Raman Research on Bleomycin-Induced DNA Strand Breaks and Repair Processes in Living Cells
Abstract
:1. Introduction
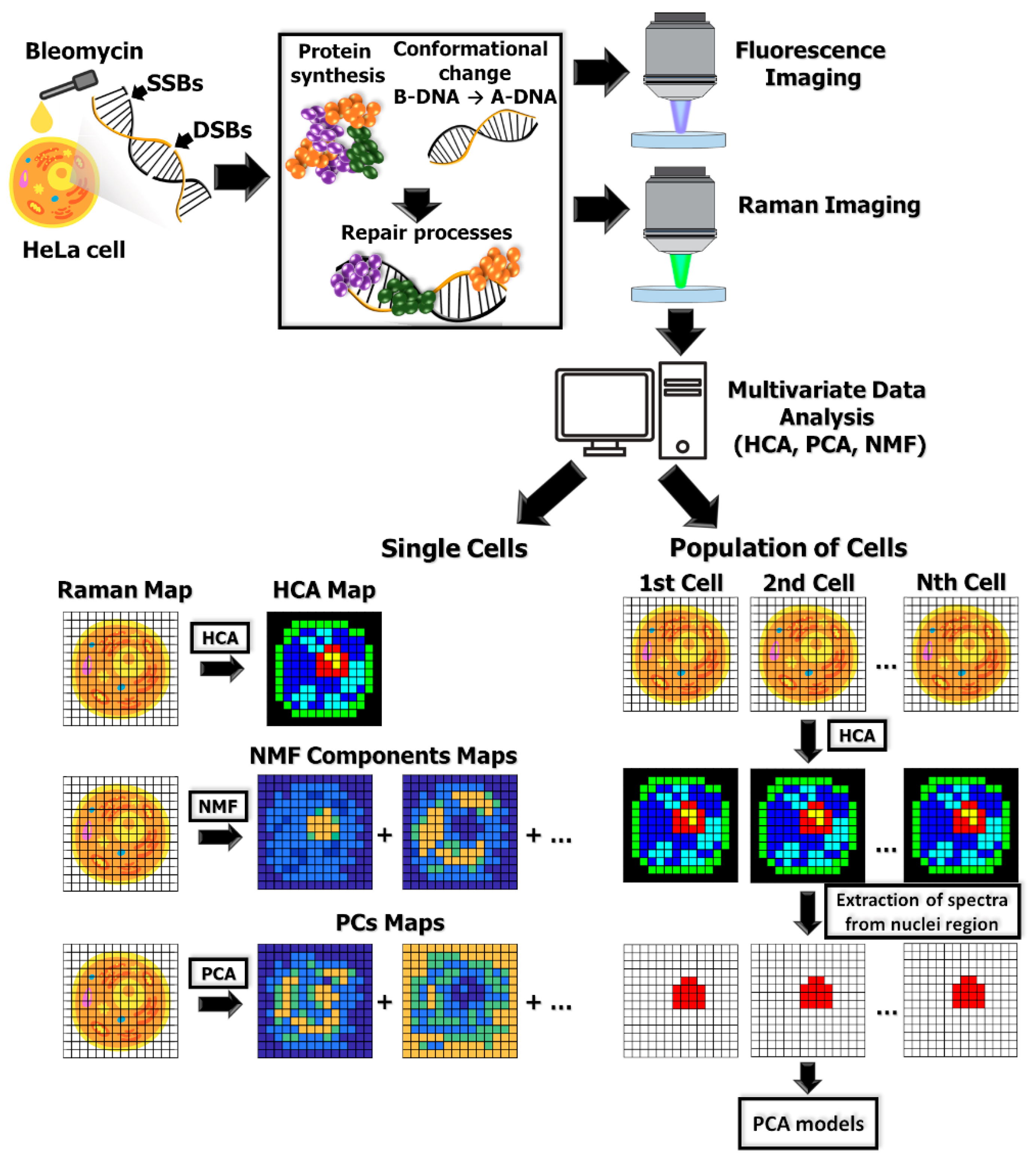
2. Results
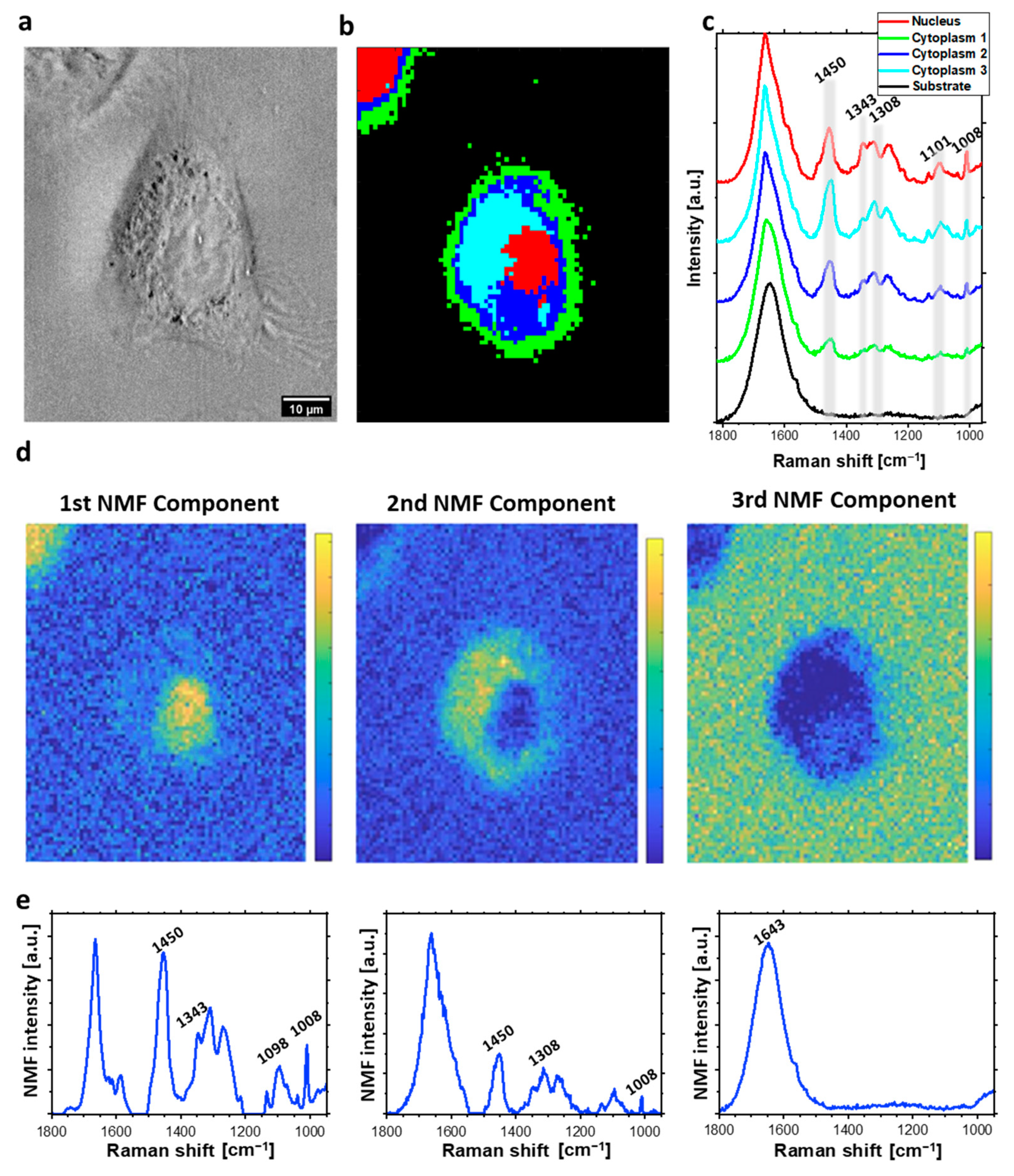
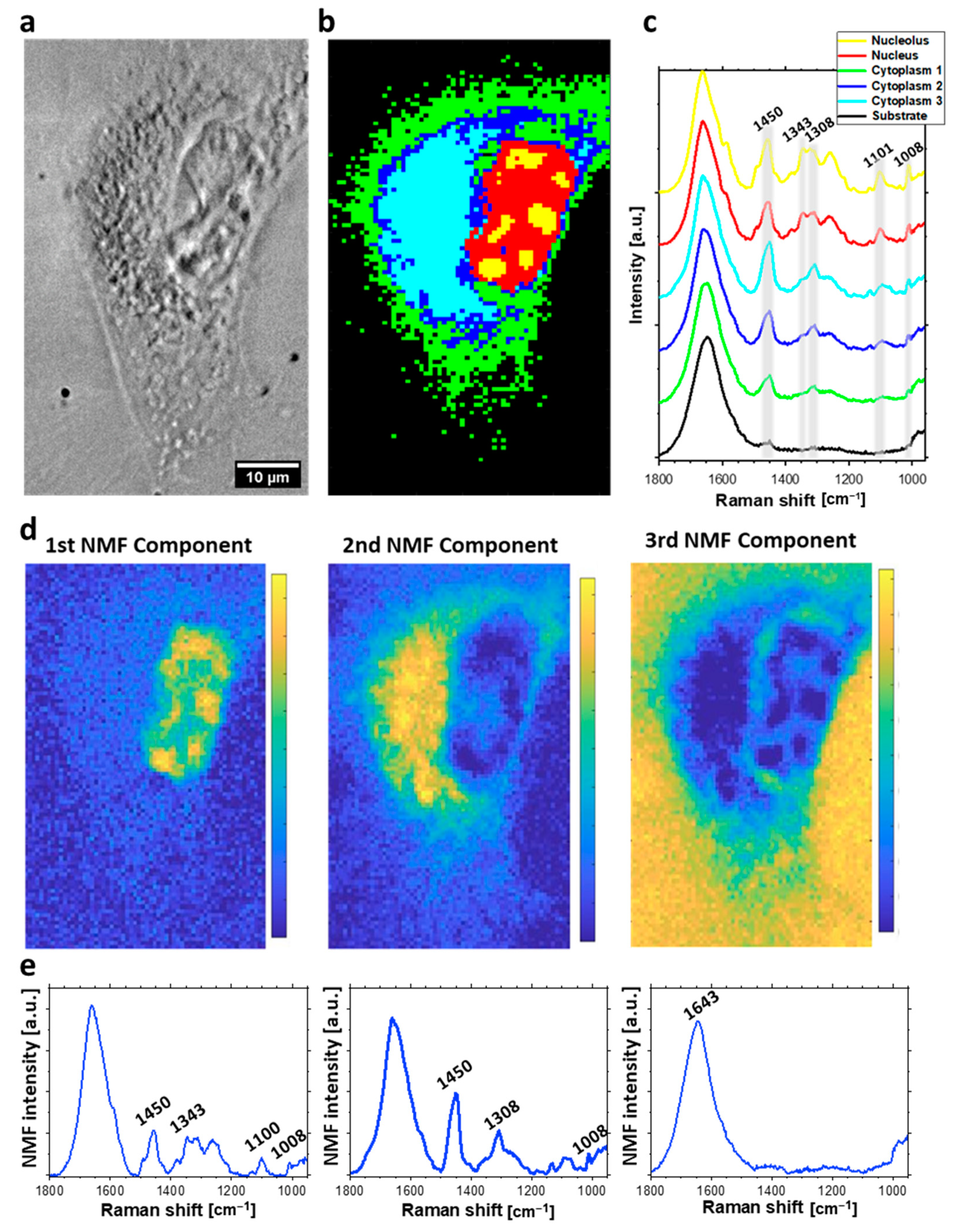
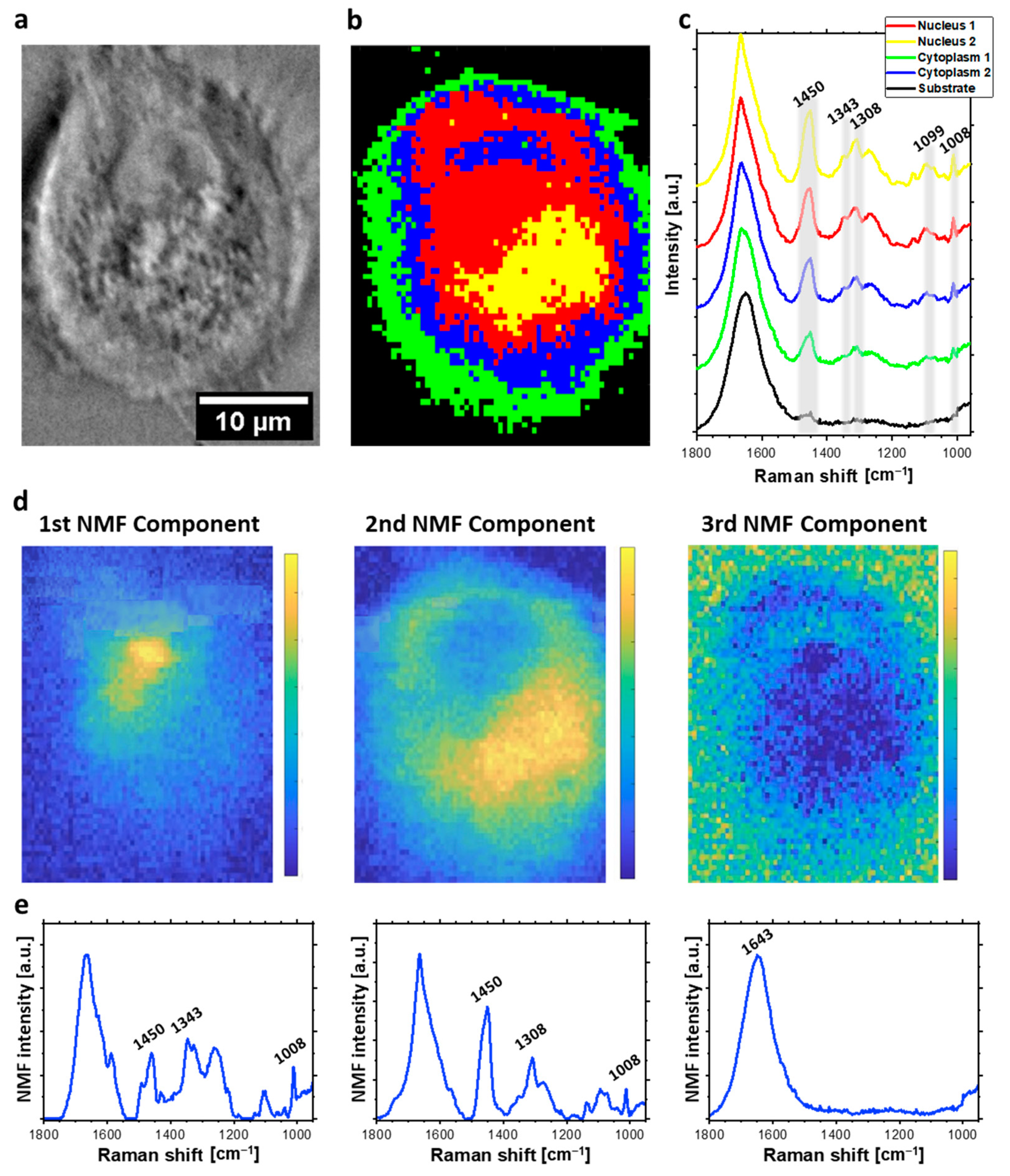
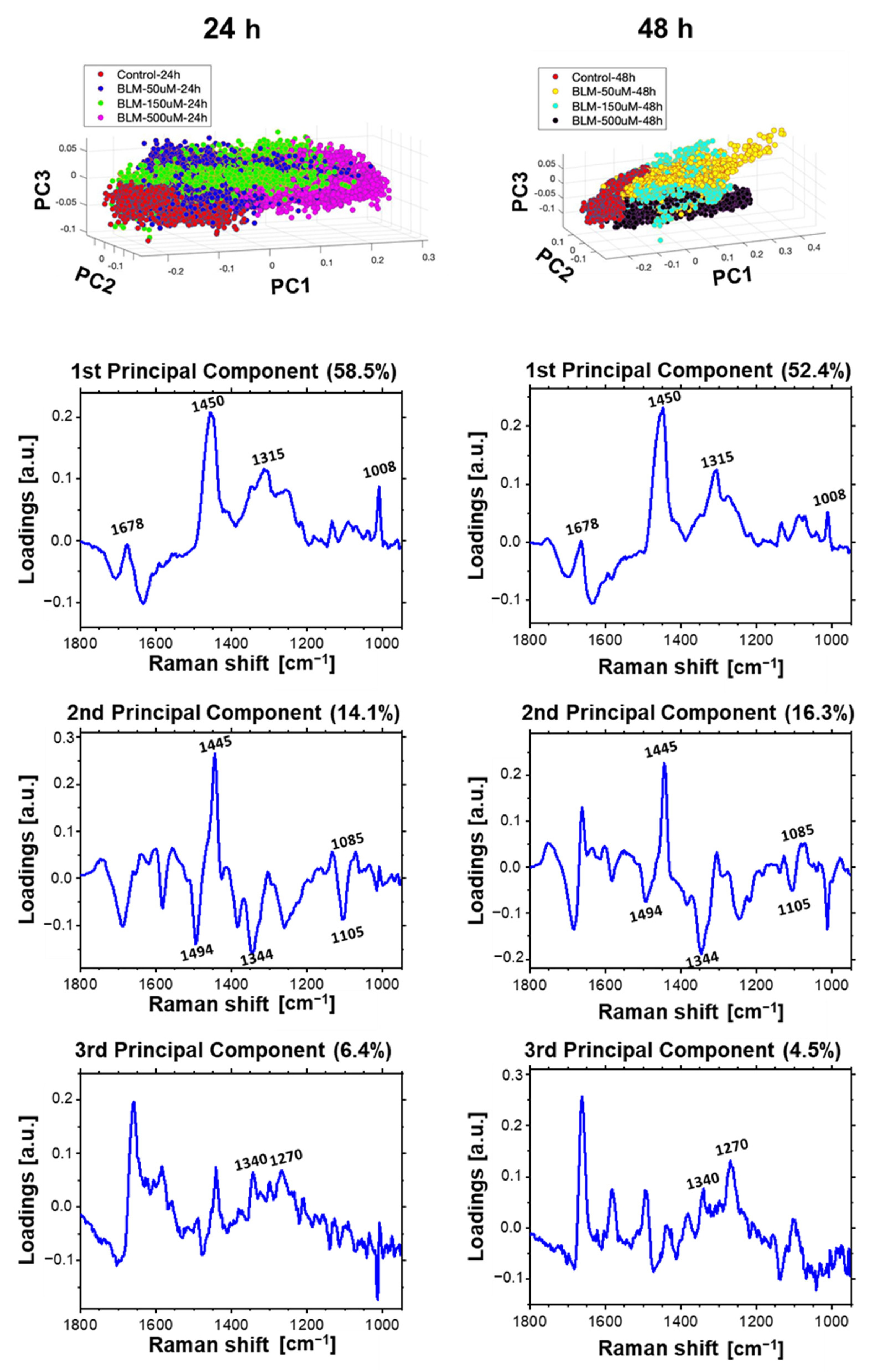
2.1. Raman Imaging and Multivariate Data Analysis of Individual HeLa Cells
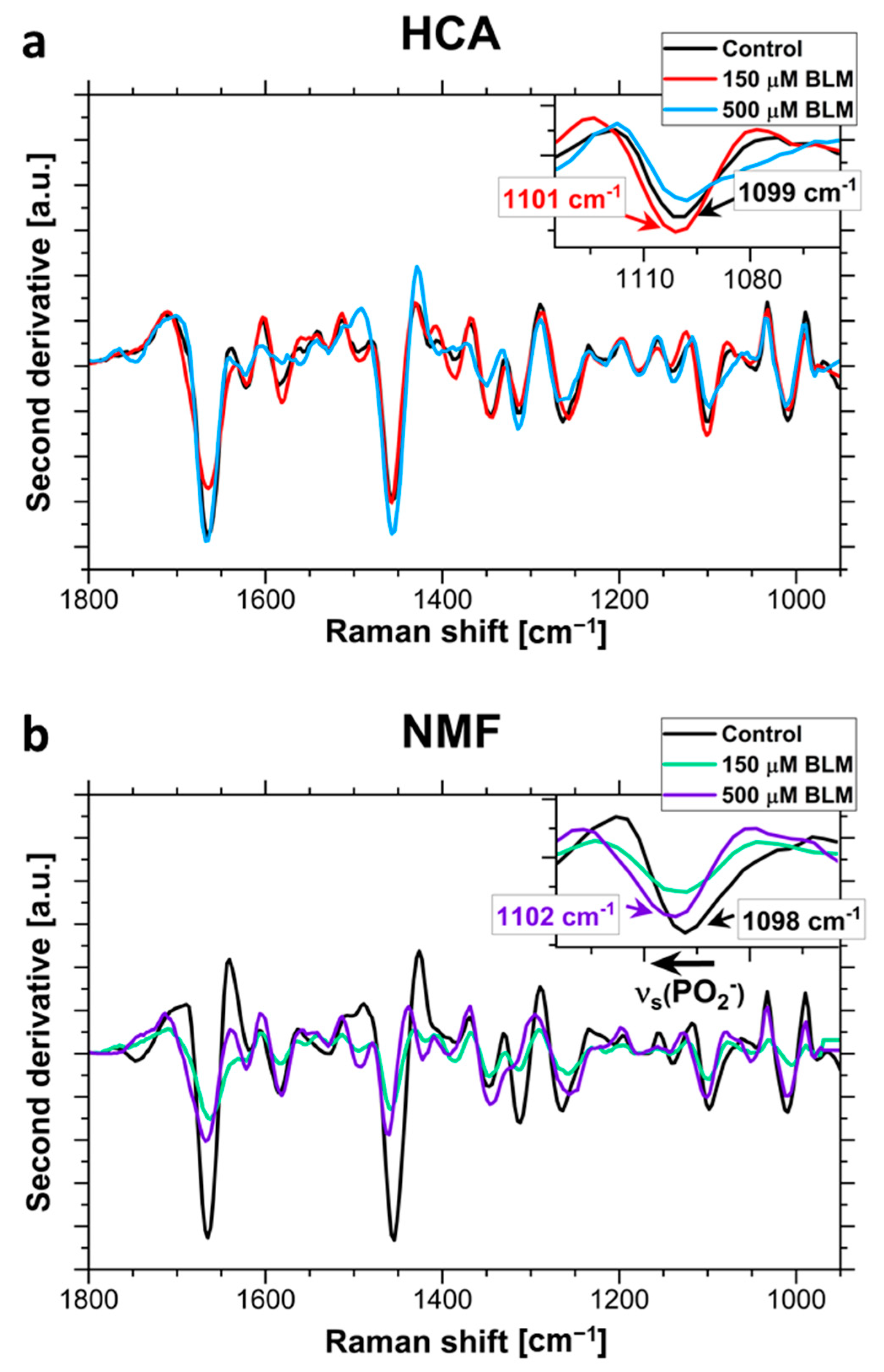
2.2. The Analysis of Bleomycin-Induced Changes in the DNA Backbone
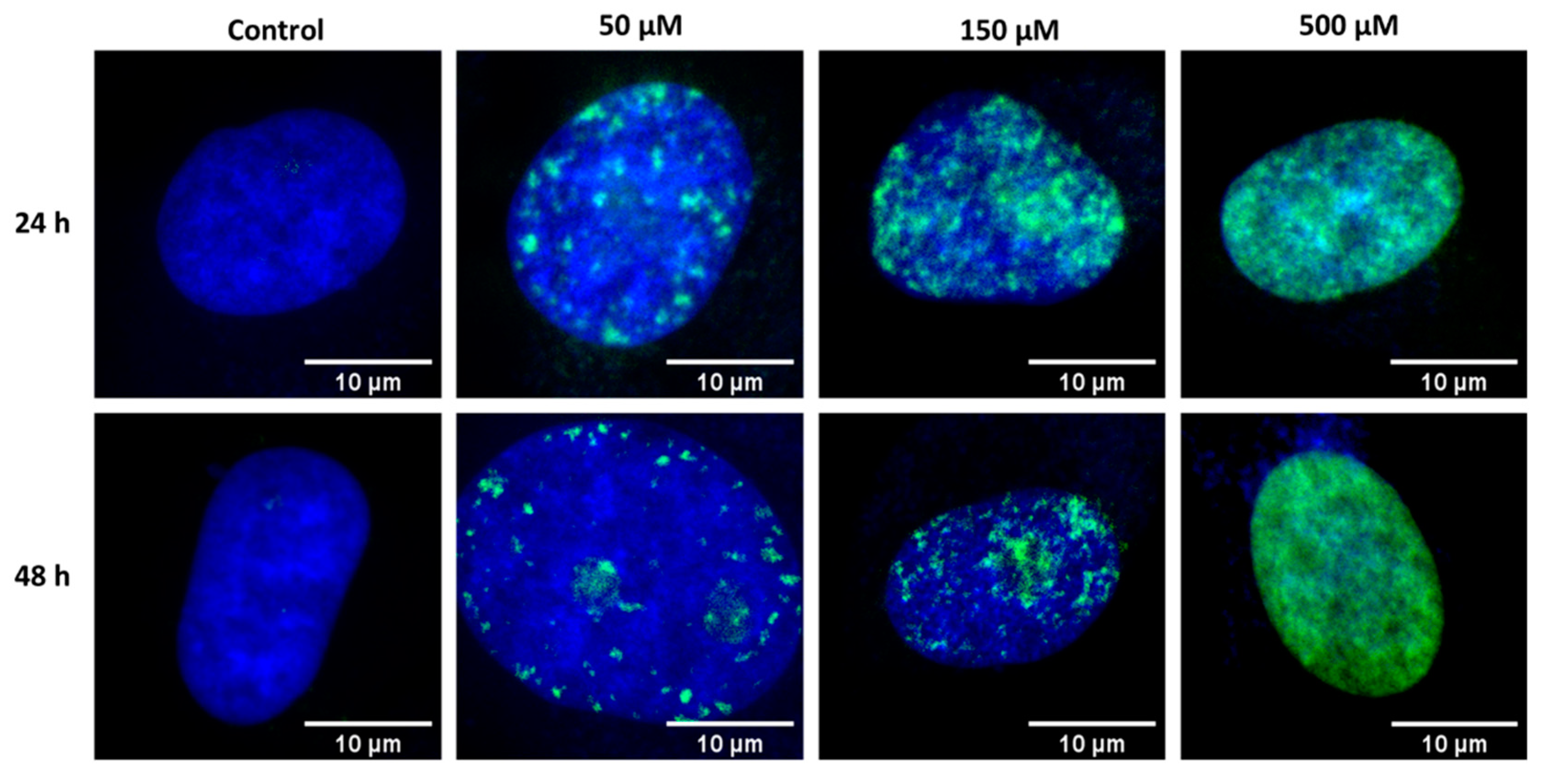
2.3. Fluorescence Imaging of Fixed HeLa Cells
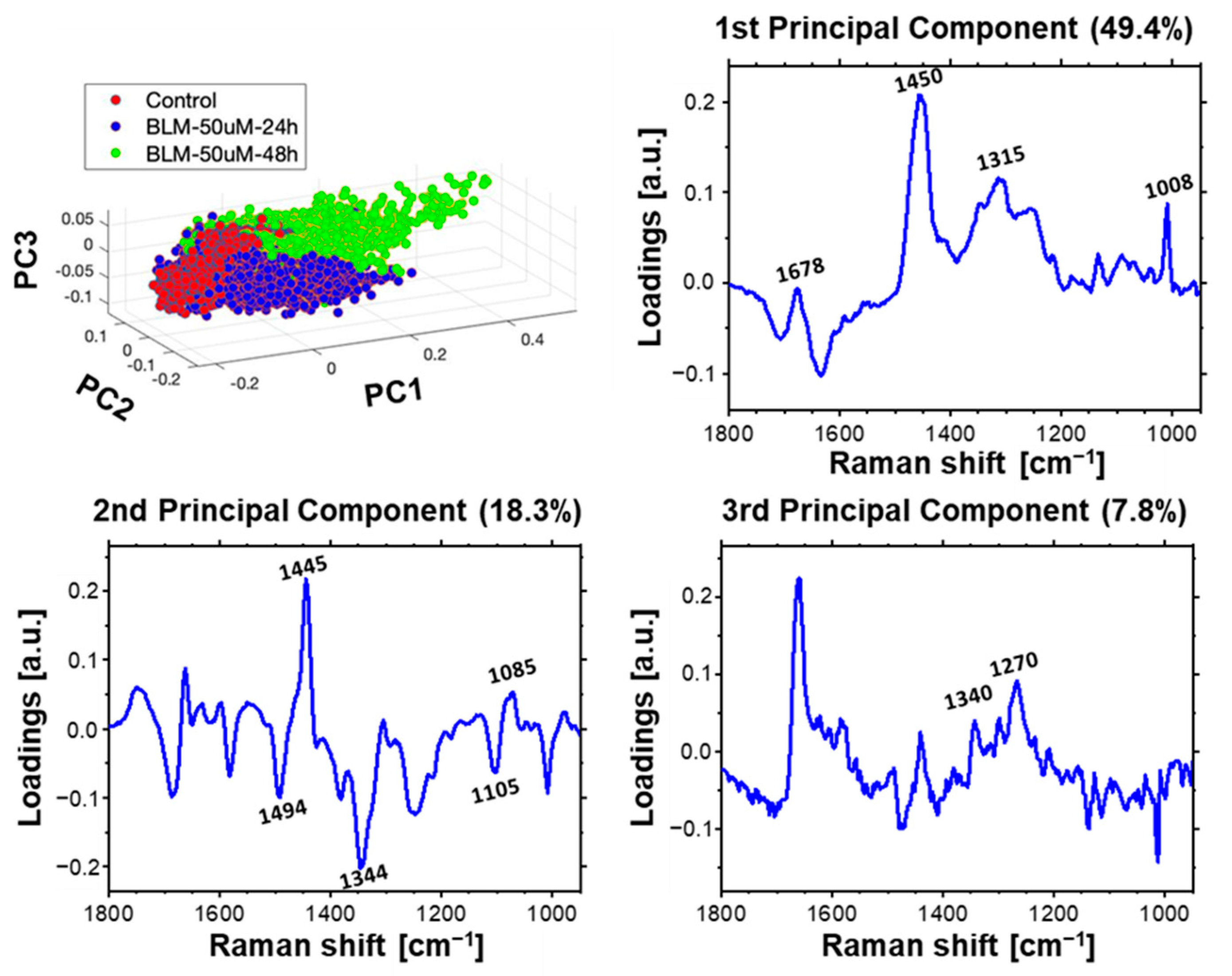
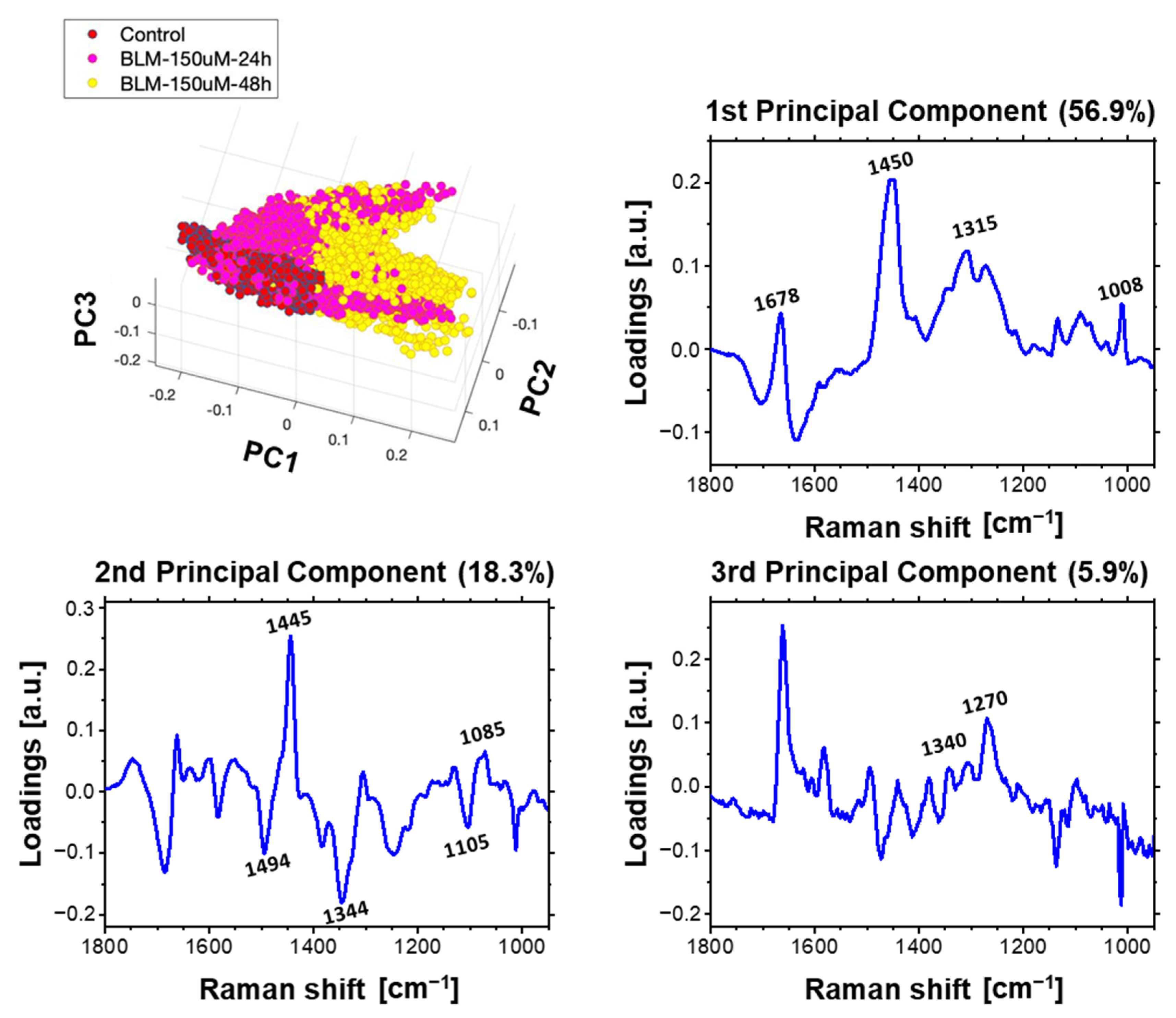
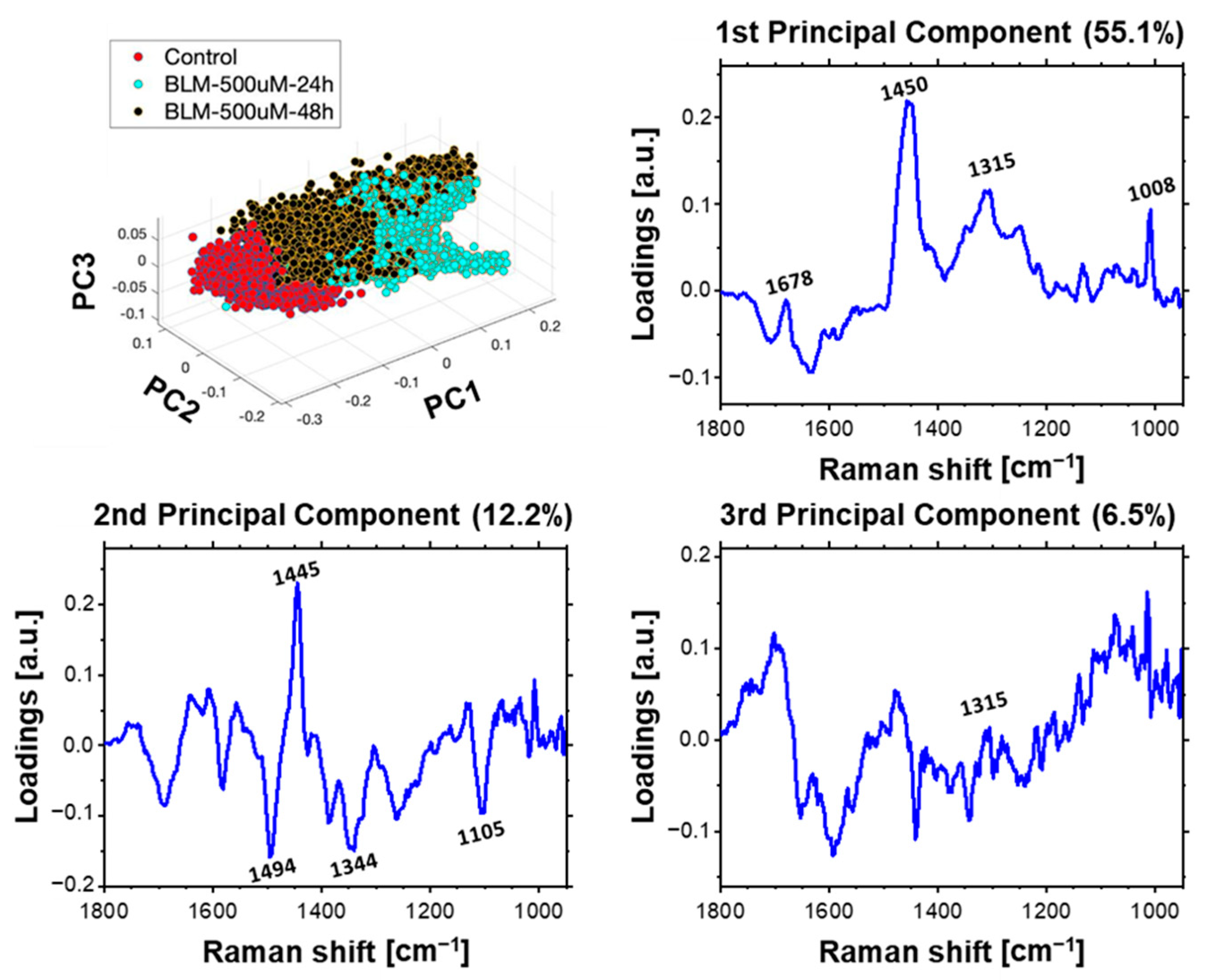
2.4. Multivariate Data Analysis of HeLa Cell Population Incubated with a Bleomycin
3. Discussion
3.1. Concentration-Dependent Effect of Bleomycin
3.2. Time-Dependent Effect of Bleomycin
4. Materials and Methods
4.1. Cell Culture
4.2. Raman Micro-Spectroscopy
4.3. Confocal Fluorescent Microscopy
4.4. Bleomycin
4.5. Data Analysis and Processing
5. Summary and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jackson, S.P.; Bartek, J. The DNA-damage response in human biology and disease. Nature 2009, 461, 1071–1078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bruner, S.D.; Norman, D.P.G.; Verdine, G.L. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature 2000, 403, 859–866. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, P. Mechanisms of DNA double-strand break repair and their potential to induce chromosomal aberrations. Mutagenesis 2000, 15, 289–302. [Google Scholar] [CrossRef]
