Roles of Nucleoporin RanBP2/Nup358 in Acute Necrotizing Encephalopathy Type 1 (ANE1) and Viral Infection
Abstract
:1. Introduction
2. RanBP2-Associated ANE1
3. Diagnosis of ANE1
4. Pathogenesis of ANE1: Roles of RanBP2 in ANE1
5. The Interplay between RanBP2 and Viruses
5.1. Herpes Simplex Viruses
5.2. Adenoviruses
5.3. Vaccinia Virus
5.4. Papillomaviruses
5.5. Severe Acute Respiratory Syndrome-Coronavirus 2
5.6. Human Rhinovirus
5.7. Hepatitis C Virus and Japanese Encephalitis Virus
5.8. Influenza Virus
5.9. Human Immunodeficiency Virus Type-1 (HIV-1)
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hoelz, A.; Debler, E.W.; Blobel, G. The Structure of the Nuclear Pore Complex. Annu. Rev. Biochem. 2011, 80, 613–643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yokoyama, N.; Hayashi, N.; Seki, T.; Panté, N.; Ohba, T.; Nishii, K.; Kuma, K.; Hayashida, T.; Miyata, T.; Aebi, U. A Giant Nucleopore Protein That Binds Ran/TC4. Nature 1995, 376, 184–188. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Matunis, M.J.; Kraemer, D.; Blobel, G.; Coutavas, E. Nup358, a Cytoplasmically Exposed Nucleoporin with Peptide Repeats, Ran-GTP Binding Sites, Zinc Fingers, a Cyclophilin A Homologous Domain, and a Leucine-Rich Region. J. Biol. Chem. 1995, 270, 14209–14213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kassube, S.A.; Stuwe, T.; Lin, D.H.; Antonuk, C.D.; Napetschnig, J.; Blobel, G.; Hoelz, A. Crystal Structure of the N-Terminal Domain of Nup358/RanBP2. J. Mol. Biol. 2012, 423, 752–765. [Google Scholar] [CrossRef] [Green Version]
- Walther, T.C.; Pickersgill, H.S.; Cordes, V.C.; Goldberg, M.W.; Allen, T.D.; Mattaj, I.W.; Fornerod, M. The Cytoplasmic Filaments of the Nuclear Pore Complex Are Dispensable for Selective Nuclear Protein Import. J. Cell Biol. 2002, 158, 63–77. [Google Scholar] [CrossRef]
- Delphin, C.; Guan, T.; Melchior, F.; Gerace, L. RanGTP Targets P97 to RanBP2, a Filamentous Protein Localized at the Cytoplasmic Periphery of the Nuclear Pore Complex. Mol. Biol. Cell 1997, 8, 2379–2390. [Google Scholar] [CrossRef] [Green Version]
- Bley, C.J.; Nie, S.; Mobbs, G.W.; Petrovic, S.; Gres, A.T.; Liu, X.; Mukherjee, S.; Harvey, S.; Huber, F.M.; Lin, D.H.; et al. Architecture of the Cytoplasmic Face of the Nuclear Pore. bioRxiv 2021. [Google Scholar] [CrossRef]
- Mahajan, R.; Delphin, C.; Guan, T.; Gerace, L.; Melchior, F. A Small Ubiquitin-Related Polypeptide Involved in Targeting RanGAP1 to Nuclear Pore Complex Protein RanBP2. Cell 1997, 88, 97–107. [Google Scholar] [CrossRef] [Green Version]
- Matunis, M.J.; Coutavas, E.; Blobel, G. A Novel Ubiquitin-like Modification Modulates the Partitioning of the Ran-GTPase-Activating Protein RanGAP1 between the Cytosol and the Nuclear Pore Complex. J. Cell Biol. 1996, 135, 1457–1470. [Google Scholar] [CrossRef]
- Matunis, M.J.; Wu, J.; Blobel, G. SUMO-1 Modification and Its Role in Targeting the Ran GTPase-Activating Protein, RanGAP1, to the Nuclear Pore Complex. J. Cell Biol. 1998, 140, 499–509. [Google Scholar] [CrossRef]
- Saitoh, H.; Pu, R.; Cavenagh, M.; Dasso, M. RanBP2 Associates with Ubc9p and a Modified Form of RanGAP1. Proc. Natl. Acad. Sci. USA 1997, 94, 3736–3741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saitoh, H.; Sparrow, D.B.; Shiomi, T.; Pu, R.T.; Nishimoto, T.; Mohun, T.J.; Dasso, M. Ubc9p and the Conjugation of SUMO-1 to RanGAP1 and RanBP2. Curr. Biol. 1998, 8, 121–124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swaminathan, S.; Kiendl, F.; Körner, R.; Lupetti, R.; Hengst, L.; Melchior, F. RanGAP1*SUMO1 Is Phosphorylated at the Onset of Mitosis and Remains Associated with RanBP2 upon NPC Disassembly. J. Cell Biol. 2004, 164, 965–971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reverter, D.; Lima, C.D. Insights into E3 Ligase Activity Revealed by a SUMO-RanGAP1-Ubc9-Nup358 Complex. Nature 2005, 435, 687–692. [Google Scholar] [CrossRef] [PubMed]
