Genomics and Epigenomics of Gestational Diabetes Mellitus: Understanding the Molecular Pathways of the Disease Pathogenesis
Abstract
1. Introduction
2. Genomic Approaches Used in Understanding Gestational Diabetes
2.1. Candidate-Gene Association Studies
2.1.1. TCF7L2
2.1.2. KCNQ1
2.1.3. CDKAL1
2.1.4. IRS1
2.1.5. MTNR1B
2.2. Genome-Wide Association Studies (GWAS)
2.3. DNA Microarrays
2.4. Next-Generation Sequencing Approaches (NGS)
2.4.1. Whole-Exome Sequencing (WES)
2.4.2. Whole-Genome Sequencing (WGS)
2.4.3. Targeted NGS
2.4.4. RNA Sequencing
3. Epigenetic Modifications in GDM
3.1. DNA Methylation
3.2. Histone Modification
3.3. MicroRNA (miRNA)
4. Molecular Pathways and Pathophysiology of GDM
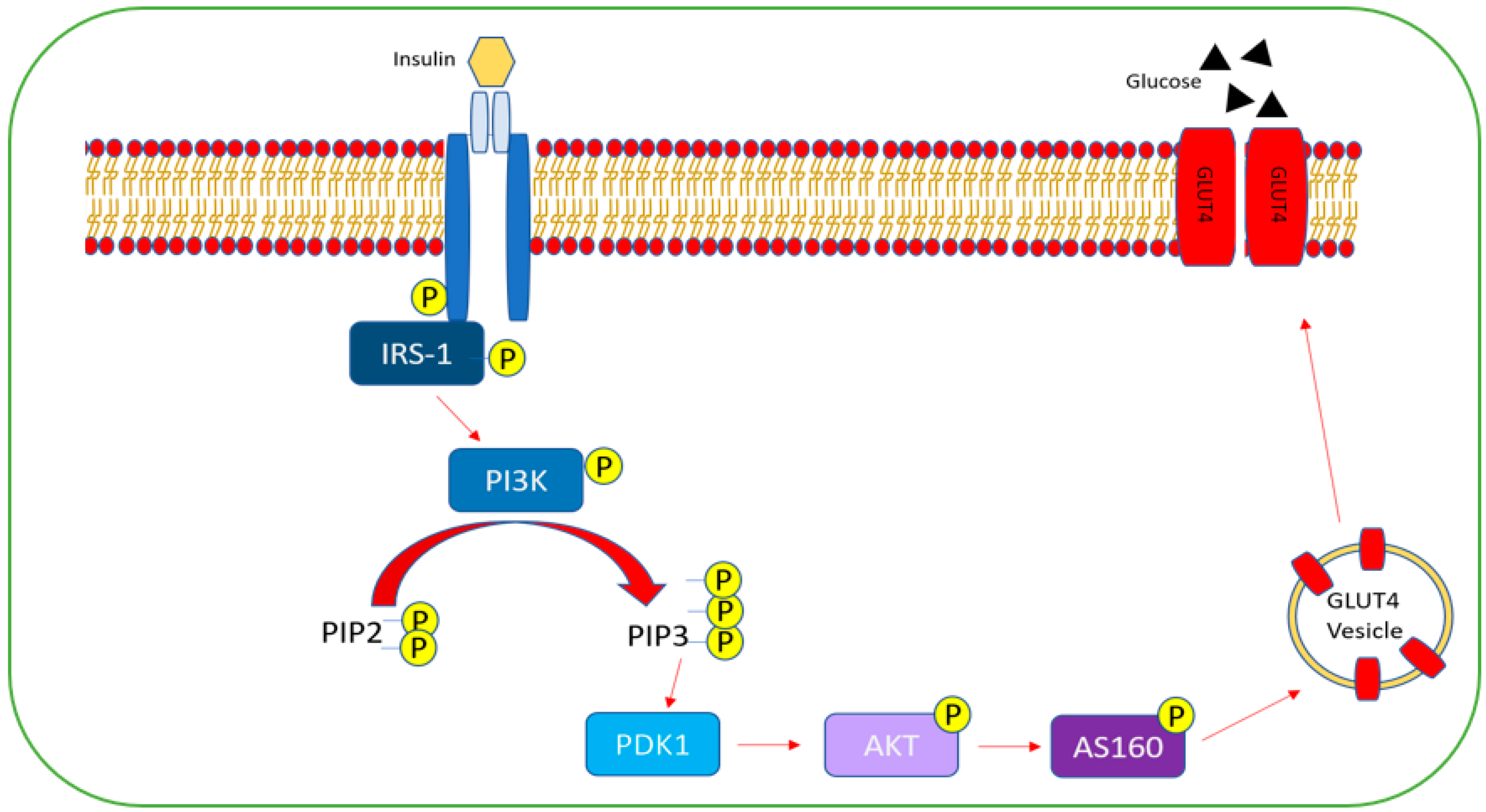
4.1. Insulin Resistance
4.2. Inflammation
4.2.1. NF-kB Pathways in Insulin Resistance
4.2.2. JNK Pathway in Insulin Resistance
4.3. Oxidative Stress
4.3.1. PKC
4.3.2. Polyol Pathway
4.3.3. Hexosamine
4.3.4. NADPH Oxidase
5. Predictive Biomarkers of GDM
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Plows, J.F.; Stanley, J.L.; Baker, P.N.; Reynolds, C.M.; Vickers, M.H. The Pathophysiology of Gestational Diabetes Mellitus. Int. J. Mol. Sci. 2018, 19, 3342. [Google Scholar] [CrossRef] [PubMed]
- Napso, T.; Yong, H.E.J.; Lopez-Tello, J.; Sferruzzi-Perri, A.N. The Role of Placental Hormones in Mediating Maternal Adaptations to Support Pregnancy and Lactation. Front. Physiol. 2018, 9, 1091. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Song, G.; Meng, T.; Zhao, G.; Guo, S. Weight retention at six weeks postpartum and the risk of gestational diabetes mellitus in a second pregnancy. BMC Pregnancy Childbirth 2019, 19, 272. [Google Scholar] [CrossRef] [PubMed]
- Valencia-Ortega, J.; Saucedo, R.; Sánchez-Rodríguez, M.A.; Cruz-Durán, J.G.; Martínez, E.G.R. Epigenetic Alterations Related to Gestational Diabetes Mellitus. Int. J. Mol. Sci. 2021, 22, 9462. [Google Scholar] [CrossRef]
- Mørkrid, K.; Jenum, A.K.; Sletner, L.; Vårdal, M.H.; Waage, C.W.; Nakstad, B.; Vangen, S.; Birkeland, K.I. Failure to increase insulin secretory capacity during pregnancy-induced insulin resistance is associated with ethnicity and gestational diabetes. Eur. J. Endocrinol. 2012, 167, 579–588. [Google Scholar] [CrossRef]
- Lau, S.M.; Lin, S.; Stokes, R.A.; Cheng, K.; Baldock, P.A.; Enriquez, R.F.; McLean, M.; Cheung, N.W.; Sainsbury, A.; Gonzalez, F.J.; et al. Synergistic effects of genetic beta cell dysfunction and maternal glucose intolerance on offspring metabolic phenotype in mice. Diabetologia 2011, 54, 910–921. [Google Scholar] [CrossRef]
- Su, F.-L.; Lu, M.-C.; Yu, S.-C.; Yang, C.-P.; Yang, C.-C.; Tseng, S.-T.; Yan, Y.-H. Increasing trend in the prevalence of gestational diabetes mellitus in Taiwan. J. Diabetes Investig. 2021, 12, 2080–2088. [Google Scholar] [CrossRef]
- IDF Diabetes Atlas, 10th ed. 2021. Available online: https://diabetesatlas.org/idfawp/resource-files/2021/07/IDF_Atlas_10th_Edition_2021.pdf (accessed on 26 January 2022).