- Chatterjee, N.; Walker, G.C. Mechanisms of DNA damage, repair, and mutagenesis. Environ. Mol. Mutagen. 2017, 58, 235–263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelms, B.E.; Maser, R.S.; MacKay, J.F.; Lagally, M.G.; Petrini, J.H.J. In situ visualization of DNA double-strand break repair in human fibroblasts. Science 1998, 280, 590–592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chapman, J.R.; Taylor, M.R.G.; Boulton, S.J. Playing the End Game: DNA Double-Strand Break Repair Pathway Choice. Mol. Cell 2012, 47, 497–510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vignard, J.; Mirey, G.; Salles, B. Ionizing-radiation induced DNA double-strand breaks: A direct and indirect lighting up. Radiother. Oncol. 2013, 108, 362–369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henner, W.D.; Grunberg, S.M.; Haseltine, W.A. Sites and structure of γ radiation-induced DNA strand breaks. J. Biol. Chem. 1982, 257, 11750–11754. [Google Scholar] [CrossRef]
- Henle, E.S.; Linn, S. Formation, prevention, and repair of DNA damage by iron/hydrogen peroxide. J. Biol. Chem. 1997, 272, 19095–19098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Stubbe, J.A. Bleomycins: Towards better therapeutics. Nat. Rev. Cancer 2005, 5, 102–112. [Google Scholar] [CrossRef] [PubMed]
- Umezawa, H.; Maeda, K.; Takeuchi, T.; Okami, Y. New antibiotics, bleomycin A and B. J. Antibiot. 1966, 19, 200–209. [Google Scholar] [CrossRef]
- Murray, V.; Martin, R.F. The sequence specificity of bleomycin-induced DNA damage in intact cells. J. Biol. Chem. 1985, 260, 10389–10391. [Google Scholar] [CrossRef]
- Hecht, S.M. Bleomycin: New perspectives on the mechanism of action. J. Nat. Prod. 2000, 63, 158–168. [Google Scholar] [CrossRef] [PubMed]
- Wood, B.R. The importance of hydration and DNA conformation in interpreting infrared spectra of cells and tissues. Chem. Soc. Rev. 2016, 45, 1980–1998. [Google Scholar] [CrossRef] [PubMed]