- Ben-Efraim, I.; Gerace, L. Gradient of Increasing Affinity of Importin Beta for Nucleoporins along the Pathway of Nuclear Import. J. Cell Biol. 2001, 152, 411–417. [Google Scholar] [CrossRef] [Green Version]
- Hutten, S.; Flotho, A.; Melchior, F.; Kehlenbach, R.H. The Nup358-RanGAP Complex Is Required for Efficient Importin Alpha/Beta-Dependent Nuclear Import. Mol. Biol. Cell 2008, 19, 2300–2310. [Google Scholar] [CrossRef] [Green Version]
- Saitoh, N.; Sakamoto, C.; Hagiwara, M.; Agredano-Moreno, L.T.; Jiménez-García, L.F.; Nakao, M. The Distribution of Phosphorylated SR Proteins and Alternative Splicing Are Regulated by RANBP2. Mol. Biol. Cell 2012, 23, 1115–1128. [Google Scholar] [CrossRef]
- Forler, D.; Rabut, G.; Ciccarelli, F.D.; Herold, A.; Köcher, T.; Niggeweg, R.; Bork, P.; Ellenberg, J.; Izaurralde, E. RanBP2/Nup358 Provides a Major Binding Site for NXF1-P15 Dimers at the Nuclear Pore Complex and Functions in Nuclear MRNA Export. Mol. Cell Biol. 2004, 24, 1155–1167. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, P.A.; Nakayama, T.A.; Pak, W.L.; Travis, G.H. Cyclophilin-Related Protein RanBP2 Acts as Chaperone for Red/Green Opsin. Nature 1996, 383, 637–640. [Google Scholar] [CrossRef]
- Ferreira, P.A.; Nakayama, T.A.; Travis, G.H. Interconversion of Red Opsin Isoforms by the Cyclophilin-Related Chaperone Protein Ran-Binding Protein 2. Proc. Natl. Acad. Sci. USA 1997, 94, 1556–1561. [Google Scholar] [CrossRef] [Green Version]
- Aslanukov, A.; Bhowmick, R.; Guruju, M.; Oswald, J.; Raz, D.; Bush, R.A.; Sieving, P.A.; Lu, X.; Bock, C.B.; Ferreira, P.A. RanBP2 Modulates Cox11 and Hexokinase I Activities and Haploinsufficiency of RanBP2 Causes Deficits in Glucose Metabolism. PLoS Genet 2006, 2, e177. [Google Scholar] [CrossRef]
- Joseph, J.; Liu, S.-T.; Jablonski, S.A.; Yen, T.J.; Dasso, M. The RanGAP1-RanBP2 Complex Is Essential for Microtubule-Kinetochore Interactions in Vivo. Curr. Biol. 2004, 14, 611–617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dawlaty, M.M.; Malureanu, L.; Jeganathan, K.B.; Kao, E.; Sustmann, C.; Tahk, S.; Shuai, K.; Grosschedl, R.; van Deursen, J.M. Resolution of Sister Centromeres Requires RanBP2-Mediated SUMOylation of Topoisomerase IIalpha. Cell 2008, 133, 103–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, Y.; Singh, B.B.; Aslanukov, A.; Zhao, H.; Ferreira, P.A. The Docking of Kinesins, KIF5B and KIF5C, to Ran-Binding Protein 2 (RanBP2) Is Mediated via a Novel RanBP2 Domain. J. Biol. Chem. 2001, 276, 41594–41602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, D.J.-K.; Baffet, A.D.; Nayak, T.; Akhmanova, A.; Doye, V.; Vallee, R.B. Dynein Recruitment to Nuclear Pores Activates Apical Nuclear Migration and Mitotic Entry in Brain Progenitor Cells. Cell 2013, 154, 1300–1313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asally, M.; Yasuda, Y.; Oka, M.; Otsuka, S.; Yoshimura, S.H.; Takeyasu, K.; Yoneda, Y. Nup358, a Nucleoporin, Functions as a Key Determinant of the Nuclear Pore Complex Structure Remodeling during Skeletal Myogenesis. FEBS J. 2011, 278, 610–621. [Google Scholar] [CrossRef]
- Um, J.W.; Min, D.S.; Rhim, H.; Kim, J.; Paik, S.R.; Chung, K.C. Parkin Ubiquitinates and Promotes the Degradation of RanBP2. J. Biol. Chem. 2006, 281, 3595–3603. [Google Scholar] [CrossRef] [Green Version]
- Grünwald, D.; Singer, R.H. In Vivo Imaging of Labelled Endogenous β-Actin MRNA during Nucleocytoplasmic Transport. Nature 2010, 467, 604–607. [Google Scholar] [CrossRef] [Green Version]
- Mahadevan, K.; Zhang, H.; Akef, A.; Cui, X.A.; Gueroussov, S.; Cenik, C.; Roth, F.P.; Palazzo, A.F. RanBP2/Nup358 Potentiates the Translation of a Subset of MRNAs Encoding Secretory Proteins. PLoS Biol. 2013, 11, e1001545. [Google Scholar] [CrossRef] [Green Version]
- Shen, Q.; Wang, Y.E.; Truong, M.; Mahadevan, K.; Wu, J.J.; Zhang, H.; Li, J.; Smith, H.W.; Smibert, C.A.; Palazzo, A.F. RanBP2/Nup358 Enhances MiRNA Activity by Sumoylating Argonautes. PLoS Genet 2021, 17, e1009378. [Google Scholar] [CrossRef]