- Catalano, P.M.; Ehrenberg, H.M. The short- and long-term implications of maternal obesity on the mother and her offspring. Bjog 2006, 113, 1126–1133. [Google Scholar] [CrossRef]
- Alejandro, E.U.; Mamerto, T.P.; Chung, G.; Villavieja, A.; Gaus, N.L.; Morgan, E.; Pineda-Cortel, M.R.B. Gestational Diabetes Mellitus: A Harbinger of the Vicious Cycle of Diabetes. Int. J. Mol. Sci. 2020, 21, 5003. [Google Scholar] [CrossRef]
- Damm, P.; Houshmand-Oeregaard, A.; Kelstrup, L.; Lauenborg, J.; Mathiesen, E.R.; Clausen, T.D. Gestational diabetes mellitus and long-term consequences for mother and offspring: A view from Denmark. Diabetologia 2016, 59, 1396–1399. [Google Scholar] [CrossRef]
- Shou, C.; Wei, Y.-M.; Wang, C.; Yang, H.-X. Updates in Long-term Maternal and Fetal Adverse Effects of Gestational Diabetes Mellitus. Matern.-Fetal Med. 2019, 1, 91–94. [Google Scholar] [CrossRef]
- Carroll, X.; Liang, X.; Zhang, W.; Zhang, W.; Liu, G.; Turner, N.; Leeper-Woodford, S. Socioeconomic, environmental and lifestyle factors associated with gestational diabetes mellitus: A matched case-control study in Beijing, China. Sci. Rep. 2018, 8, 8103. [Google Scholar] [CrossRef] [PubMed]
- Słupecka-Ziemilska, M.; Wychowański, P.; Puzianowska-Kuznicka, M. Gestational Diabetes Mellitus Affects Offspring’s Epigenome. Is There a Way to Reduce the Negative Consequences? Nutrients 2020, 12, 2792. [Google Scholar] [CrossRef]
- Lowe, W.L., Jr.; Scholtens, D.M.; Sandler, V.; Hayes, M.G. Genetics of Gestational Diabetes Mellitus and Maternal Metabolism. Curr. Diabetes Rep. 2016, 16, 15. [Google Scholar] [CrossRef] [PubMed]
- Kwon, J.M.; Goate, A.M. The candidate gene approach. Alcohol Res. Health 2000, 24, 164–168. [Google Scholar]
- Patnala, R.; Clements, J.; Batra, J. Candidate gene association studies: A comprehensive guide to useful in silicotools. BMC Genetics 2013, 14, 39. [Google Scholar] [CrossRef]
- Zhu, M.; Zhao, S. Candidate gene identification approach: Progress and challenges. Int. J. Biol. Sci. 2007, 3, 420–427. [Google Scholar] [CrossRef]
- Mishra, S.; Rao, C.R.; Shetty, A. Trends in the Diagnosis of Gestational Diabetes Mellitus. Scientifica 2016, 2016, 5489015. [Google Scholar] [CrossRef][Green Version]
- Robitaille, J.; Grant, A.M. The genetics of gestational diabetes mellitus: Evidence for relationship with type 2 diabetes mellitus. Genet. Med. 2008, 10, 240–250. [Google Scholar] [CrossRef]
- Buchanan, T.A. Pancreatic B-cell defects in gestational diabetes: Implications for the pathogenesis and prevention of type 2 diabetes. J. Clin. Endocrinol. Metab. 2001, 86, 989–993. [Google Scholar] [CrossRef]
- Ben-Haroush, A.; Yogev, Y.; Hod, M. Epidemiology of gestational diabetes mellitus and its association with Type 2 diabetes. Diabet Med. 2004, 21, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Dabelea, D.; Hanson, R.L.; Bennett, P.H.; Roumain, J.; Knowler, W.C.; Pettitt, D.J. Increasing prevalence of Type II diabetes in American Indian children. Diabetologia 1998, 41, 904–910. [Google Scholar] [CrossRef] [PubMed]
- Dabelea, D.; Snell-Bergeon, J.K.; Hartsfield, C.L.; Bischoff, K.J.; Hamman, R.F.; McDuffie, R.S. Increasing Prevalence of Gestational Diabetes Mellitus (GDM) Over Time and by Birth Cohort: Kaiser Permanente of Colorado GDM Screening Program. Diabetes Care 2005, 28, 579–584. [Google Scholar] [CrossRef] [PubMed]
- Kwak, S.H.; Jang, H.C.; Park, K.S. Finding genetic risk factors of gestational diabetes. Genom. Inform. 2012, 10, 239–243. [Google Scholar] [CrossRef]
- Rhee, S.Y.; Kim, J.Y.; Woo, J.-T.; Kim, Y.S.; Kim, S.-H. Familial clustering of type 2 diabetes in Korean women with gestational diabetes mellitus. Korean J. Intern. Med. 2010, 25, 269–272. [Google Scholar] [CrossRef]
- Yahaya, T.O.; Salisu, T.; Abdulrahman, Y.B.; Umar, A.K. Update on the genetic and epigenetic etiology of gestational diabetes mellitus: A review. Egypt. J. Med. Hum. Genet. 2020, 21, 13. [Google Scholar] [CrossRef]
- Wei, W.; He, Y.; Wang, X.; Tan, G.; Zhou, F.; Zheng, G.; Tian, D.; Ma, X.; Yu, H. Gestational Diabetes Mellitus: The Genetic Susceptibility behind the Disease. Horm. Metab. Res. 2021, 53, 489–498. [Google Scholar] [CrossRef]
- Shaat, N.; Lernmark, Å.; Karlsson, E.; Ivarsson, S.; Parikh, H.; Berntorp, K.; Groop, L. A variant in the transcription factor 7-like 2 (TCF7L2) gene is associated with an increased risk of gestational diabetes mellitus. Diabetologia 2007, 50, 972–979. [Google Scholar] [CrossRef]
- Pappa, K.I.; Gazouli, M.; Economou, K.; Daskalakis, G.; Anastasiou, E.; Anagnou, N.P.; Antsaklis, A. Gestational diabetes mellitus shares polymorphisms of genes associated with insulin resistance and type 2 diabetes in the Greek population. Gynecol. Endocrinol. 2011, 27, 267–272. [Google Scholar] [CrossRef]
- Freathy, R.M.; Hayes, M.G.; Urbanek, M.; Lowe, L.P.; Lee, H.; Ackerman, C.; Frayling, T.M.; Cox, N.J.; Dunger, D.B.; Dyer, A.R.; et al. Hyperglycemia and Adverse Pregnancy Outcome (HAPO) study: Common genetic variants in GCK and TCF7L2 are associated with fasting and postchallenge glucose levels in pregnancy and with the new consensus definition of gestational diabetes mellitus from the International Association of Diabetes and Pregnancy Study Groups. Diabetes 2010, 59, 2682–2689. [Google Scholar] [CrossRef]
- Lauenborg, J.; Grarup, N.; Damm, P.; Borch-Johnsen, K.; Jørgensen, T.; Pedersen, O.; Hansen, T. Common type 2 diabetes risk gene variants associate with gestational diabetes. J. Clin. Endocrinol. Metab. 2009, 94, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.M.; Kim, T.H.; Lim, S.; Choi, S.H.; Shin, H.D.; Lee, H.K.; Park, K.S.; Jang, H.C. Type 2 diabetes-associated genetic variants discovered in the recent genome-wide association studies are related to gestational diabetes mellitus in the Korean population. Diabetologia 2009, 52, 253–261. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulou, A.; Lynch, K.F.; Shaat, N.; Håkansson, R.; Ivarsson, S.A.; Berntorp, K.; Agardh, C.D.; Lernmark, Å.; DiPi, S.S.G. Gestational diabetes mellitus is associated with TCF7L2 gene polymorphisms independent of HLA-DQB1*0602 genotypes and islet cell autoantibodies. Diabet. Med. J. Br. Diabet. Assoc. 2011, 28, 1018–1027. [Google Scholar] [CrossRef] [PubMed]
- Franzago, M.; Fraticelli, F.; Marchetti, D.; Celentano, C.; Liberati, M.; Stuppia, L.; Vitacolonna, E. Nutrigenetic variants and cardio-metabolic risk in women with or without gestational diabetes. Diabetes Res. Clin. Pract. 2018, 137, 64–71. [Google Scholar] [CrossRef]
- Huopio, H.; Cederberg, H.; Vangipurapu, J.; Hakkarainen, H.; Pääkkönen, M.; Kuulasmaa, T.; Heinonen, S.; Laakso, M. Association of risk variants for type 2 diabetes and hyperglycemia with gestational diabetes. Eur. J. Endocrinol. 2013, 169, 291–297. [Google Scholar] [CrossRef]
- Huerta-Chagoya, A.; Vázquez-Cárdenas, P.; Moreno-Macías, H.; Tapia-Maruri, L.; Rodríguez-Guillén, R.; López-Vite, E.; García-Escalante, G.; Escobedo-Aguirre, F.; Parra-Covarrubias, A.; Cordero-Brieño, R.; et al. Genetic Determinants for Gestational Diabetes Mellitus and Related Metabolic Traits in Mexican Women. PLoS ONE 2015, 10, e0126408. [Google Scholar] [CrossRef]
- Ding, M.; Chavarro, J.; Olsen, S.; Lin, Y.; Ley, S.H.; Bao, W.; Rawal, S.; Grunnet, L.G.; Thuesen, A.C.B.; Mills, J.L.; et al. Genetic variants of gestational diabetes mellitus: A study of 112 SNPs among 8722 women in two independent populations. Diabetologia 2018, 61, 1758–1768. [Google Scholar] [CrossRef]
- Stuebe, A.M.; Wise, A.; Nguyen, T.; Herring, A.; North, K.E.; Siega-Riz, A.M. Maternal genotype and gestational diabetes. Am. J. Perinatol. 2014, 31, 69–76. [Google Scholar] [CrossRef]