- Whelan, D.R.; Bambery, K.R.; Heraud, P.; Tobin, M.J.; Diem, M.; McNaughton, D.; Wood, B.R. Monitoring the reversible B to A-like transition of DNA in eukaryotic cells using Fourier transform infrared spectroscopy. Nucleic Acids Res. 2011, 39, 5439–5448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lipiec, E.; Bambery, K.R.; Heraud, P.; Kwiatek, W.M.; McNaughton, D.; Tobin, M.J.; Vogel, C.; Wood, B.R. Monitoring UVR induced damage in single cells and isolated nuclei using SR-FTIR microspectroscopy and 3D confocal Raman imaging. Analyst 2014, 139, 4200–4209. [Google Scholar] [CrossRef] [PubMed]
- LeBlanc, S.J.; Gauer, J.W.; Hao, P.; Case, B.C.; Hingorani, M.M.; Weninger, K.R.; Erie, D.A. Coordinated protein and DNA conformational changes govern mismatch repair initiation by MutS. Nucleic Acids Res. 2018, 46, 10782–10795. [Google Scholar] [CrossRef] [PubMed]
- Hashiguchi, K.; Matsumoto, Y.; Yasui, A. Recruitment of DNA repair synthesis machinery to sites of DNA damage/repair in living human cells. Nucleic Acids Res. 2007, 35, 2913–2923. [Google Scholar] [CrossRef] [PubMed]
- Rogakou, E.P.; Boon, C.; Redon, C.; Bonner, W.M. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 1999, 146, 905–915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kodaira, S.; Konishi, T.; Kobayashi, A.; Maeda, T.; Ahmad, T.A.F.T.; Yang, G.; Akselrod, M.S.; Furusawa, Y.; Uchihori, Y. Co-visualization of DNA damage and ion traversals in live mammalian cells using a fluorescent nuclear track detector. J. Radiat. Res. 2015, 56, 360–365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, R.; Wright, K.L.; Ashton, L. Raman spectroscopy: An evolving technique for live cell studies. Analyst 2016, 141, 3590–3600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palonpon, A.F.; Sodeoka, M.; Fujita, K. Molecular imaging of live cells by Raman microscopy. Curr. Opin. Chem. Biol. 2013, 17, 708–715. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-P. Practical Statistics for Data Scientists: 50+ Essential Concepts Using R and Python; O’Reilly Media: Sebastopol, CA, USA, 2021; Volume 63, ISBN 1491952938. [Google Scholar]