- Sahoo, M.R.; Gaikwad, S.; Khuperkar, D.; Ashok, M.; Helen, M.; Yadav, S.K.; Singh, A.; Magre, I.; Deshmukh, P.; Dhanvijay, S.; et al. Nup358 Binds to AGO Proteins through Its SUMO-Interacting Motifs and Promotes the Association of Target MRNA with MiRISC. EMBO Rep. 2017, 18, 241–263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deshmukh, P.; Singh, A.; Khuperkar, D.; Joseph, J. Acute Necrotizing Encephalopathy-Linked Mutations in Nup358 Impair Interaction of Nup358 with TNRC6/GW182 and MiRNA Function. Biochem. Biophys. Res. Commun. 2021, 559, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Deshmukh, P.; Markande, S.; Fandade, V.; Ramtirtha, Y.; Madhusudhan, M.S.; Joseph, J. The MiRISC Component AGO2 Has Multiple Binding Sites for Nup358 SUMO-Interacting Motif. Biochem. Biophys. Res. Commun. 2021, 556, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Mavlyutov, T.A.; Cai, Y.; Ferreira, P.A. Identification of RanBP2- and Kinesin-Mediated Transport Pathways with Restricted Neuronal and Subcellular Localization. Traffic 2002, 3, 630–640. [Google Scholar] [CrossRef] [PubMed]
- Hampoelz, B.; Schwarz, A.; Ronchi, P.; Bragulat-Teixidor, H.; Tischer, C.; Gaspar, I.; Ephrussi, A.; Schwab, Y.; Beck, M. Nuclear Pores Assemble from Nucleoporin Condensates During Oogenesis. Cell 2019, 179, 671–686.e17. [Google Scholar] [CrossRef] [PubMed]
- Marmor-Kollet, H.; Siany, A.; Kedersha, N.; Knafo, N.; Rivkin, N.; Danino, Y.M.; Moens, T.G.; Olender, T.; Sheban, D.; Cohen, N.; et al. Spatiotemporal Proteomic Analysis of Stress Granule Disassembly Using APEX Reveals Regulation by SUMOylation and Links to ALS Pathogenesis. Mol. Cell 2020, 80, 876–891.e6. [Google Scholar] [CrossRef] [PubMed]
- Neilson, D.E.; Adams, M.D.; Orr, C.M.D.; Schelling, D.K.; Eiben, R.M.; Kerr, D.S.; Anderson, J.; Bassuk, A.G.; Bye, A.M.; Childs, A.-M.; et al. Infection-Triggered Familial or Recurrent Cases of Acute Necrotizing Encephalopathy Caused by Mutations in a Component of the Nuclear Pore, RANBP2. Am. J. Hum. Genet. 2009, 84, 44–51. [Google Scholar] [CrossRef] [Green Version]
- Sell, K.; Storch, K.; Hahn, G.; Lee-Kirsch, M.A.; Ramantani, G.; Jackson, S.; Neilson, D.; von der Hagen, M.; Hehr, U.; Smitka, M. Variable Clinical Course in Acute Necrotizing Encephalopathy and Identification of a Novel RANBP2 Mutation. Brain Dev. 2016, 38, 777–780. [Google Scholar] [CrossRef]
- Iyer, G.; Utage, P.; Bailur, S.; Utage, A.; Srirambhatla, A.; Hasan, Q. Familial Acute Necrotizing Encephalopathy: Evidence From Next Generation Sequencing of Digenic Inheritance. J. Child Neurol. 2020, 35, 393–397. [Google Scholar] [CrossRef]
- Mizuguchi, M.; Abe, J.; Mikkaichi, K.; Noma, S.; Yoshida, K.; Yamanaka, T.; Kamoshita, S. Acute Necrotising Encephalopathy of Childhood: A New Syndrome Presenting with Multifocal, Symmetric Brain Lesions. J. Neurol. Neurosurg. Psychiatry 1995, 58, 555–561. [Google Scholar] [CrossRef] [Green Version]
- Sondhi, V.; Chakrabarty, B.; Kumar, A.; Kohli, S.; Saxena, R.; Verma, I.C.; Gulati, S. RANBP2 Mutation in an Indian Child with Recurrent Acute Necrotizing Encephalopathy. Brain Dev. 2016, 38, 937–942. [Google Scholar] [CrossRef] [PubMed]
- Neilson, D. Susceptibility to Infection-Induced Acute Encephalopathy 3—Retired Chapter, FOR Historical Reference only. In GeneReviews®; Adam, M.P., Ardinger, H.H., Pagon, R.A., Wallace, S.E., Bean, L.J., Mirzaa, G., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Bashiri, F.A.; Al Johani, S.; Hamad, M.H.; Kentab, A.Y.; Alwadei, A.H.; Hundallah, K.; Hasan, H.H.; Alshuaibi, W.; Jad, L.; Alrifai, M.T.; et al. Acute Necrotizing Encephalopathy of Childhood: A Multicenter Experience in Saudi Arabia. Front. Pediatr. 2020, 8, 526. [Google Scholar] [CrossRef]
- Hartley, M.; Sinha, A.; Kumar, A.; Aliu, E.; Mainali, G.; Paudel, S. Acute Necrotizing Encephalopathy: 2 Case Reports on RANBP2 Mutation. Child Neurol. Open 2021, 8, 2329048X211030751. [Google Scholar] [CrossRef] [PubMed]