- Wang, B.; Xue, X. Investigations of Associations between Seven Gene Polymorphisms and Gestational Diabetes Mellitus: Evidence from a Meta-Analysis. Gynecol. Obstet. Investig. 2020, 85, 229–236. [Google Scholar] [CrossRef]
- Shin, H.D.; Park, B.L.; Shin, H.J.; Kim, J.Y.; Park, S.; Kim, B.; Kim, S.H. Association of KCNQ1 polymorphisms with the gestational diabetes mellitus in Korean women. J. Clin. Endocrinol. Metab. 2010, 95, 445–449. [Google Scholar] [CrossRef]
- Kwak, S.H.; Kim, T.H.; Cho, Y.M.; Choi, S.H.; Jang, H.C.; Park, K.S. Polymorphisms in KCNQ1 are associated with gestational diabetes in a Korean population. Horm. Res. Paediatr. 2010, 74, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Ao, D.; Wang, H.J.; Wang, L.F.; Song, J.Y.; Yang, H.X.; Wang, Y. The rs2237892 Polymorphism in KCNQ1 Influences Gestational Diabetes Mellitus and Glucose Levels: A Case-Control Study and Meta-Analysis. PLoS ONE 2015, 10, e0128901. [Google Scholar] [CrossRef] [PubMed]
- Fatima, S.S.; Chaudhry, B.; Khan, T.; Farooq, S. KCNQ1 rs2237895 polymorphism is associated with Gestational Diabetes in Pakistani Women. Pak. J. Med. Sci. Online 2016, 32, 1380. [Google Scholar] [CrossRef]
- Jespersen, T.; Grunnet, M.; Olesen, S.-P. The KCNQ1 Potassium Channel: From Gene to Physiological Function. Physiology 2005, 20, 408–416. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.A.; Jahan, P.; Hasan, Q.; Rao, P. Genetic confirmation of T2DM meta-analysis variants studied in gestational diabetes mellitus in an Indian population. Diabetes Metab. Syndr. 2019, 13, 688–694. [Google Scholar] [CrossRef]
- Wang, K.; Chen, Q.; Feng, Y.; Yang, H.; Wu, W.; Zhang, P.; Wang, Y.; Ko, J.; Zhao, F.; Du, W.; et al. Single Nucleotide Polymorphisms in CDKAL1 Gene Are Associated with Risk of Gestational Diabetes Mellitus in Chinese Population. J. Diabetes Res. 2019, 2019, 3618103. [Google Scholar] [CrossRef]
- Guo, F.; Long, W.; Zhou, W.; Zhang, B.; Liu, J.; Yu, B. FTO, GCKR, CDKAL1 and CDKN2A/B gene polymorphisms and the risk of gestational diabetes mellitus: A meta-analysis. Arch. Gynecol. Obstet. 2018, 298, 705–715. [Google Scholar] [CrossRef]
- Alharbi, K.K.; Khan, I.A.; Abotalib, Z.; Al-Hakeem, M.M. Insulin receptor substrate-1 (IRS-1) Gly927Arg: Correlation with gestational diabetes mellitus in Saudi women. Biomed. Res. Int. 2014, 2014, 146495. [Google Scholar] [CrossRef]
- Shaat, N.; Ekelund, M.; Lernmark, Å.; Ivarsson, S.; Almgren, P.; Berntorp, K.; Groop, L. Association of the E23K polymorphism in the KCNJ11 gene with gestational diabetes mellitus. Diabetologia 2005, 48, 2544–2551. [Google Scholar] [CrossRef]
- Xie, K.; Chen, T.; Zhang, Y.; Wen, J.; Cui, X.; You, L.; Zhu, L.; Xu, B.; Ji, C.; Guo, X. Association of rs10830962 polymorphism with gestational diabetes mellitus risk in a Chinese population. Sci. Rep. 2019, 9, 5357. [Google Scholar] [CrossRef]
- Wu, L.; Cui, L.; Tam, W.H.; Ma, R.C.; Wang, C.C. Genetic variants associated with gestational diabetes mellitus: A meta-analysis and subgroup analysis. Sci. Rep. 2016, 6, 30539. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, M.C.; da Silva, M.E.R.; Fukui, R.T.; Arruda-Marques, M.d.C.; dos Santos, R.F. TCF7L2 correlation in both insulin secretion and postprandial insulin sensitivity. Diabetol. Metab. Syndr. 2018, 10, 37. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.; Wang, Z.; Wu, L.; Lu, X.; Shangguan, S.; Xin, Y.; Li, L.; Wang, L. Association between TCF7L2 polymorphisms and gestational diabetes mellitus: A meta-analysis. J. Diabetes Investig. 2017, 8, 560–570. [Google Scholar] [CrossRef]
- Perez-Martinez, P.; Perez-Caballero, A.I.; Garcia-Rios, A.; Yubero-Serrano, E.M.; Camargo, A.; Gomez-Luna, M.J.; Marin, C.; Gomez-Luna, P.; Dembinska-Kiec, A.; Rodriguez-Cantalejo, F.; et al. Effects of rs7903146 variation in the Tcf7l2 gene in the lipid metabolism of three different populations. PLoS ONE 2012, 7, e43390. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ayala, I.; Shannon, C.; Fourcaudot, M.; Acharya, N.K.; Jenkinson, C.P.; Heikkinen, S.; Norton, L. The Diabetes Gene and Wnt Pathway Effector TCF7L2 Regulates Adipocyte Development and Function. Diabetes 2018, 67, 554–568. [Google Scholar] [CrossRef]
- Wu, H.H.; Li, Y.L.; Liu, N.J.; Yang, Z.; Tao, X.M.; Du, Y.P.; Wang, X.C.; Lu, B.; Zhang, Z.Y.; Hu, R.M.; et al. TCF7L2 regulates pancreatic β-cell function through PI3K/AKT signal pathway. Diabetol. Metab. Syndr. 2019, 11, 55. [Google Scholar] [CrossRef]
- Lin, P.C.; Lin, W.T.; Yeh, Y.H.; Wung, S.F. Transcription Factor 7-Like 2 (TCF7L2) rs7903146 Polymorphism as a Risk Factor for Gestational Diabetes Mellitus: A Meta-Analysis. PLoS ONE 2016, 11, e0153044. [Google Scholar] [CrossRef]
- Včelák, J.; Vejražková, D.; Vaňková, M.; Lukášová, P.; Bradnová, O.; Hálková, T.; Bešťák, J.; Andělová, K.; Kvasničková, H.; Hoskovcová, P.; et al. T2D risk haplotypes of the TCF7L2 gene in the Czech population sample: The association with free fatty acids composition. Physiol. Res. 2012, 61, 229–240. [Google Scholar] [CrossRef]
- Pagán, A.; Sabater-Molina, M.; Olza, J.; Prieto-Sánchez, M.T.; Blanco-Carnero, J.E.; Parrilla, J.J.; Gil, Á.; Larqué, E. A gene variant in the transcription factor 7-like 2 (TCF7L2) is associated with an increased risk of gestational diabetes mellitus. Eur. J. Obstet. Gynecol. Reprod. Biol. 2014, 180, 77–82. [Google Scholar] [CrossRef]
- Watanabe, R.M.; Allayee, H.; Xiang, A.H.; Trigo, E.; Hartiala, J.; Lawrence, J.M.; Buchanan, T.A. Transcription factor 7-like 2 (TCF7L2) is associated with gestational diabetes mellitus and interacts with adiposity to alter insulin secretion in Mexican Americans. Diabetes 2007, 56, 1481–1485. [Google Scholar] [CrossRef]
- Klein, K.; Haslinger, P.; Bancher-Todesca, D.; Leipold, H.; Knöfler, M.; Handisurya, A.; Kautzky-Willer, A.; Worda, C. Transcription factor 7-like 2 gene polymorphisms and gestational diabetes mellitus. J. Matern.-Fetal Neonatal Med. 2012, 25, 1783–1786. [Google Scholar] [CrossRef] [PubMed]
- Reyes-López, R.; Pérez-Luque, E.; Malacara, J.M. Metabolic, hormonal characteristics and genetic variants of TCF7L2 associated with development of gestational diabetes mellitus in Mexican women. Diabetes Metab. Res. Rev. 2014, 30, 701–706. [Google Scholar] [CrossRef] [PubMed]
- Ye, D.; Fei, Y.; Ling, Q.; Xu, W.; Zhang, Z.; Shu, J.; Li, C.; Dong, F. Polymorphisms in TCF7L2 gene are associated with gestational diabetes mellitus in Chinese Han population. Sci. Rep. 2016, 6, 30686. [Google Scholar] [CrossRef] [PubMed]
- Donley, V.R.; Hiskett, E.K.; Kidder, A.C.; Schermerhorn, T. ATP-sensitive potassium channel (KATPchannel) expression in the normal canine pancreas and in canine insulinomas. BMC Vet. Res. 2005, 1, 8. [Google Scholar] [CrossRef]
- Chon, S.J.; Kim, S.Y.; Cho, N.R.; Min, D.L.; Hwang, Y.J.; Mamura, M. Association of variants in PPARγ², IGF2BP2, and KCNQ1 with a susceptibility to gestational diabetes mellitus in a Korean population. Yonsei Med. J. 2013, 54, 352–357. [Google Scholar] [CrossRef]
- Cao, M.; Zhang, L.; Chen, T.; Shi, A.; Xie, K.; Li, Z.; Xu, J.; Chen, Z.; Ji, C.; Wen, J. Genetic Susceptibility to Gestational Diabetes Mellitus in a Chinese Population. Front. Endocrinol. 2020, 11, 247. [Google Scholar] [CrossRef]
- Zhou, Q.; Zhang, K.; Li, W.; Liu, J.T.; Hong, J.; Qin, S.W.; Ping, F.; Sun, M.L.; Nie, M. Association of KCNQ1 gene polymorphism with gestational diabetes mellitus in a Chinese population. Diabetologia 2009, 52, 2466–2468. [Google Scholar] [CrossRef]
- Palmer, C.J.; Bruckner, R.J.; Paulo, J.A.; Kazak, L.; Long, J.Z.; Mina, A.I.; Deng, Z.; LeClair, K.B.; Hall, J.A.; Hong, S.; et al. Cdkal1, a type 2 diabetes susceptibility gene, regulates mitochondrial function in adipose tissue. Mol. Metab. 2017, 6, 1212–1225. [Google Scholar] [CrossRef]
