. Biol. 2007, 44, 504–516. [Google Scholar] [CrossRef] [PubMed]
- Calo, S.; Nicolás, F.E.; Lee, S.C.; Vila, A.; Cervantes, M.; Torres-Martinez, S.; Ruiz-Vazquez, R.M.; Cardenas, M.E.; Heitman, J. A non-canonical RNA degradation pathway suppresses RNAi-dependent epimutations in the human fungal pathogen Mucor circinelloides. PLoS Genet. 2017, 13, e1006686. [Google Scholar] [CrossRef]
- Nicolas, F.E.; Moxon, S.; de Haro, J.P.; Calo, S.; Grigoriev, I.V.; Torres-Martínez, S.; Moulton, V.; Ruiz-Vázquez, R.M.; Dalmay, T. Endogenous short RNAs generated by Dicer 2 and RNA-dependent RNA polymerase 1 regulate mRNAs in the basal fungus Mucor circinelloides. Nucleic Acids Res. 2010, 38, 5535–5541. [Google Scholar] [CrossRef]
- Nicolás, F.E.; Torres-Martínez, S.; Ruiz-Vázquez, R.M. Transcriptional activation increases RNA silencing efficiency and stability in the fungus Mucor circinelloides. J. Biotechnol. 2009, 142, 123–126. [Google Scholar] [CrossRef]
- Nicolás, F.E.; Torres-Martínez, S.; Ruiz-Vázquez, R.M. Loss and Retention of RNA Interference in Fungi and Parasites. PLoS Pathog. 2013, 9, e1003089. [Google Scholar] [CrossRef]
- Nicolás, F.E.; Vila, A.; Moxon, S.; Cascales, M.D.; Torres-Martínez, S.; Ruiz-Vázquez, R.M.; Garre, V. The RNAi machinery controls distinct responses to environmental signals in the basal fungus Mucor circinelloides. BMC Genom. 2015, 16, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Chang, Z.; Heitman, J. Drug-Resistant Epimutants Exhibit Organ-Specific Stability and Induction during Murine Infections Caused by the Human Fungal Pathogen Mucor circinelloides. mBio 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Chang, Z.; Billmyre, R.B.; Lee, S.C.; Heitman, J. Broad antifungal resistance mediated by RNAi-dependent epimutation in the basal human fungal pathogen Mucor circinelloides. PLoS Genet. 2019, 15, e1007957. [Google Scholar] [CrossRef] [PubMed]
- Cánovas-Márquez, J.T.; Falk, S.; E Nicolás, F.; Padmanabhan, S.; Zapata-Pérez, R.; Sánchez-Ferrer, Á.; Navarro, E.; Garre, V. A ribonuclease III involved in virulence of Mucorales fungi has evolved to cut exclusively single-stranded RNA. Nucleic Acids Res. 2021, 49, 5294–5307. [Google Scholar] [CrossRef]
- Osorio-Concepción, M.; Lax, C.; Navarro, E.; Nicolás, F.E.; Garre, V. DNA Methylation on N6-Adenine Regulates the Hyphal Development during Dimorphism in the Early-Diverging Fungus Mucor lusitanicus. J. Fungi 2021, 7, 738. [Google Scholar] [CrossRef]
- Lee, S.C.; Li, A.; Calo, S.; Inoue, M.; Tonthat, N.K.; Bain, J.M.; Louw, J.; Shinohara, M.L.; Erwig, L.P.; Schumacher, M.A.; et al. Calcineurin orchestrates dimorphic transitions, antifungal drug responses and host-pathogen interactions of the pathogenic mucoralean fungus Mucor circinelloides. Mol. Microbiol. 2015, 97, 844–865. [Google Scholar] [CrossRef]
- Ocampo, J.; Nuñez, L.F.; Silva, F.; Pereyra, E.; Moreno, S.; Garre, V.; Rossi, S. A Subunit of Protein Kinase A Regulates Growth and Differentiation in the Fungus Mucor circinelloides. Eukaryot. Cell 2009, 8, 933–944. [Google Scholar] [CrossRef]
- Patiño-Medina, J.A.; Maldonado-Herrera, G.; Pérez-Arques, C.; Alejandre-Castañeda, V.; Reyes-Mares, N.Y.; Valle-Maldonado, M.I.; Campos-García, J.; Ortiz-Alvarado, R.; Jácome-Galarza, I.E.; Ramírez-Díaz, M.I.; et al. Control of morphology and virulence by ADP-ribosylation factors (Arf) in Mucor circinelloides. Curr. Genet. 2017, 64, 853–869. [Google Scholar] [CrossRef]
- Nicolás, F.E.; Navarro-Mendoza, M.I.; Pérez-Arques, C.; López-García, S.; Navarro, E.; Torres-Martínez, S.; Garre, V. Molecular tools for carotenogenesis analysis in the mucoral Mucor circinelloides. In Methods in Molecular Biology; Humana Press Inc.: New York, NY, USA, 2018; Volume 1852, pp. 221–237. [Google Scholar]