- Alawadhi, A.; Saint-Martin, C.; Bhanji, F.; Srour, M.; Atkinson, J.; Sébire, G. Acute Hemorrhagic Encephalitis Responding to Combined Decompressive Craniectomy, Intravenous Immunoglobulin, and Corticosteroid Therapies: Association with Novel RANBP2 Variant. Front. Neurol. 2018, 9, 130. [Google Scholar] [CrossRef] [Green Version]
- Neilson, D.E.; Eiben, R.M.; Waniewski, S.; Hoppel, C.L.; Varnes, M.E.; Bangert, B.A.; Wiznitzer, M.; Warman, M.L.; Kerr, D.S. Autosomal Dominant Acute Necrotizing Encephalopathy. Neurology 2003, 61, 226–230. [Google Scholar] [CrossRef]
- Neilson, D.E. The Interplay of Infection and Genetics in Acute Necrotizing Encephalopathy. Curr. Opin. Pediatr. 2010, 22, 751–757. [Google Scholar] [CrossRef]
- Mastroyianni, S.D.; Gionnis, D.; Voudris, K.; Skardoutsou, A.; Mizuguchi, M. Acute Necrotizing Encephalopathy of Childhood in Non-Asian Patients: Report of Three Cases and Literature Review. J. Child Neurol. 2006, 21, 872–879. [Google Scholar] [CrossRef]
- Campistol, J.; Gassió, R.; Pineda, M.; Fernandez-Alvarez, E. Acute Necrotizing Encephalopathy of Childhood (Infantile Bilateral Thalamic Necrosis): Two Non-Japanese Cases. Dev. Med. Child Neurol. 1998, 40, 771–774. [Google Scholar] [CrossRef]
- Saji, N.; Yamamoto, N.; Yoda, J.; Tadano, M.; Yamasaki, H.; Shimizu, H.; Kawarai, T.; Kita, Y. Adult case of acute encephalopathy associated with bilateral thalamic lesions and peripheral neuropathy. No To Shinkei 2006, 58, 1009–1014. [Google Scholar]
- Miyata, E. An adult case of acute necrotizing encephalopathy. No To Shinkei 2002, 54, 354–355. [Google Scholar]
- Nakamura, Y.; Miura, K.; Yamada, I.; Ino, H.; Mizobata, T. A novel adult case of acute necrotizing encephalopathy of childhood with bilateral symmetric thalamic lesions. Rinsho Shinkeigaku 2000, 40, 827–831. [Google Scholar] [PubMed]
- Fasano, A.; Natoli, G.F.; Cianfoni, A.; Ferraro, D.; Loria, G.; Bentivoglio, A.R.; Servidei, S. Acute Necrotizing Encephalopathy: A Relapsing Case in a European Adult. J. Neurol. Neurosurg. Psychiatry 2008, 79, 227–228. [Google Scholar] [CrossRef] [PubMed]
- Kato, H.; Hasegawa, H.; Iijima, M.; Uchigata, M.; Terada, T.; Okada, Y. Brain Magnetic Resonance Imaging of an Adult Case of Acute Necrotizing Encephalopathy. J. Neurol. 2007, 254, 1135–1137. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.R.; Sedani, S.; Lim, M.; Wassmer, E.; Absoud, M. RANBP2 Mutation and Acute Necrotizing Encephalopathy: 2 Cases and a Literature Review of the Expanding Clinico-Radiological Phenotype. Eur. J. Paediatr. Neurol. 2015, 19, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Gika, A.D.; Rich, P.; Gupta, S.; Neilson, D.E.; Clarke, A. Recurrent Acute Necrotizing Encephalopathy Following Influenza A in a Genetically Predisposed Family. Dev. Med. Child Neurol. 2010, 52, 99–102. [Google Scholar] [CrossRef]
- Loh, N.-R.; Appleton, D.B. Untreated Recurrent Acute Necrotising Encephalopathy Associated with RANBP2 Mutation, and Normal Outcome in a Caucasian Boy. Eur. J. Pediatr. 2010, 169, 1299–1302. [Google Scholar] [CrossRef]
- Marco, E.J.; Anderson, J.E.; Neilson, D.E.; Strober, J.B. Acute Necrotizing Encephalopathy in 3 Brothers. Pediatrics 2010, 125, e693–e698. [Google Scholar] [CrossRef] [Green Version]
- Gilson, C.; McFarland, R.; Forsyth, R. Autosomal Dominant Acute Necrotising Encephalopathy: A Case Report with Possible Disease-Expression Modification by Coincidental Homocysteinuria. Eur. J. Paediatr. Neurol. 2011, 15, 174–176. [Google Scholar] [CrossRef]
- Howayyer, E.; Mhanni, A.A.; Wrogemann, J.; Salman, M.S. Recurrent Acute Necrotizing Encephalopathy in a Canadian Aboriginal Child. Can J. Neurol. Sci. 2011, 38, 925–928. [Google Scholar] [CrossRef] [Green Version]
- Lönnqvist, T.; Isohanni, P.; Valanne, L.; Olli-Lähdesmäki, T.; Suomalainen, A.; Pihko, H. Dominant Encephalopathy Mimicking Mitochondrial Disease. Neurology 2011, 76, 101–103. [Google Scholar] [CrossRef]