- Dias, S.; Pheiffer, C.; Abrahams, Y.; Rheeder, P.; Adam, S. Molecular Biomarkers for Gestational Diabetes Mellitus. Int. J. Mol. Sci. 2018, 19, 2926. [Google Scholar] [CrossRef]
- Baroni, M.G.; D’Andrea, M.P.; Montali, A.; Pannitteri, G.; Barillà, F.; Campagna, F.; Mazzei, E.; Lovari, S.; Seccareccia, F.; Campa, P.P.; et al. A common mutation of the insulin receptor substrate-1 gene is a risk factor for coronary artery disease. Arterioscler. Thromb. Vasc. Biol. 1999, 19, 2975–2980. [Google Scholar] [CrossRef][Green Version]
- Popova, P.V.; Klyushina, A.A.; Vasilyeva, L.B.; Tkachuk, A.S.; Bolotko, Y.A.; Gerasimov, A.S.; Pustozerov, E.A.; Kravchuk, E.N.; Predeus, A.; Kostareva, A.A.; et al. Effect of gene-lifestyle interaction on gestational diabetes risk. Oncotarget 2017, 8, 112024–112035. [Google Scholar] [CrossRef] [PubMed]
- Rosta, K.; Al-Aissa, Z.; Hadarits, O.; Harreiter, J.; Nádasdi, Á.; Kelemen, F.; Bancher-Todesca, D.; Komlósi, Z.; Németh, L.; Rigó, J., Jr.; et al. Association Study with 77 SNPs Confirms the Robust Role for the rs10830963/G of MTNR1B Variant and Identifies Two Novel Associations in Gestational Diabetes Mellitus Development. PLoS ONE 2017, 12, e0169781. [Google Scholar] [CrossRef] [PubMed]
- Rosik, J.; Szostak, B.; Machaj, F.; Pawlik, A. The role of genetics and epigenetics in the pathogenesis of gestational diabetes mellitus. Ann. Hum. Genet. 2020, 84, 114–124. [Google Scholar] [CrossRef] [PubMed]
- Tam, V.; Patel, N.; Turcotte, M.; Bossé, Y.; Paré, G.; Meyre, D. Benefits and limitations of genome-wide association studies. Nat. Rev. Genet. 2019, 20, 467–484. [Google Scholar] [CrossRef]
- Kwak, S.H.; Kim, S.-H.; Cho, Y.M.; Go, M.J.; Cho, Y.S.; Choi, S.H.; Moon, M.K.; Jung, H.S.; Shin, H.D.; Kang, H.M.; et al. A Genome-Wide Association Study of Gestational Diabetes Mellitus in Korean Women. Diabetes 2012, 61, 531–541. [Google Scholar] [CrossRef]
- Wu, N.N.; Zhao, D.; Ma, W.; Lang, J.N.; Liu, S.M.; Fu, Y.; Wang, X.; Wang, Z.W.; Li, Q. A genome-wide association study of gestational diabetes mellitus in Chinese women. J. Matern. Fetal Neonatal Med. 2021, 34, 1557–1564. [Google Scholar] [CrossRef]
- Hayes, M.G.; Urbanek, M.; Hivert, M.-F.; Armstrong, L.L.; Morrison, J.; Guo, C.; Lowe, L.P.; Scheftner, D.A.; Pluzhnikov, A.; Levine, D.M.; et al. Identification of HKDC1 and BACE2 as genes influencing glycemic traits during pregnancy through genome-wide association studies. Diabetes 2013, 62, 3282–3291. [Google Scholar] [CrossRef]
- Ludvik, A.E.; Pusec, C.M.; Priyadarshini, M.; Angueira, A.R.; Guo, C.; Lo, A.; Hershenhouse, K.S.; Yang, G.-Y.; Ding, X.; Reddy, T.E.; et al. HKDC1 Is a Novel Hexokinase Involved in Whole-Body Glucose Use. Endocrinology 2016, 157, 3452–3461. [Google Scholar] [CrossRef]
- Khan, M.W.; Priyadarshini, M.; Cordoba-Chacon, J.; Becker, T.C.; Layden, B.T. Hepatic hexokinase domain containing 1 (HKDC1) improves whole body glucose tolerance and insulin sensitivity in pregnant mice. Biochim. Biophys. Acta Mol. Basis Dis. 2019, 1865, 678–687. [Google Scholar] [CrossRef]
- Powe, C.E.; Kwak, S.H. Genetic Studies of Gestational Diabetes and Glucose Metabolism in Pregnancy. Curr. Diabetes Rep. 2020, 20, 69. [Google Scholar] [CrossRef]
- Dyrskjøt, L.; Dalsgaard-Sørensen, K.; Stampe-Ostenfeld, M.; Birkenkamp-Demtroder, K.; Thorsen, K.; Andersen, C.L.; Kruhøffer, M.; Jensen, J.L.; Ørntoft, T.F. Chapter 16—DNA Microarrays and Genetic Testing. In Molecular Diagnostics, 2nd ed.; Patrinos, G.P., Ansorge, W.J., Eds.; Academic Press: San Diego, CA, USA, 2010; pp. 247–265. [Google Scholar] [CrossRef]
- Jares, P. DNA microarray applications in functional genomics. Ultrastruct. Pathol. 2006, 30, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Govindarajan, R.; Duraiyan, J.; Kaliyappan, K.; Palanisamy, M. Microarray and its applications. J. Pharm. Bioallied Sci. 2012, 4, S310–S312. [Google Scholar] [CrossRef] [PubMed]
- Radaelli, T.; Varastehpour, A.; Catalano, P.; Hauguel-de Mouzon, S. Gestational diabetes induces placental genes for chronic stress and inflammatory pathways. Diabetes 2003, 52, 2951–2958. [Google Scholar] [CrossRef] [PubMed]
- Enquobahrie, D.A.; Williams, M.A.; Qiu, C.; Meller, M.; Sorensen, T.K. Global placental gene expression in gestational diabetes mellitus. Am. J. Obstet. Gynecol. 2009, 200, e201–e206. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wang, C.T.; Forsyth, N.R.; Wu, P. Transcriptional profiling reveals altered biological characteristics of chorionic stem cells from women with gestational diabetes. Stem Cell Res. Ther. 2020, 11, 319. [Google Scholar] [CrossRef] [PubMed]
- Jatavan, P. Chapter 8—Oxidative stress in gestational diabetes mellitus. In Diabetes, 2nd ed.; Preedy, V.R., Ed.; Academic Press: San Diego, CA, USA, 2020; pp. 79–85. [Google Scholar] [CrossRef]
- Baynes, J.W.; Thorpe, S.R. Role of oxidative stress in diabetic complications: A new perspective on an old paradigm. Diabetes 1999, 48, 1–9. [Google Scholar] [CrossRef]
- Viana, M.; Herrera, E.; Bonet, B. Teratogenic effects of diabetes mellitus in the rat. Prevention by vitamin E. Diabetologia 1996, 39, 1041–1046. [Google Scholar] [CrossRef]
- Qian, Y.; Sun, H.; Xiao, H.; Ma, M.; Xiao, X.; Qu, Q. Microarray analysis of differentially expressed genes and their functions in omental visceral adipose tissues of pregnant women with vs. without gestational diabetes mellitus. Biomed. Rep. 2017, 6, 503–512. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, H.; Liu, F.; Song, X. Analysis of key genes and their functions in placental tissue of patients with gestational diabetes mellitus. Reprod. Biol. Endocrinol. 2019, 17, 104. [Google Scholar] [CrossRef]
- Steinborn, A.; Saran, G.; Schneider, A.; Fersis, N.; Sohn, C.; Schmitt, E. The presence of gestational diabetes is associated with increased detection of anti-HLA-class II antibodies in the maternal circulation. Am. J. Reprod. Immunol. 2006, 56, 124–134. [Google Scholar] [CrossRef]
- Meera Krishna, B.; Khan, M.A.; Khan, S.T. Next-Generation Sequencing (NGS) Platforms: An Exciting Era of Genome Sequence Analysis. In Microbial Genomics in Sustainable Agroecosystems; Springer: Singapore, 2019; pp. 89–109. [Google Scholar]
- Behjati, S.; Tarpey, P.S. What is next generation sequencing? Arch. Dis. Child. Educ. Pract. Ed. 2013, 98, 236–238. [Google Scholar] [CrossRef] [PubMed]
- Dou, J.; Wu, D.; Ding, L.; Wang, K.; Jiang, M.; Chai, X.; Reilly, D.F.; Tai, E.S.; Liu, J.; Sim, X.; et al. Using off-target data from whole-exome sequencing to improve genotyping accuracy, association analysis and polygenic risk prediction. Brief Bioinform. 2021, 22. [Google Scholar] [CrossRef] [PubMed]
- Pace, N.P.; Vella, B.; Craus, J.; Caruana, R.; Savona-Ventura, C.; Vassallo, J. Screening for monogenic subtypes of gestational diabetes in a high prevalence island population—A whole exome sequencing study. Diabetes Metab. Res. Rev. 2021, 38, e3486. [Google Scholar] [CrossRef] [PubMed]
- WES vs. WGS vs. Custom Panels. Available online: https://sequencing.roche.com/en-us/science-education/education/articles/wes-wgs-custom.html (accessed on 26 January 2022).
- Kozarewa, I.; Armisen, J.; Gardner, A.F.; Slatko, B.E.; Hendrickson, C.L. Overview of Target Enrichment Strategies. Curr. Protoc. Mol. Biol. 2015, 112, 7–21. [Google Scholar] [CrossRef]
- Zubkova, N.; Petrukhin, V.; Tiulpakov, A.; Budyikina, T.; Panov, A.; Burumkulova, F.; Ulyatovskaya, V.; Makretskaya, N.; Plechanova, M. Targeted Next-Generation Sequencing Demonstrates High Frequency of Monogenic Forms in Gestational Diabetes. Available online: https://www.easd.org/virtualmeeting/home.html#!resources/targeted-next-generation-sequencing-demonstrates-high-frequency-of-monogenic-forms-in-gestational-diabetes (accessed on 4 October 2021).