- Barzan, G.; Sacco, A.; Mandrile, L.; Giovannozzi, A.M.; Portesi, C.; Rossi, A.M. Hyperspectral chemical imaging of single bacterial cell structure by raman spectroscopy and machine learning. Appl. Sci. 2021, 11, 3409. [Google Scholar] [CrossRef]
- Sinjab, F.; Sicilia, G.; Shipp, D.W.; Marlow, M.; Notingher, I. Label-Free Raman Hyperspectral Imaging of Single Cells Cultured on Polymer Substrates. Appl. Spectrosc. 2017, 71, 2595–2607. [Google Scholar] [CrossRef] [PubMed]
- Szymańska-Chargot, M.; Pieczywek, P.M.; Chylińska, M.; Zdunek, A. Hyperspectral image analysis of Raman maps of plant cell walls for blind spectra characterization by nonnegative matrix factorization algorithm. Chemom. Intell. Lab. Syst. 2016, 151, 136–145. [Google Scholar] [CrossRef]
- Kniggendorf, A.K.; Gaul, T.W.; Meinhardtwollweber, M. Hierarchical cluster analysis (HCA) of microorganisms: An assessment of algorithms for resonance Raman spectra. Appl. Spectrosc. 2011, 65, 165–173. [Google Scholar] [CrossRef]
- Luce, R.; Hildebrandt, P.; Kuhlmann, U.; Liesen, J. Using separable nonnegative matrix factorization techniques for the analysis of time-resolved Raman spectra. Appl. Spectrosc. 2016, 70, 1464–1475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graham, D.J.; Castner, D.G. Multivariate analysis of ToF-SIMS data from multicomponent systems: The why, when, and how. Biointerphases 2012, 7, 49. [Google Scholar] [CrossRef] [PubMed]
- Holzweber, M.; Heinrich, T.; Kunz, V.; Richter, S.; Traulsen, C.H.-H.; Schalley, C.A.; Unger, W.E.S. Principal component analysis (PCA)-assisted time-of-flight secondary-ion mass spectrometry (ToF-SIMS): A versatile method for the investigation of self-assembled monolayers and multilayers as precursors for the bottom-up approach of nanoscaled devices. Anal. Chem. 2014, 86, 5740–5748. [Google Scholar] [CrossRef] [PubMed]