- Bergamino, L.; Capra, V.; Biancheri, R.; Rossi, A.; Tacchella, A.; Ambrosini, L.; Mizuguchi, M.; Saitoh, M.; Marazzi, M.G. Immunomodulatory Therapy in Recurrent Acute Necrotizing Encephalopathy ANE1: Is It Useful? Brain Dev. 2012, 34, 384–391. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Lee, M.; Lee, J. Recurrent Acute Necrotizing Encephalopathy in a Korean Child: The First Non-Caucasian Case. J. Child Neurol. 2012, 27, 1343–1347. [Google Scholar] [CrossRef] [PubMed]
- Wolf, K.; Schmitt-Mechelke, T.; Kollias, S.; Curt, A. Acute Necrotizing Encephalopathy (ANE1): Rare Autosomal-Dominant Disorder Presenting as Acute Transverse Myelitis. J. Neurol. 2013, 260, 1545–1553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Denier, C.; Balu, L.; Husson, B.; Nasser, G.; Burglen, L.; Rodriguez, D.; Labauge, P.; Chevret, L. Familial Acute Necrotizing Encephalopathy Due to Mutation in the RANBP2 Gene. J. Neurol. Sci. 2014, 345, 236–238. [Google Scholar] [CrossRef] [PubMed]
- McSwiney, P.; Purnama, J.; Kornberg, A.; Danchin, M. A Severe Neurological Complication of Influenza in a Previously Well Child. Case Rep. 2014, 2014, bcr2014206930. [Google Scholar] [CrossRef]
- Di Meglio, C.; Cano, A.; Milh, M.; Girard, N.; Burglen, L.; Chabrol, B. Postinfectious family case of acute necrotizing encephalopathy caused by RANBP2 gene mutation. Arch. Pediatr. 2014, 21, 73–77. [Google Scholar] [CrossRef]
- Anand, G.; Visagan, R.; Chandratre, S.; Segal, S.; Nemeth, A.H.; Squier, W.; Sheerin, F.; Neilson, D.; Jayawant, S. H1N1 Triggered Recurrent Acute Necrotizing Encephalopathy in a Family with a T653I Mutation in the RANBP2 Gene. Pediatr. Infect. Dis. J. 2015, 34, 318–320. [Google Scholar] [CrossRef]
- Bloch, C.; Suter, B.; Fischmann, A.; Gensicke, H.; Rüegg, S.; Weisser, M. Only a Touch of the Flu? The Simultaneous Manifestation of Acute Necrotizing Encephalopathy in Two Consanguineous Patients. Open Forum. Infect. Dis. 2015, 2, ofv013. [Google Scholar] [CrossRef] [Green Version]
- Nishimura, N.; Higuchi, Y.; Kimura, N.; Nozaki, F.; Kumada, T.; Hoshino, A.; Saitoh, M.; Mizuguchi, M. Familial Acute Necrotizing Encephalopathy without RANBP2 Mutation: Poor Outcome. Pediatr. Int. 2016, 58, 1215–1218. [Google Scholar] [CrossRef]
- Lee, Y.-J.; Hwang, S.-K.; Lee, S.M.; Kwon, S. Familial Acute Necrotizing Encephalopathy with RANBP2 Mutation: The First Report in Northeast Asia. Brain Dev. 2017, 39, 625–628. [Google Scholar] [CrossRef]
- Howard, A.; Uyeki, T.M.; Fergie, J. Influenza-Associated Acute Necrotizing Encephalopathy in Siblings. J. Pediatr. Infect. Dis. Soc. 2018, 7, e172–e177. [Google Scholar] [CrossRef] [PubMed]
- Işıkay, S.; Şahin, Y. RANBP2 Mutation in Clinically Undiagnosed Acute Necrotizing Encephalopathy. Indian J. Pediatr. 2018, 85, 820–821. [Google Scholar] [CrossRef]
- Soriano-Ramos, M.; Navarro-Abia, V.; Enamorado, N.N.; Camacho-Salas, A.; De Aragón, A.M.; García-Hoyos, M.; de Las Heras, R.S. Steroids for Familial Acute Necrotizing Encephalopathy: A Future Investment? Clin. Neurol. Neurosurg. 2018, 174, 134–136. [Google Scholar] [CrossRef] [PubMed]
- Kelly, E.; Harvey, J.; Brion, K.; Fletcher, J.; Slee, M. Relapsing Necrotising Encephalomyelopathy Due to RANBP2 Mutation. Pract. Neurol. 2019, 19, 360–363. [Google Scholar] [CrossRef]
- Chew, H.B.; Ngu, L.H. RANBP2 Susceptibility to Infection-Induced Encephalopathy: Clinicoradiologic and Molecular Description in a Malaysian Family. Mol. Genet. Metab. Rep. 2020, 24, 100627. [Google Scholar] [CrossRef]
- Chow, C.K.; Ma, C.K.L. Presentation and Outcome of Acute Necrotizing Encephalopathy of Childhood: A 10-Year Single-Center Retrospective Study From Hong Kong. J. Child Neurol. 2020, 35, 674–680. [Google Scholar] [CrossRef]
- Huang, X.; Li, Y.; Yu, Y.; Yang, S.; Li, M.; Li, T.; Huang, L.; Tao, J.; Zhang, M.; Delwart, E.; et al. Human Herpesvirus 6-Associated Acute Necrotizing Encephalopathy in an Infant with a Mutation in the RANBP2 Gene. J. Paediatr. Child Health 2020, 56, 1308–1310. [Google Scholar] [CrossRef]