- Zubkova, N.; Burumkulova, F.; Plechanova, M.; Petrukhin, V.; Petrov, V.; Vasilyev, E.; Panov, A.; Sorkina, E.; Ulyatovskaya, V.; Makretskaya, N.; et al. High frequency of pathogenic and rare sequence variants in diabetes-related genes among Russian patients with diabetes in pregnancy. Acta Diabetol. 2019, 56, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Kukurba, K.R.; Montgomery, S.B. RNA Sequencing and Analysis. Cold Spring Harb. Protoc. 2015, 2015, 951–969. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Hu, D.; Gong, Y. Identification of potential lncRNAs and co-expressed mRNAs in gestational diabetes mellitus by RNA sequencing. J. Matern. Fetal Neonatal Med. 2021, 1–15. [Google Scholar] [CrossRef]
- Wang, H.; She, G.; Zhou, W.; Liu, K.; Miao, J.; Yu, B. Expression profile of circular RNAs in placentas of women with gestational diabetes mellitus. Endocr. J. 2019, 66, 431–441. [Google Scholar] [CrossRef]
- Wang, H.; Zhou, W.; She, G.; Yu, B.; Sun, L. Downregulation of hsa_circ_0005243 induces trophoblast cell dysfunction and inflammation via the β-catenin and NF-κB pathways. Reprod. Biol. Endocrinol. 2020, 18, 51. [Google Scholar] [CrossRef]
- Yang, Y.; Guo, F.; Peng, Y.; Chen, R.; Zhou, W.; Wang, H.; OuYang, J.; Yu, B.; Xu, Z. Transcriptomic Profiling of Human Placenta in Gestational Diabetes Mellitus at the Single-Cell Level. Front. Endocrinol. 2021, 12, 679582. [Google Scholar] [CrossRef]
- Felsenfeld, G. A brief history of epigenetics. Cold Spring Harb. Perspect. Biol. 2014, 6, a018200. [Google Scholar] [CrossRef] [PubMed]
- Alegría-Torres, J.A.; Baccarelli, A.; Bollati, V. Epigenetics and lifestyle. Epigenomics 2011, 3, 267–277. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; von Meyenn, F.; Peleg-Raibstein, D.; Wolfrum, C. Environmental and Nutritional Effects Regulating Adipose Tissue Function and Metabolism Across Generations. Adv. Sci. 2019, 6, 1900275. [Google Scholar] [CrossRef] [PubMed]
- Lumey, L.; Stein, A.D.; Kahn, H.S.; van der Pal-de Bruin, K.M.; Blauw, G.; Zybert, P.A.; Susser, E.S. Cohort Profile: The Dutch Hunger Winter Families Study. Int. J. Epidemiol. 2007, 36, 1196–1204. [Google Scholar] [CrossRef]
- Heijmans, B.T.; Tobi, E.W.; Stein, A.D.; Putter, H.; Blauw, G.J.; Susser, E.S.; Slagboom, P.E.; Lumey, L.H. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc. Natl. Acad. Sci. USA 2008, 105, 17046–17049. [Google Scholar] [CrossRef]
- Lussana, F.; Painter, R.C.; Ocke, M.C.; Buller, H.R.; Bossuyt, P.M.; Roseboom, T.J. Prenatal exposure to the Dutch famine is associated with a preference for fatty foods and a more atherogenic lipid profile. Am. J. Clin. Nutr. 2008, 88, 1648–1652. [Google Scholar] [CrossRef]
- Lumey, L.H.; Van Poppel, F.W. The Dutch famine of 1944-45: Mortality and morbidity in past and present generations. Soc. Hist. Med. 1994, 7, 229–246. [Google Scholar] [CrossRef]
- Dłuski, D.F.; Wolińska, E.; Skrzypczak, M. Epigenetic Changes in Gestational Diabetes Mellitus. Int. J. Mol. Sci. 2021, 22, 7649. [Google Scholar] [CrossRef]
- Ornoy, A.; Wolf, A.; Ratzon, N.; Greenbaum, C.; Dulitzky, M. Neurodevelopmental outcome at early school age of children born to mothers with gestational diabetes. Arch. Dis. Child. Fetal Neonatal Ed. 1999, 81, F10–F14. [Google Scholar] [CrossRef]
- Pavlinkova, G.; Salbaum, J.M.; Kappen, C. Maternal diabetes alters transcriptional programs in the developing embryo. BMC Genom. 2009, 10, 274. [Google Scholar] [CrossRef]
- Kappen, C.; Kruger, C.; MacGowan, J.; Salbaum, J.M. Maternal diet modulates the risk for neural tube defects in a mouse model of diabetic pregnancy. Reprod. Toxicol. 2011, 31, 41–49. [Google Scholar] [CrossRef]
- Salbaum, J.M.; Kappen, C. Neural tube defect genes and maternal diabetes during pregnancy. Birth Defects Res. Part A Clin. Mol. Teratol. 2010, 88, 601–611. [Google Scholar] [CrossRef] [PubMed]
- Elliott, H.R.; Sharp, G.C.; Relton, C.L.; Lawlor, D.A. Epigenetics and gestational diabetes: A review of epigenetic epidemiology studies and their use to explore epigenetic mediation and improve prediction. Diabetologia 2019, 62, 2171–2178. [Google Scholar] [CrossRef] [PubMed]
- Moore, L.D.; Le, T.; Fan, G. DNA methylation and its basic function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef]
- Handy, D.E.; Castro, R.; Loscalzo, J. Epigenetic modifications: Basic mechanisms and role in cardiovascular disease. Circulation 2011, 123, 2145–2156. [Google Scholar] [CrossRef]
- Cardenas, A.; Gagné-Ouellet, V.; Allard, C.; Brisson, D.; Perron, P.; Bouchard, L.; Hivert, M.-F. Placental DNA Methylation Adaptation to Maternal Glycemic Response in Pregnancy. Diabetes 2018, 67, 1673–1683. [Google Scholar] [CrossRef] [PubMed]
- Weng, X.; Liu, F.; Zhang, H.; Kan, M.; Wang, T.; Dong, M.; Liu, Y. Genome-wide DNA methylation profiling in infants born to gestational diabetes mellitus. Diabetes Res. Clin. Pract. 2018, 142, 10–18. [Google Scholar] [CrossRef]
- Kang, J.; Lee, C.N.; Li, H.Y.; Hsu, K.H.; Lin, S.Y. Genome-wide DNA methylation variation in maternal and cord blood of gestational diabetes population. Diabetes Res. Clin. Pract. 2017, 132, 127–136. [Google Scholar] [CrossRef]
- Awamleh, Z.; Butcher, D.T.; Hanley, A.; Retnakaran, R.; Haertle, L.; Haaf, T.; Hamilton, J.; Weksberg, R. Exposure to Gestational Diabetes Mellitus (GDM) alters DNA methylation in placenta and fetal cord blood. Diabetes Res. Clin. Pract. 2021, 174, 108690. [Google Scholar] [CrossRef]
- Deng, X.; Yang, Y.; Sun, H.; Qi, W.; Duan, Y.; Qian, Y. Analysis of whole genome-wide methylation and gene expression profiles in visceral omental adipose tissue of pregnancies with gestational diabetes mellitus. J. Chin. Med. Assoc. 2018, 81, 623–630. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, T.; Chen, Y. Comprehensive Analysis of Gene Expression Profiles and DNA Methylome reveals Oas1, Ppie, Polr2g as Pathogenic Target Genes of Gestational Diabetes Mellitus. Sci. Rep. 2018, 8, 16244. [Google Scholar] [CrossRef] [PubMed]
- West, N.A.; Kechris, K.; Dabelea, D. Exposure to Maternal Diabetes in Utero and DNA Methylation Patterns in the Offspring. Immunometabolism 2013, 1, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Huang, J.; Zheng, Y.; Long, S.; Lin, H.; Zhang, N.; Tian, M.; Wu, X.; An, R.; Ma, S.; et al. Study on the relationship between DNA methylation of target CpG sites in peripheral blood and gestational diabetes during early pregnancy. Sci. Rep. 2021, 11, 20455. [Google Scholar] [CrossRef]
- Allard, C.; Desgagné, V.; Patenaude, J.; Lacroix, M.; Guillemette, L.; Battista, M.C.; Doyon, M.; Ménard, J.; Ardilouze, J.L.; Perron, P.; et al. Mendelian randomization supports causality between maternal hyperglycemia and epigenetic regulation of leptin gene in newborns. Epigenetics 2015, 10, 342–351. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Piaggi, P.; Traurig, M.; Bogardus, C.; Knowler, W.C.; Baier, L.J.; Hanson, R.L. Differential methylation of genes in individuals exposed to maternal diabetes in utero. Diabetologia 2017, 60, 645–655. [Google Scholar] [CrossRef]
- Jin, S.L.C.; Conti, M. Induction of the cyclic nucleotide phosphodiesterase PDE4B is essential for LPS-activated TNF-alpha responses. Proc. Natl. Acad. Sci. USA 2002, 99, 7628–7633. [Google Scholar] [CrossRef] [PubMed]
- Dalfrà, M.G.; Burlina, S.; Del Vescovo, G.G.; Lapolla, A. Genetics and Epigenetics: New Insight on Gestational Diabetes Mellitus. Front. Endocrinol. 2020, 11, 602477. [Google Scholar] [CrossRef]
- Dabelea, D.; Knowler, W.C.; Pettitt, D.J. Effect of diabetes in pregnancy on offspring: Follow-up research in the Pima Indians. J. Matern. Fetal Med. 2000, 9, 83–88. [Google Scholar] [CrossRef]
- Zhu, Z.; Chen, X.; Xiao, Y.; Wen, J.; Chen, J.; Wang, K.; Chen, G. Gestational diabetes mellitus alters DNA methylation profiles in pancreas of the offspring mice. J. Diabetes Complicat. 2019, 33, 15–22. [Google Scholar] [CrossRef]
- Dueva, R.; Akopyan, K.; Pederiva, C.; Trevisan, D.; Dhanjal, S.; Lindqvist, A.; Farnebo, M. Neutralization of the Positive Charges on Histone Tails by RNA Promotes an Open Chromatin Structure. Cell Chem. Biol. 2019, 26, 1436–1449.e5. [Google Scholar] [CrossRef]
- Smith, M.M. Histone structure and function. Curr. Opin. Cell Biol. 1991, 3, 429–437. [Google Scholar] [CrossRef]
- Shahid, Z.; Simpson, B.; Miao, K.H.; Singh, G. Genetics, Histone Code. Available online: https://www.ncbi.nlm.nih.gov/books/NBK538477/ (accessed on 6 October 2021).