- Muramoto, S.; Graham, D.J.; Wagner, M.S.; Lee, T.G.; Moon, D.W.; Castner, D.G. ToF-SIMS analysis of adsorbed proteins: Principal component analysis of the primary ion species effect on the protein fragmentation patterns. J. Phys. Chem. C 2011, 115, 24247–24255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milligan, K.; Deng, X.; Shreeves, P.; Ali-Adeeb, R.; Matthews, Q.; Brolo, A.; Lum, J.J.; Andrews, J.L.; Jirasek, A. Raman spectroscopy and group and basis-restricted non negative matrix factorisation identifies radiation induced metabolic changes in human cancer cells. Sci. Rep. 2021, 11, 3853. [Google Scholar] [CrossRef] [PubMed]
- Naumann, D. Infrared and NIR Raman spectroscopy in medical microbiology. In Infrared Spectroscopy: New Tool in Medicine; SPIE: Philadelphia, PA, USA, 1998; Volume 3257, pp. 245–257. [Google Scholar]
- Shetty, G.; Kendall, C.; Shepherd, N.; Stone, N.; Barr, H. Raman spectroscopy: Elucidation of biochemical changes in carcinogenesis of oesophagus. Br. J. Cancer 2006, 94, 1460–1464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lakshmi, R.J.; Kartha, V.B.; Krishna, C.M.; Solomon, J.G.R.; Ullas, G.; Devi, P.U. Tissue Raman spectroscopy for the study of radiation damage: Brain irradiation of mice. Radiat. Res. 2002, 157, 175–182. [Google Scholar] [CrossRef]
- Rygula, A.; Majzner, K.; Marzec, K.M.; Kaczor, A.; Pilarczyk, M.; Baranska, M. Raman spectroscopy of proteins: A review. J. Raman Spectrosc. 2013, 44, 1061–1076. [Google Scholar] [CrossRef]
- Czamara, K.; Majzner, K.; Pacia, M.Z.; Kochan, K.; Kaczor, A.; Baranska, M. Raman spectroscopy of lipids: A review. J. Raman Spectrosc. 2015, 46, 4–20. [Google Scholar] [CrossRef]