- Xavier, C.; Boncquet Vieira, M.; Ferreira, C.; Tavares Ferreira, J. Neuro-Ophthalmological Consequences of Acute Influenza A Encephalitis in a Genetically Predisposed Child. BMJ Case Rep. 2020, 13, e235636. [Google Scholar] [CrossRef]
- Ohashi, E.; Hayakawa, I.; Murofushi, Y.; Kawai, M.; Suzuki-Muromoto, S.; Abe, Y.; Yoshida, M.; Kono, N.; Kosaki, R.; Hoshino, A.; et al. Recurrent Acute Necrotizing Encephalopathy in a Boy with RANBP2 Mutation and Thermolabile CPT2 Variant: The First Case of ANE1 in Japan. Brain Dev. 2021, 43, 873–878. [Google Scholar] [CrossRef]
- Paktinat, M.; Hessami, K.; Inaloo, S.; Nemati, H.; Katibeh, P.; Nejabat, M.; Darabi, M.H.; Bereshneh, A.H. Case Report of RANBP2 Mutation and Familial Acute Necrotizing Encephalopathy. Int. J. Pediatr. 2021, 2021, 6695119. [Google Scholar] [CrossRef]
- Chatur, N.; Yea, C.; Ertl-Wagner, B.; Yeh, E.A. Outcomes in Influenza and RANBP2 Mutation-Associated Acute Necrotizing Encephalopathy of Childhood. Dev. Med. Child Neurol. 2022, 1–9. [Google Scholar] [CrossRef]
- Shukla, P.; Mandalla, A.; Elrick, M.J.; Venkatesan, A. Clinical Manifestations and Pathogenesis of Acute Necrotizing Encephalopathy: The Interface Between Systemic Infection and Neurologic Injury. Front. Neurol. 2022, 12, 628811. [Google Scholar] [CrossRef] [PubMed]
- Joseph, J.; Dasso, M. The Nucleoporin Nup358 Associates with and Regulates Interphase Microtubules. FEBS Lett. 2008, 582, 190–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, K.; Cai, Y.; Yi, H.; Yeh, A.; Aslanukov, A.; Ferreira, P.A. Association of the Kinesin-Binding Domain of RanBP2 to KIF5B and KIF5C Determines Mitochondria Localization and Function. Traffic 2007, 8, 1722–1735. [Google Scholar] [CrossRef]
- Shinohara, M.; Saitoh, M.; Takanashi, J.; Yamanouchi, H.; Kubota, M.; Goto, T.; Kikuchi, M.; Shiihara, T.; Yamanaka, G.; Mizuguchi, M. Carnitine Palmitoyl Transferase II Polymorphism Is Associated with Multiple Syndromes of Acute Encephalopathy with Various Infectious Diseases. Brain Dev. 2011, 33, 512–517. [Google Scholar] [CrossRef]
- Shibata, A.; Kasai, M.; Hoshino, A.; Tanaka, T.; Mizuguchi, M. RANBP2 Mutation Causing Autosomal Dominant Acute Necrotizing Encephalopathy Attenuates Its Interaction with COX11. Neurosci. Lett. 2021, 763, 136173. [Google Scholar] [CrossRef]
- Ichiyama, T.; Nishikawa, M.; Yoshitomi, T.; Hayashi, T.; Furukawa, S. Tumor Necrosis Factor-Alpha, Interleukin-1 Beta, and Interleukin-6 in Cerebrospinal Fluid from Children with Prolonged Febrile Seizures. Comparison with Acute Encephalitis/Encephalopathy. Neurology 1998, 50, 407–411. [Google Scholar] [CrossRef]
- Ichiyama, T.; Endo, S.; Kaneko, M.; Isumi, H.; Matsubara, T.; Furukawa, S. Serum Cytokine Concentrations of Influenza-Associated Acute Necrotizing Encephalopathy. Pediatr. Int. 2003, 45, 734–736. [Google Scholar] [CrossRef]
- Ichiyama, T.; Isumi, H.; Ozawa, H.; Matsubara, T.; Morishima, T.; Furukawa, S. Cerebrospinal Fluid and Serum Levels of Cytokines and Soluble Tumor Necrosis Factor Receptor in Influenza Virus-Associated Encephalopathy. Scand. J. Infect. Dis. 2003, 35, 59–61. [Google Scholar] [CrossRef]
- Ito, Y.; Ichiyama, T.; Kimura, H.; Shibata, M.; Ishiwada, N.; Kuroki, H.; Furukawa, S.; Morishima, T. Detection of Influenza Virus RNA by Reverse Transcription-PCR and Proinflammatory Cytokines in Influenza-Virus-Associated Encephalopathy. J. Med. Virol. 1999, 58, 420–425. [Google Scholar] [CrossRef]
- Akiyoshi, K.; Hamada, Y.; Yamada, H.; Kojo, M.; Izumi, T. Acute Necrotizing Encephalopathy Associated with Hemophagocytic Syndrome. Pediatr. Neurol. 2006, 34, 315–318. [Google Scholar] [CrossRef]
- Tabarki, B.; Thabet, F.; Al Shafi, S.; Al Adwani, N.; Chehab, M.; Al Shahwan, S. Acute Necrotizing Encephalopathy Associated with Enterovirus Infection. Brain Dev. 2013, 35, 454–457. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Wu, W.; Pan, W.; Wu, L.; Liu, K.; Zhang, H.-L. Acute Necrotizing Encephalopathy: An Underrecognized Clinicoradiologic Disorder. Mediat. Inflamm. 2015, 2015, 792578. [Google Scholar] [CrossRef]