- Gahl, W. Histone. Available online: https://www.genome.gov/genetics-glossary/histone (accessed on 28 October 2021).
- Vaquero, A.; Loyola, A.; Reinberg, D. The constantly changing face of chromatin. Sci. Aging Knowl. Environ. 2003, 2003, Re4. [Google Scholar] [CrossRef] [PubMed]
- Ramazi, S.; Allahverdi, A.; Zahiri, J. Evaluation of post-translational modifications in histone proteins: A review on histone modification defects in developmental and neurological disorders. J. Biosci. 2020, 45, 135. [Google Scholar] [CrossRef] [PubMed]
- Görisch, S.M.; Wachsmuth, M.; Tóth, K.F.; Lichter, P.; Rippe, K. Histone acetylation increases chromatin accessibility. J. Cell Sci. 2005, 118, 5825–5834. [Google Scholar] [CrossRef]
- Hepp, P.; Hutter, S.; Knabl, J.; Hofmann, S.; Kuhn, C.; Mahner, S.; Jeschke, U. Histone H3 Lysine 9 Acetylation is Downregulated in GDM Placentas and Calcitriol Supplementation Enhanced This Effect. Int. J. Mol. Sci. 2018, 19, 4061. [Google Scholar] [CrossRef]
- Michalczyk, A.A.; Dunbar, J.A.; Janus, E.D.; Best, J.D.; Ebeling, P.R.; Ackland, M.J.; Asproloupos, D.; Ackland, M.L. Epigenetic Markers to Predict Conversion from Gestational Diabetes to Type 2 Diabetes. J. Clin. Endocrinol. Metab. 2016, 101, 2396–2404. [Google Scholar] [CrossRef]
- Yao, Q.; Chen, Y.; Zhou, X. The roles of microRNAs in epigenetic regulation. Curr. Opin. Chem. Biol. 2019, 51, 11–17. [Google Scholar] [CrossRef]
- Filardi, T.; Catanzaro, G.; Mardente, S.; Zicari, A.; Santangelo, C.; Lenzi, A.; Morano, S.; Ferretti, E. Non-Coding RNA: Role in Gestational Diabetes Pathophysiology and Complications. Int. J. Mol. Sci. 2020, 21, 4020. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Feng, Y.; Qu, X.; Chen, Y.; Feng, Q.; Zhang, Y.; Hu, J.; Li, X. MicroRNA-33a-5p sponges to inhibit pancreatic β-cell function in gestational diabetes mellitus LncRNA DANCR. Reprod. Biol. Endocrinol. 2020, 18, 61. [Google Scholar] [CrossRef]
- Holley, C.L.; Topkara, V.K. An introduction to small non-coding RNAs: miRNA and snoRNA. Cardiovasc. Drugs Ther. 2011, 25, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Ibarra, A.; Vega-Guedes, B.; Brito-Casillas, Y.; Wägner, A.M. Diabetes in Pregnancy and MicroRNAs: Promises and Limitations in Their Clinical Application. Noncoding RNA 2018, 4, 32. [Google Scholar] [CrossRef] [PubMed]
- Yoffe, L.; Polsky, A.; Gilam, A.; Raff, C.; Mecacci, F.; Ognibene, A.; Crispi, F.; Gratacós, E.; Kanety, H.; Mazaki-Tovi, S.; et al. Early diagnosis of gestational diabetes mellitus using circulating microRNAs. Eur. J. Endocrinol. 2019, 181, 565–577. [Google Scholar] [CrossRef] [PubMed]
- Tagoma, A.; Alnek, K.; Kirss, A.; Uibo, R.; Haller-Kikkatalo, K. MicroRNA profiling of second trimester maternal plasma shows upregulation of miR-195-5p in patients with gestational diabetes. Gene 2018, 672, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Pan, Y.; Dai, F.; Wang, F.; Qiu, H.; Huang, X. Serum miR-195-5p is upregulated in gestational diabetes mellitus. J. Clin. Lab. Anal. 2020, 34, e23325. [Google Scholar] [CrossRef] [PubMed]
- Sebastiani, G.; Guarino, E.; Grieco, G.E.; Formichi, C.; Delli Poggi, C.; Ceccarelli, E.; Dotta, F. Circulating microRNA (miRNA) Expression Profiling in Plasma of Patients with Gestational Diabetes Mellitus Reveals Upregulation of miRNA miR-330-3p. Front. Endocrinol. 2017, 8, 345. [Google Scholar] [CrossRef]
- Lee, K.H.; Chen, Y.L.; Yeh, S.D.; Hsiao, M.; Lin, J.T.; Goan, Y.G.; Lu, P.J. MicroRNA-330 acts as tumor suppressor and induces apoptosis of prostate cancer cells through E2F1-mediated suppression of Akt phosphorylation. Oncogene 2009, 28, 3360–3370. [Google Scholar] [CrossRef]
- Li, Y.; Zhu, X.; Xu, W.; Wang, D.; Yan, J. miR-330 regulates the proliferation of colorectal cancer cells by targeting Cdc42. Biochem Biophys. Res. Commun. 2013, 431, 560–565. [Google Scholar] [CrossRef]
- Tryggestad, J.B.; Vishwanath, A.; Jiang, S.; Mallappa, A.; Teague, A.M.; Takahashi, Y.; Thompson, D.M.; Chernausek, S.D. Influence of gestational diabetes mellitus on human umbilical vein endothelial cell miRNA. Clin. Sci. 2016, 130, 1955–1967. [Google Scholar] [CrossRef]
- Short, K.R.; Teague, A.M.; Fields, D.A.; Lyons, T.; Chernausek, S.D. Lower Resting Energy Expenditure and Fat Oxidation in Native American and Hispanic Infants Born to Mothers with Diabetes. J. Pediatr. 2015, 166, 884–889. [Google Scholar] [CrossRef]
- Joshi, A.; Azuma, R.; Akumuo, R.; Goetzl, L.; Pinney, S.E. Gestational diabetes and maternal obesity are associated with sex-specific changes in miRNA and target gene expression in the fetus. Int. J. Obes. 2020, 44, 1497–1507. [Google Scholar] [CrossRef] [PubMed]
- Houshmand-Oeregaard, A.; Schrölkamp, M.; Kelstrup, L.; Hansen, N.S.; Hjort, L.; Thuesen, A.C.B.; Broholm, C.; Mathiesen, E.R.; Clausen, T.D.; Vaag, A.; et al. Increased expression of microRNA-15a and microRNA-15b in skeletal muscle from adult offspring of women with diabetes in pregnancy. Hum. Mol. Genet. 2018, 27, 1763–1771. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.-L.; Jiang, B.-G.; Li, W.-T.; Zou, J.-J.; Shi, Y.-Q.; Liu, Z.-M. MicroRNA-15a positively regulates insulin synthesis by inhibiting uncoupling protein-2 expression. Diabetes Res. Clin. Pract. 2011, 91, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Bork-Jensen, J.; Scheele, C.; Christophersen, D.V.; Nilsson, E.; Friedrichsen, M.; Fernandez-Twinn, D.S.; Grunnet, L.G.; Litman, T.; Holmstrøm, K.; Vind, B.; et al. Glucose tolerance is associated with differential expression of microRNAs in skeletal muscle: Results from studies of twins with and without type 2 diabetes. Diabetologia 2015, 58, 363–373. [Google Scholar] [CrossRef]
- Wander, P.L.; Boyko, E.J.; Hevner, K.; Parikh, V.J.; Tadesse, M.G.; Sorensen, T.K.; Williams, M.A.; Enquobahrie, D.A. Circulating early- and mid-pregnancy microRNAs and risk of gestational diabetes. Diabetes Res. Clin. Pract. 2017, 132, 1–9. [Google Scholar] [CrossRef]
- Zhao, C.; Dong, J.; Jiang, T.; Shi, Z.; Yu, B.; Zhu, Y.; Chen, D.; Xu, J.; Huo, R.; Dai, J.; et al. Early second-trimester serum miRNA profiling predicts gestational diabetes mellitus. PLoS ONE 2011, 6, e23925. [Google Scholar] [CrossRef] [PubMed]