- El-Mashtoly, S.F.; Niedieker, D.; Petersen, D.; Krauss, S.D.; Freier, E.; Maghnouj, A.; Mosig, A.; Hahn, S.; Kötting, C.; Gerwert, K. Automated identification of subcellular organelles by coherent anti-stokes raman scattering. Biophys. J. 2014, 106, 1910–1920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuzmin, A.N.; Pliss, A.; Rzhevskii, A.; Lita, A.; Larion, M. BCAbox algorithm expands capabilities of Raman microscope for single organelles assessment. Biosensors 2018, 8, 106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Notingher, I.; Green, C.; Dyer, C.; Perkins, E.; Hopkins, N.; Lindsay, C.; Hench, L.L. Discrimination between ricin and sulphur mustard toxicity in vitro using Raman spectroscopy. J. R. Soc. Interface 2004, 1, 79–90. [Google Scholar] [CrossRef] [Green Version]
- Weitzman, M.D.; Wang, J.Y.J. Cell Cycle: DNA Damage Checkpoints. In Encyclopedia of Biological Chemistry, 2nd ed.; Academic Press: Cambridge, MA, USA, 2013; pp. 410–416. ISBN 9780123786319. [Google Scholar]
- Bortner, C.D.; Cidlowski, J.A. Ions, the Movement of Water and the Apoptotic Volume Decrease. Front. Cell Dev. Biol. 2020, 8, 1415. [Google Scholar] [CrossRef]
- Bortner, C.D.; Cidlowski, J.A. Apoptotic volume decrease and the incredible shrinking cell. Cell Death Differ. 2002, 9, 1307–1310. [Google Scholar] [CrossRef]
- Wang, L.; Wang, Y.; Liang, Y.; Li, J.; Liu, Y.; Zhang, J.; Zhang, A.; Fu, J.; Jiang, G. Specific accumulation of lipid droplets in hepatocyte nuclei of PFOA-exposed BALB/c mice. Sci. Rep. 2013, 3, 2174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carey, D.M. Measurement of the Raman spectrum of liquid water. J. Chem. Phys. 1998, 108, 2669–2675. [Google Scholar] [CrossRef]
- Thomas, G.J.; Benevides, J.M.; Overman, S.A.; Ueda, T.; Ushizawa, K.; Saitoh, M.; Tsuboi, M. Polarized Raman spectra of oriented fibers of A DNA and B DNA: Anisotropic and isotropic local Raman tensors of base and backbone vibrations. Biophys. J. 1995, 68, 1073–1088. [Google Scholar] [CrossRef] [Green Version]