- Yang, J.; Cánovas-Márquez, J.T.; Li, P.; Li, S.; Niu, J.; Wang, X.; Nazir, Y.; López-García, S.; Garre, V.; Song, Y. Deletion of Plasma Membrane Malate Transporters Increased Lipid Accumulation in the Oleaginous Fungus Mucor circinelloides WJ11. J. Agric. Food Chem. 2021, 69, 9632–9641. [Google Scholar] [CrossRef]
- Rodríguez-Frómeta, R.A.; Gutiérrez, A.; Torres-Martínez, S.; Garre, V. Malic enzyme activity is not the only bottleneck for lipid accumulation in the oleaginous fungus Mucor circinelloides. Appl. Microbiol. Biotechnol. 2012, 97, 3063–3072. [Google Scholar] [CrossRef]
- Hussain, S.A.; Hameed, A.; Khan, A.K.; Zhang, Y.; Zhang, H.; Garre, V.; Song, Y. Engineering of Fatty Acid Synthases (FASs) to Boost the Production of Medium-Chain Fatty Acids (MCFAs) in Mucor circinelloides. Int. J. Mol. Sci. 2019, 20, 786. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.K.; Yang, J.; Hussain, S.A.; Zhang, H.; Garre, V.; Song, Y. Genetic Modification of Mucor circinelloides to Construct Stearidonic Acid Producing Cell Factory. Int. J. Mol. Sci. 2019, 20, 1683. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.K.; Yang, J.; Hussain, S.A.; Zhang, H.; Liang, L.; Garre, V.; Song, Y. Construction of DGLA producing cell factory by genetic modification of Mucor circinelloides. Microb. Cell Factories 2019, 18, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Mondo, S.J.; Lastovetsky, O.; Gaspar, M.L.; Schwardt, N.H.; Barber, C.C.; Riley, R.; Sun, H.; Grigoriev, I.V.; Pawlowska, T.E. Bacterial endosymbionts influence host sexuality and reveal reproductive genes of early divergent fungi. Nat. Commun. 2017, 8, 1–9. [Google Scholar] [CrossRef]
- Lastovetsky, O.A.; Gaspar, M.L.; Mondo, S.J.; LaButti, K.M.; Sandor, L.; Grigoriev, I.V.; Henry, S.A.; Pawlowska, T.E. Lipid metabolic changes in an early divergent fungus govern the establishment of a mutualistic symbiosis with endobacteria. Proc. Natl. Acad. Sci. USA 2016, 113, 15102–15107. [Google Scholar] [CrossRef] [PubMed]
- Aanen, D.K.; Hoekstra, R.F. The evolution of obligate mutualism: If you can’t beat ’em, join ’em. Trends Ecol. Evol. 2007, 22, 506–509. [Google Scholar] [CrossRef]
- Partida-Martinez, L.P.; Hertweck, C. Pathogenic fungus harbours endosymbiotic bacteria for toxin production. Nature 2005, 437, 884–888. [Google Scholar] [CrossRef]
- Ibrahim, A.S.; Gebremariam, T.; Liu, M.; Chamilos, G.; Kontoyiannis, D.P.; Mink, R.; Kwon-Chung, K.J.; Fu, Y.; Skory, C.D.; Edwards, J.E., Jr.; et al. Bacterial Endosymbiosis Is Widely Present among Zygomycetes but Does Not Contribute to the Pathogenesis of Mucormycosis. J. Infect. Dis. 2008, 198, 1083–1090. [Google Scholar] [CrossRef]
- Nicolás, F.E.; Murcia, L.; Navarro, E.; Navarro-Mendoza, M.I.; Pérez-Arques, C.; Garre, V. Mucorales Species and Macrophages. J. Fungi 2020, 6, 94. [Google Scholar] [CrossRef]
- Al Abdallah, Q.; Ge, W.; Fortwendel, J.R. A Simple and Universal System for Gene Manipulation in Aspergillus fumigatus: In Vitro -Assembled Cas9-Guide RNA Ribonucleoproteins Coupled with Microhomology Repair Templates. mSphere 2017, 2, e00446-17. [Google Scholar] [CrossRef]
- Yuzbashev, T.V.; Larina, A.S.; Vybornaya, T.V.; Yuzbasheva, E.Y.; Gvilava, I.T.; Sineoky, S.P. Repetitive genomic sequences as a substrate for homologous integration in the Rhizopus oryzae genome. Fungal Biol. 2015, 119, 494–502. [Google Scholar] [CrossRef] [PubMed]
- Roncero, M.I.G.; Jepsen, L.P.; Strøman, P.; van Heeswijck, R. Characterization of a leuA gene and an ARS element from Mucor circinelloides. Gene 1989, 84, 335–343. [Google Scholar] [CrossRef]