- Guan, C.-Y.; Tian, S.; Cao, J.-L.; Wang, X.-Q.; Ma, X.; Xia, H.-F. Down-Regulated miR-21 in Gestational Diabetes Mellitus Placenta Induces PPAR-α to Inhibit Cell Proliferation and Infiltration. Diabetes Metab. Syndr. Obes. Targets Ther. 2020, 13, 3009–3034. [Google Scholar] [CrossRef]
- Qiu, H.; Liu, X.; Yao, S.; Zhou, J.; Zhang, X.; Du, J. Regulation and Mechanism of miR-518d through the PPARα-Mediated NF-κB Pathway in the Development of Gestational Diabetes Mellitus. J. Diabetes Res. 2020, 2020, 7019597. [Google Scholar] [CrossRef]
- Floris, I.; Descamps, B.; Vardeu, A.; Mitić, T.; Posadino, A.M.; Shantikumar, S.; Sala-Newby, G.; Capobianco, G.; Mangialardi, G.; Howard, L.; et al. Gestational diabetes mellitus impairs fetal endothelial cell functions through a mechanism involving microRNA-101 and histone methyltransferase enhancer of zester homolog-2. Arterioscler. Thromb. Vasc. Biol. 2015, 35, 664–674. [Google Scholar] [CrossRef]
- Li, L.; Wang, S.; Li, H.; Wan, J.; Zhou, Q.; Zhou, Y.; Zhang, C. microRNA-96 protects pancreatic β-cell function by targeting PAK1 in gestational diabetes mellitus. BioFactors 2018, 44, 539–547. [Google Scholar] [CrossRef]
- Pan, X.; Jin, X.; Wang, J.; Hu, Q.; Dai, B. Placenta inflammation is closely associated with gestational diabetes mellitus. Am. J. Transl. Res. 2021, 13, 4068–4079. [Google Scholar] [PubMed]
- Alexander, E. Omu. Pro-Inflammatory Cytokines, Lipid Metabolism and Inflammation in Gestational Diabetes Mellitus as Cause of Insulin Resistance. In Gestational Diabetes–Causes, Diagnosis and Treatment; InTech: Rijeka, Croatia, 2013; pp. 79–102. [Google Scholar]
- Copps, K.D.; White, M.F. Regulation of insulin sensitivity by serine/threonine phosphorylation of insulin receptor substrate proteins IRS1 and IRS2. Diabetologia 2012, 55, 2565–2582. [Google Scholar] [CrossRef]
- Olmos-Ortiz, A.; Flores-Espinosa, P.; Díaz, L.; Velázquez, P.; Ramírez-Isarraraz, C.; Zaga-Clavellina, V. Immunoendocrine Dysregulation during Gestational Diabetes Mellitus: The Central Role of the Placenta. Int. J. Mo.l Sci. 2021, 22, 8087. [Google Scholar] [CrossRef]
- Alonso, A.; Del Rey, C.G.; Navarro, A.; Tolivia, J.; González, C.G. Effects of gestational diabetes mellitus on proteins implicated in insulin signaling in human placenta. Gynecol. Endocrinol. 2006, 22, 526–535. [Google Scholar] [CrossRef] [PubMed]
- Colomiere, M.; Permezel, M.; Riley, C.; Desoye, G.; Lappas, M. Defective insulin signaling in placenta from pregnancies complicated by gestational diabetes mellitus. Eur. J. Endocrinol. 2009, 160, 567–578. [Google Scholar] [CrossRef] [PubMed]
- De Meyts, P. The Insulin Receptor and Its Signal Transduction Network. In Endotext; Feingold, K.R., Anawalt, B., Boyce, A., Chrousos, G., de Herder, W.W., Dhatariya, K., Dungan, K., Hershman, J.M., Hofland, J., Kalra, S., et al., Eds.; MDText.com, Inc.: South Dartmouth, MA, USA, 2000. [Google Scholar]
- Nguyen-Ngo, C.; Jayabalan, N.; Salomon, C.; Lappas, M. Molecular pathways disrupted by gestational diabetes mellitus. J. Mol. Endocrinol. 2019, 63, R51–R72. [Google Scholar] [CrossRef]
- Abell, S.K.; De Courten, B.; Boyle, J.A.; Teede, H.J. Inflammatory and Other Biomarkers: Role in Pathophysiology and Prediction of Gestational Diabetes Mellitus. Int. J. Mol. Sci. 2015, 16, 13442–13473. [Google Scholar] [CrossRef]
- Ma, Q.; Fan, J.; Wang, J.; Yang, S.; Cong, Q.; Wang, R.; Lv, Q.; Liu, R.; Ning, G. High levels of chorionic gonadotrophin attenuate insulin sensitivity and promote inflammation in adipocytes. J. Mol. Endocrinol. 2015, 54, 161–170. [Google Scholar] [CrossRef]
- Fu, Z.; Gilbert, E.R.; Liu, D. Regulation of insulin synthesis and secretion and pancreatic Beta-cell dysfunction in diabetes. Curr. Diabetes Rev. 2013, 9, 25–53. [Google Scholar] [CrossRef]
- Phoswa, W.N.; Khaliq, O.P. The Role of Oxidative Stress in Hypertensive Disorders of Pregnancy (Preeclampsia, Gestational Hypertension) and Metabolic Disorder of Pregnancy (Gestational Diabetes Mellitus). Oxidative Med. Cell. Longev. 2021, 2021, 5581570. [Google Scholar] [CrossRef]
- Chen, Y.; Tang, J.; Zhang, Y.; Du, J.; Wang, Y.; Yu, H.; He, Y. Astaxanthin alleviates gestational diabetes mellitus in mice through suppression of oxidative stress. Naunyn Schmiedebergs Arch. Pharm. 2020, 393, 2517–2527. [Google Scholar] [CrossRef] [PubMed]
- Turek, I.; Wozniak, L.; Cypryk, K.; Wojcik, M. Hyperglycaemia-induced oxidative stress in gestational diabetes mellitus (GDM). Diabetol. Klin. 2015, 4, 189–198. [Google Scholar] [CrossRef]
- de Mendonça, E.L.S.S.; Fragoso, M.B.T.; de Oliveira, J.M.; Xavier, J.A.; Goulart, M.O.F.; de Oliveira, A.C.M. Gestational Diabetes Mellitus: The Crosslink among Inflammation, Nitroxidative Stress, Intestinal Microbiota and Alternative Therapies. Antioxidants 2022, 11, 129. [Google Scholar] [CrossRef] [PubMed]
- Tu, D.; Gao, Y.; Yang, R.; Guan, T.; Hong, J.-S.; Gao, H.-M. The pentose phosphate pathway regulates chronic neuroinflammation and dopaminergic neurodegeneration. J. Neuroinflamm. 2019, 16, 255. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo-Almorós, A.; Hang, T.; Peiró, C.; Soriano-Guillén, L.; Egido, J.; Tuñón, J.; Lorenzo, Ó. Predictive and diagnostic biomarkers for gestational diabetes and its associated metabolic and cardiovascular diseases. Cardiovasc. Diabetol. 2019, 18, 140. [Google Scholar] [CrossRef]
- Powe, C.E. Early Pregnancy Biochemical Predictors of Gestational Diabetes Mellitus. Curr. Diab. Rep. 2017, 17, 12. [Google Scholar] [CrossRef]
- Ma, D.; Luque-Fernandez, M.A.; Bogdanet, D.; Desoye, G.; Dunne, F.; Halperin, J.A.; Group, D.C.I. Plasma Glycated CD59 Predicts Early Gestational Diabetes and Large for Gestational Age Newborns. J. Clin. Endocrinol. Metab. 2020, 105. [Google Scholar] [CrossRef]
- Pramodkumar, T.A.; Jayashri, R.; Gokulakrishnan, K.; Velmurugan, K.; Pradeepa, R.; Venkatesan, U.; Saravanan, P.; Uma, R.; Anjana, R.M.; Mohan, V. 1,5 Anhydroglucitol in gestational diabetes mellitus. J. Diabetes Its Complicat. 2019, 33, 231–235. [Google Scholar] [CrossRef]

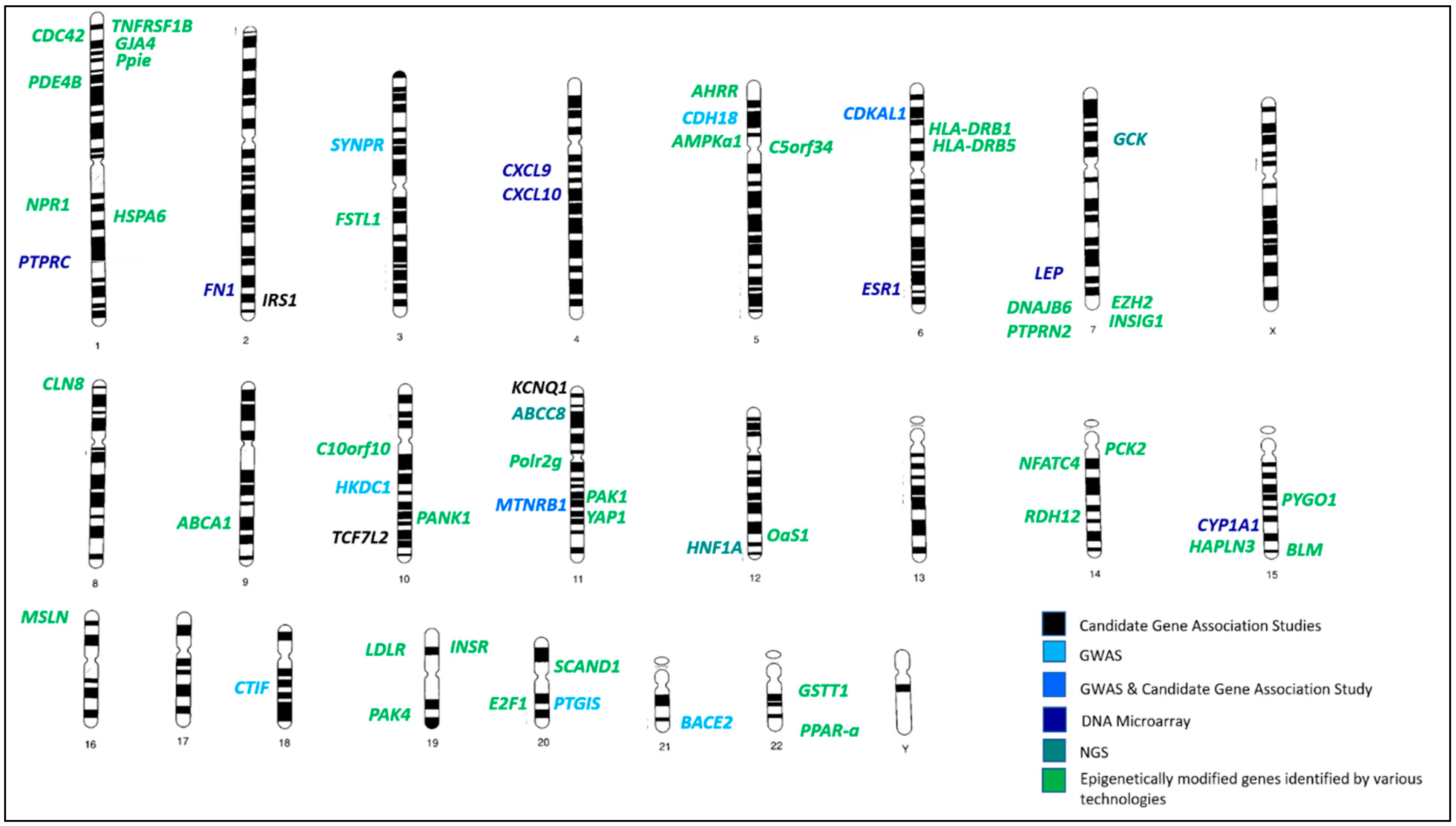
 refers to hypermethylated genes;
refers to hypermethylated genes;  refers to hypomethylated genes;
refers to hypomethylated genes;  refers to upregulated genes;
refers to upregulated genes;  refers to downregulated genes. Differentially methylated genes are in Bold.