- Ichiyama, T.; Morishima, T.; Isumi, H.; Matsufuji, H.; Matsubara, T.; Furukawa, S. Analysis of Cytokine Levels and NF-KappaB Activation in Peripheral Blood Mononuclear Cells in Influenza Virus-Associated Encephalopathy. Cytokine 2004, 27, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Sato, N.; Goto, T.; Haranaka, K.; Satomi, N.; Nariuchi, H.; Mano-Hirano, Y.; Sawasaki, Y. Actions of Tumor Necrosis Factor on Cultured Vascular Endothelial Cells: Morphologic Modulation, Growth Inhibition, and Cytotoxicity. J. Natl. Cancer Inst. 1986, 76, 1113–1121. [Google Scholar] [PubMed]
- Selmaj, K.W.; Raine, C.S. Tumor Necrosis Factor Mediates Myelin and Oligodendrocyte Damage in Vitro. Ann. Neurol. 1988, 23, 339–346. [Google Scholar] [CrossRef]
- Takahashi, M.; Yamada, T.; Nakashita, Y.; Saikusa, H.; Deguchi, M.; Kida, H.; Tashiro, M.; Toyoda, T. Influenza Virus-Induced Encephalopathy: Clinicopathologic Study of an Autopsied Case. Pediatr. Int. 2000, 42, 204–214. [Google Scholar] [CrossRef]
- Yoshikawa, H.; Yamazaki, S.; Watanabe, T.; Abe, T. Study of Influenza-Associated Encephalitis/Encephalopathy in Children during the 1997 to 2001 Influenza Seasons. J. Child Neurol. 2001, 16, 885–890. [Google Scholar] [CrossRef]
- Kawada, J.; Kimura, H.; Ito, Y.; Hara, S.; Iriyama, M.; Yoshikawa, T.; Morishima, T. Systemic Cytokine Responses in Patients with Influenza-Associated Encephalopathy. J. Infect. Dis. 2003, 188, 690–698. [Google Scholar] [CrossRef]
- Nakai, Y.; Itoh, M.; Mizuguchi, M.; Ozawa, H.; Okazaki, E.; Kobayashi, Y.; Takahashi, M.; Ohtani, K.; Ogawa, A.; Narita, M.; et al. Apoptosis and Microglial Activation in Influenza Encephalopathy. Acta Neuropathol. 2003, 105, 233–239. [Google Scholar] [CrossRef]
- Yokota, S.; Imagawa, T.; Miyamae, T.; Ito, S.; Nakajima, S.; Nezu, A.; Mori, M. Hypothetical Pathophysiology of Acute Encephalopathy and Encephalitis Related to Influenza Virus Infection and Hypothermia Therapy. Pediatr. Int. 2000, 42, 197–203. [Google Scholar] [CrossRef]
- Nunoi, H.; Mercado, M.R.; Mizukami, T.; Okajima, K.; Morishima, T.; Sakata, H.; Nakayama, S.; Mori, S.; Hayashi, M.; Mori, H.; et al. Apoptosis under Hypercytokinemia Is a Possible Pathogenesis in Influenza-Associated Encephalopathy. Pediatr. Int. 2005, 47, 175–179. [Google Scholar] [CrossRef]
- Cho, K.; Yoon, D.; Yu, M.; Peachey, N.S.; Ferreira, P.A. Microglial Activation in an Amyotrophic Lateral Sclerosis-like Model Caused by Ranbp2 Loss and Nucleocytoplasmic Transport Impairment in Retinal Ganglion Neurons. Cell. Mol. Life Sci. 2019, 76, 3407–3432. [Google Scholar] [CrossRef]
- Ohn, T.; Kedersha, N.; Hickman, T.; Tisdale, S.; Anderson, P. A Functional RNAi Screen Links O-GlcNAc Modification of Ribosomal Proteins to Stress Granule and Processing Body Assembly. Nat. Cell Biol. 2008, 10, 1224–1231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, Q.; Wang, Y.E.; Palazzo, A.F. Crosstalk between Nucleocytoplasmic Trafficking and the Innate Immune Response to Viral Infection. J. Biol. Chem. 2021, 297, 100856. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Trnka, M.J.; Guan, S.; Kwon, D.; Kim, D.-H.; Chen, J.-J.; Greer, P.A.; Burlingame, A.L.; Correia, M.A. A Novel Mechanism for NF-ΚB-Activation via IκB-Aggregation: Implications for Hepatic Mallory-Denk-Body Induced Inflammation. Mol. Cell Proteom. 2020, 19, 1968–1986. [Google Scholar] [CrossRef]
- Rogers, R.S.; Horvath, C.M.; Matunis, M.J. SUMO Modification of STAT1 and Its Role in PIAS-Mediated Inhibition of Gene Activation. J. Biol. Chem. 2003, 278, 30091–30097. [Google Scholar] [CrossRef] [Green Version]
- Song, L.; Bhattacharya, S.; Yunus, A.A.; Lima, C.D.; Schindler, C. Stat1 and SUMO Modification. Blood 2006, 108, 3237–3244. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Bridges, R.; Wortham, A.; Kulesz-Martin, M. NF-ΚB Repression by PIAS3 Mediated RelA SUMOylation. PLoS ONE 2012, 7, e37636. [Google Scholar] [CrossRef] [Green Version]