- Nicolás-Molina, F.E.; Navarro, E.; Ruiz-Vázquez, R.M. Lycopene over-accumulation by disruption of the negative regulator gene crgA in Mucor circinelloides. Appl. Microbiol. Biotechnol. 2007, 78, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Nicolã¡s, F.E.; Calo, S.; Murcia-Flores, L.; Garre, V.; Ruiz-Vã¡zquez, R.M.; Torres-Martãnez, S. A RING-finger photocarotenogenic repressor involved in asexual sporulation inMucor circinelloides. FEMS Microbiol. Lett. 2008, 280, 81–88. [Google Scholar] [CrossRef]
- D’Enfert, C.; Diaquin, M.; Delit, A.; Wuscher, N.; Debeaupuis, J.P.; Huerre, M.; Latge, J.P. Attenuated virulence of uridine-uracil auxotrophs of Aspergillus fumigatus. Infect. Immun. 1996, 64, 4401–4405. [Google Scholar] [CrossRef]
- Bain, J.M.; Stubberfield, C.; Gow, N.A.R. Ura-status-dependent adhesion of Candida albicans mutants. FEMS Microbiol. Lett. 2006, 204, 323–328. [Google Scholar] [CrossRef][Green Version]
- Brand, A.; MacCallum, D.M.; Brown, A.J.P.; Gow, N.A.R.; Odds, F.C. Ectopic Expression of URA3 Can Influence the Virulence Phenotypes and Proteome of Candida albicans but Can Be Overcome by Targeted Reintegration of URA3 at the RPS10 Locus. Eukaryot. Cell 2004, 3, 900–909. [Google Scholar] [CrossRef]
- Petrikkos, G.; Skiada, A.; Lortholary, O.; Roilides, E.; Walsh, T.J.; Kontoyiannis, D.P. Epidemiology and Clinical Manifestations of Mucormycosis. Clin. Infect. Dis. 2012, 54 (Suppl. 1), S23–S34. [Google Scholar] [CrossRef]
- Ma, L.-J.; Ibrahim, A.S.; Skory, C.; Grabherr, M.G.; Burger, G.; Butler, M.; Elias, M.; Idnurm, A.; Lang, B.F.; Sone, T.; et al. Genomic Analysis of the Basal Lineage Fungus Rhizopus oryzae Reveals a Whole-Genome Duplication. PLoS Genet. 2009, 5, e1000549. [Google Scholar] [CrossRef]
- Bruni, G.O.; Zhong, K.; Lee, S.C.; Wang, P. CRISPR-Cas9 induces point mutation in the mucormycosis fungus Rhizopus delemar. Fungal Genet. Biol. 2018, 124, 1–7. [Google Scholar] [CrossRef]
- Ibragimova, S.; Szebenyi, C.; Sinka, R.; Alzyoud, E.I.; Homa, M.; Vágvölgyi, C.; Nagy, G.; Papp, T. CRISPR-Cas9-Based Mutagenesis of the Mucormycosis-Causing Fungus lichtheimia corymbifera. Int. J. Mol. Sci. 2020, 21, 3727. [Google Scholar] [CrossRef] [PubMed]
- Boeke, J.D.; Trueheart, J.; Natsoulis, G.; Fink, G.R. [10] 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987, 154, 164–175. [Google Scholar] [CrossRef] [PubMed]
- Patiño-Medina, J.A.; Reyes-Mares, N.Y.; Valle-Maldonado, M.I.; Jácome-Galarza, I.E.; Pérez-Arques, C.; Nuñez-Anita, R.E.; Campos-García, J.; Anaya-Martínez, V.; Ortiz-Alvarado, R.; Ramírez-Díaz, M.I.; et al. Heterotrimeric G-alpha subunits Gpa11 and Gpa12 define a transduction pathway that control spore size and virulence in Mucor circinelloides. PLoS ONE 2019, 14, e0226682. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lax, C.; Cánovas-Márquez, J.T.; Tahiri, G.; Navarro, E.; Garre, V.; Nicolás, F.E. Genetic Manipulation in Mucorales and New Developments to Study Mucormycosis. Int. J. Mol. Sci. 2022, 23, 3454. https://doi.org/10.3390/ijms23073454
Lax C, Cánovas-Márquez JT, Tahiri G, Navarro E, Garre V, Nicolás FE. Genetic Manipulation in Mucorales and New Developments to Study Mucormycosis. International Journal of Molecular Sciences. 2022; 23(7):3454. https://doi.org/10.3390/ijms23073454
Chicago/Turabian StyleLax, Carlos, José Tomás Cánovas-Márquez, Ghizlane Tahiri, Eusebio Navarro, Victoriano Garre, and Francisco Esteban Nicolás. 2022. "Genetic Manipulation in Mucorales and New Developments to Study Mucormycosis" International Journal of Molecular Sciences 23, no. 7: 3454. https://doi.org/10.3390/ijms23073454
APA StyleLax, C., Cánovas-Márquez, J. T., Tahiri, G., Navarro, E., Garre, V., & Nicolás, F. E. (2022). Genetic Manipulation in Mucorales and New Developments to Study Mucormycosis. International Journal of Molecular Sciences, 23(7), 3454. https://doi.org/10.3390/ijms23073454