refers to downregulated genes. Differentially methylated genes are in Bold.
 refers to hypermethylated genes;
refers to hypermethylated genes;  refers to hypomethylated genes;
refers to hypomethylated genes;  refers to upregulated genes;
refers to upregulated genes;  refers to downregulated genes. Differentially methylated genes are in Bold.
refers to downregulated genes. Differentially methylated genes are in Bold.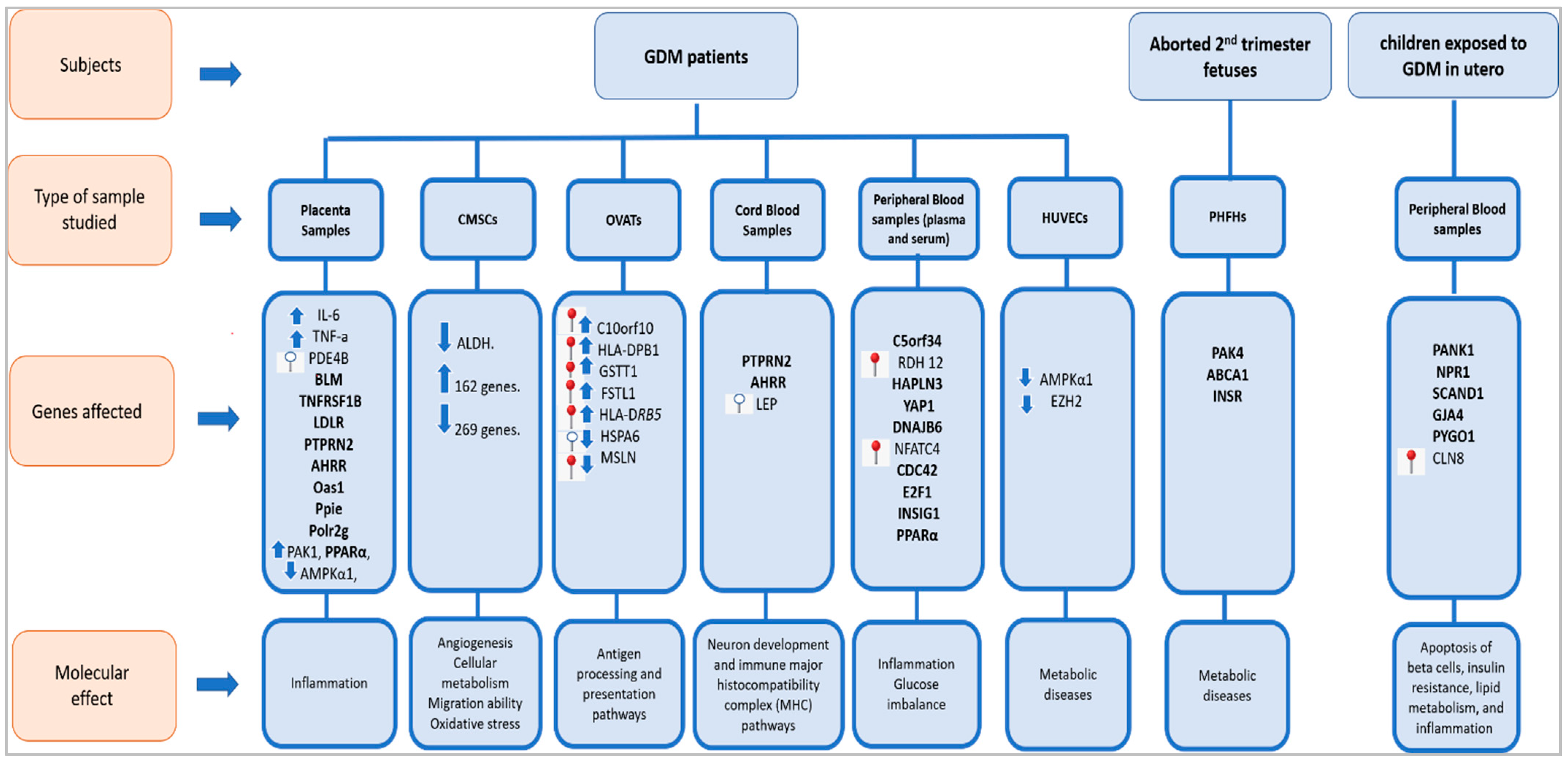

| Gene | SNP | Population/Ethnicity | Reference |
|---|---|---|---|
| TCF7L2 | rs7903146 | Scandinavian | [29] |
| Greek | [30] | ||
| Australian and British | [31] | ||
| Danish | [32] | ||
| Korean | [33] | ||
| Swedish | [34] | ||
| Italian | [35] | ||
| Finnish | [36] | ||
| Mexican | [37] | ||
| rs4506565 | Mexican | [37] | |
| Danish | [38] | ||
| rs7901695 | American Caucasian | [39] | |
| Swedish | [34] | ||
| Mexican | [37] | ||
| rs12243326 | Mexican | [37] | |
| rs12255372 | Caucasian | [40] | |
| rs34872471 | Danish | [38] | |
| rs290487 | Chinese | [40] | |
| KCNQ1 | rs2237892 | Korean | [41,42] |
| Chinese | [43] | ||
| Asian | [43] | ||
| rs2074196 | Korean | [42] | |
| rs2237895 | Pakistani | [44,45] | |
| Korean | [41] | ||
| rs2283228 | Indian | [46] | |
| CDKAL1 | rs9295478 | Chinese | [47] |
| rs6935599 | Chinese | [47] | |
| rs7747752 | Chinese | [47] | |
| rs7754840 | Asian and Caucasian | [48] | |
| rs7756992 | Asian and Caucasian | [48] | |
| IRS1 | rs1801278 | Saudi Arabian | [49] |
| Greek | [30] | ||
| Scandinavian | [50] | ||
| MTNR1B | rs10830962 | Chinese | [51] |
| rs10830963 | Asian and Caucasian | [52] | |
| Danish | [38] | ||
| Finnish | [36] | ||
| Saudi Arabian | [49] | ||
| rs1387153 | Danish | [38] | |
| Saudi Arabian | [49] | ||
| Mexican | [37] | ||
| Finnish | [36] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abu Samra, N.; Jelinek, H.F.; Alsafar, H.; Asghar, F.; Seoud, M.; Hussein, S.M.; Mubarak, H.M.; Anwar, S.; Memon, M.; Afify, N.; et al. Genomics and Epigenomics of Gestational Diabetes Mellitus: Understanding the Molecular Pathways of the Disease Pathogenesis. Int. J. Mol. Sci. 2022, 23, 3514. https://doi.org/10.3390/ijms23073514
Abu Samra N, Jelinek HF, Alsafar H, Asghar F, Seoud M, Hussein SM, Mubarak HM, Anwar S, Memon M, Afify N, et al. Genomics and Epigenomics of Gestational Diabetes Mellitus: Understanding the Molecular Pathways of the Disease Pathogenesis. International Journal of Molecular Sciences. 2022; 23(7):3514. https://doi.org/10.3390/ijms23073514
Chicago/Turabian StyleAbu Samra, Nadia, Herbert F. Jelinek, Habiba Alsafar, Farah Asghar, Muhieddine Seoud, Shahad M. Hussein, Hisham M. Mubarak, Siddiq Anwar, Mashal Memon, Nariman Afify, and et al. 2022. "Genomics and Epigenomics of Gestational Diabetes Mellitus: Understanding the Molecular Pathways of the Disease Pathogenesis" International Journal of Molecular Sciences 23, no. 7: 3514. https://doi.org/10.3390/ijms23073514
APA StyleAbu Samra, N., Jelinek, H. F., Alsafar, H., Asghar, F., Seoud, M., Hussein, S. M., Mubarak, H. M., Anwar, S., Memon, M., Afify, N., Manzoor, R., Al-Homedi, Z., & Osman, W. (2022). Genomics and Epigenomics of Gestational Diabetes Mellitus: Understanding the Molecular Pathways of the Disease Pathogenesis. International Journal of Molecular Sciences, 23(7), 3514. https://doi.org/10.3390/ijms23073514