- Benevides, J.M.; Thomas, G.J. Characterization of DNA structures by Raman spectroscopy: High-salt and low-salt forms of double helical poly(dG-dC) in H2O and D2O solutions and application to B, Z and A-DNA. Nucleic Acids Res. 1983, 11, 5747–5761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Majtnerová, P.; Roušar, T. An overview of apoptosis assays detecting DNA fragmentation. Mol. Biol. Rep. 2018, 45, 1469–1478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robertson, J.D.; Orrenius, S.; Zhivotovsky, B. Review: Nuclear events in apoptosis. J. Struct. Biol. 2000, 129, 346–358. [Google Scholar] [CrossRef] [PubMed]
- Piétrement, O.; Pastré, D.; Landousy, F.; David, M.O.; Fusil, S.; Hamon, L.; Zozime, A.; Le Cam, E. Studying the effect of a charged surface on the interaction of bleomycin with DNA using an atomic force microscope. Eur. Biophys. J. 2005, 34, 200–207. [Google Scholar] [CrossRef] [PubMed]
- Roman, M.; Wrobel, T.P.; Panek, A.; Paluszkiewicz, C.; Kwiatek, W.M. Physicochemical damage and early-stage biological response to X-ray radiation studied in prostate cancer cells by Raman spectroscopy. J. Biophotonics 2020, 13, e202000252. [Google Scholar] [CrossRef]
- Wilkosz, N.; Czaja, M.; Seweryn, S.; Skirlinska-Nosek, K.; Szymonski, M.; Lipiec, E.; Sofinska, K. Molecular spectroscopic markers of abnormal protein aggregation. Molecules 2020, 25, 2498. [Google Scholar] [CrossRef] [PubMed]
- Maphanao, P.; Thanan, R.; Loilome, W.; Chio-Srichan, S.; Wongwattanakul, M.; Sakonsinsiri, C. Synchrotron FTIR microspectroscopy revealed apoptosis-induced biomolecular changes of cholangiocarcinoma cells treated with ursolic acid. Biochim. Biophys. Acta-Gen. Subj. 2020, 1864, 129708. [Google Scholar] [CrossRef]
- Ponkumar, S.; Duraisamy, P.; Iyandurai, N. Structural Analysis of DNA Interactions with Magnesium Ion Studied by Raman Spectroscopy. Am. J. Biochem. Biotechnol. 2011, 7, 135–140. [Google Scholar]
- Martin, O.A.; Ivashkevich, A.; Choo, S.; Woodbine, L.; Jeggo, P.A.; Martin, R.F.; Lobachevsky, P. Statistical analysis of kinetics, distribution and co-localisation of DNA repair foci in irradiated cells: Cell cycle effect and implications for prediction of radiosensitivity. DNA Repair 2013, 12, 844–855. [Google Scholar] [CrossRef] [PubMed]
- Dinant, C.; Houtsmuller, A.B.; Vermeulen, W. Chromatin structure and DNA damage repair. Epigenetics Chromatin 2008, 1, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rueden, C.T.; Schindelin, J.; Hiner, M.C.; DeZonia, B.E.; Walter, A.E.; Arena, E.T.; Eliceiri, K.W. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinform. 2017, 18, 529. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Czaja, M.; Skirlińska-Nosek, K.; Adamczyk, O.; Sofińska, K.; Wilkosz, N.; Rajfur, Z.; Szymoński, M.; Lipiec, E. Raman Research on Bleomycin-Induced DNA Strand Breaks and Repair Processes in Living Cells. Int. J. Mol. Sci. 2022, 23, 3524. https://doi.org/10.3390/ijms23073524
Czaja M, Skirlińska-Nosek K, Adamczyk O, Sofińska K, Wilkosz N, Rajfur Z, Szymoński M, Lipiec E. Raman Research on Bleomycin-Induced DNA Strand Breaks and Repair Processes in Living Cells. International Journal of Molecular Sciences. 2022; 23(7):3524. https://doi.org/10.3390/ijms23073524
Chicago/Turabian StyleCzaja, Michał, Katarzyna Skirlińska-Nosek, Olga Adamczyk, Kamila Sofińska, Natalia Wilkosz, Zenon Rajfur, Marek Szymoński, and Ewelina Lipiec. 2022. "Raman Research on Bleomycin-Induced DNA Strand Breaks and Repair Processes in Living Cells" International Journal of Molecular Sciences 23, no. 7: 3524. https://doi.org/10.3390/ijms23073524