- Hofemeister, H.; O’Hare, P. Nuclear Pore Composition and Gating in Herpes Simplex Virus-Infected Cells. J. Virol. 2008, 82, 8392–8399. [Google Scholar] [CrossRef] [Green Version]
- Copeland, A.M.; Newcomb, W.W.; Brown, J.C. Herpes Simplex Virus Replication: Roles of Viral Proteins and Nucleoporins in Capsid-Nucleus Attachment. J. Virol. 2009, 83, 1660–1668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pasdeloup, D.; Blondel, D.; Isidro, A.L.; Rixon, F.J. Herpesvirus Capsid Association with the Nuclear Pore Complex and Viral DNA Release Involve the Nucleoporin CAN/Nup214 and the Capsid Protein PUL25. J. Virol. 2009, 83, 6610–6623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strunze, S.; Engelke, M.F.; Wang, I.-H.; Puntener, D.; Boucke, K.; Schleich, S.; Way, M.; Schoenenberger, P.; Burckhardt, C.J.; Greber, U.F. Kinesin-1-Mediated Capsid Disassembly and Disruption of the Nuclear Pore Complex Promote Virus Infection. Cell Host Microbe 2011, 10, 210–223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carlon-Andres, I.; Lagadec, F.; Pied, N.; Rayne, F.; Lafon, M.-E.; Kehlenbach, R.H.; Wodrich, H. Nup358 and Transportin 1 Cooperate in Adenoviral Genome Import. J. Virol. 2020, 94, e00164-20. [Google Scholar] [CrossRef]
- Khuperkar, D.; Kamble, A.; Singh, A.; Ghate, A.; Nawadkar, R.; Sahu, A.; Joseph, J. Selective Recruitment of Nucleoporins on Vaccinia Virus Factories and the Role of Nup358 in Viral Infection. Virology 2017, 512, 151–160. [Google Scholar] [CrossRef]
- Rosas-Acosta, G.; Langereis, M.A.; Deyrieux, A.; Wilson, V.G. Proteins of the PIAS Family Enhance the Sumoylation of the Papillomavirus E1 Protein. Virology 2005, 331, 190–203. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.-G.; Huang, W.; Lee, H.; van de Leemput, J.; Kane, M.A.; Han, Z. Characterization of SARS-CoV-2 Proteins Reveals Orf6 Pathogenicity, Subcellular Localization, Host Interactions and Attenuation by Selinexor. Cell Biosci. 2021, 11, 58. [Google Scholar] [CrossRef]
- Bock, J.-O.; Ortea, I. Re-Analysis of SARS-CoV-2-Infected Host Cell Proteomics Time-Course Data by Impact Pathway Analysis and Network Analysis: A Potential Link with Inflammatory Response. Aging 2020, 12, 11277–11286. [Google Scholar] [CrossRef]
- Ghildyal, R.; Jordan, B.; Li, D.; Dagher, H.; Bardin, P.G.; Gern, J.E.; Jans, D.A. Rhinovirus 3C Protease Can Localize in the Nucleus and Alter Active and Passive Nucleocytoplasmic Transport. J. Virol. 2009, 83, 7349–7352. [Google Scholar] [CrossRef] [Green Version]
- Neufeldt, C.J.; Joyce, M.A.; Levin, A.; Steenbergen, R.H.; Pang, D.; Shields, J.; Tyrrell, D.L.J.; Wozniak, R.W. Hepatitis C Virus-Induced Cytoplasmic Organelles Use the Nuclear Transport Machinery to Establish an Environment Conducive to Virus Replication. PLoS Pathog. 2013, 9, e1003744. [Google Scholar] [CrossRef] [Green Version]
- Levin, A.; Neufeldt, C.J.; Pang, D.; Wilson, K.; Loewen-Dobler, D.; Joyce, M.A.; Wozniak, R.W.; Tyrrell, D.L.J. Functional Characterization of Nuclear Localization and Export Signals in Hepatitis C Virus Proteins and Their Role in the Membranous Web. PLoS ONE 2014, 9, e114629. [Google Scholar] [CrossRef]
- Zhang, L.-K.; Chai, F.; Li, H.-Y.; Xiao, G.; Guo, L. Identification of Host Proteins Involved in Japanese Encephalitis Virus Infection by Quantitative Proteomics Analysis. J. Proteome Res. 2013, 12, 2666–2678. [Google Scholar] [CrossRef] [PubMed]
- Schaller, T.; Ocwieja, K.E.; Rasaiyaah, J.; Price, A.J.; Brady, T.L.; Roth, S.L.; Hué, S.; Fletcher, A.J.; Lee, K.; KewalRamani, V.N.; et al. HIV-1 Capsid-Cyclophilin Interactions Determine Nuclear Import Pathway, Integration Targeting and Replication Efficiency. PLoS Pathog. 2011, 7, e1002439. [Google Scholar] [CrossRef]
- Bichel, K.; Price, A.J.; Schaller, T.; Towers, G.J.; Freund, S.M.V.; James, L.C. HIV-1 Capsid Undergoes Coupled Binding and Isomerization by the Nuclear Pore Protein NUP358. Retrovirology 2013, 10, 81. [Google Scholar] [CrossRef] [Green Version]
- Di Nunzio, F.; Danckaert, A.; Fricke, T.; Perez, P.; Fernandez, J.; Perret, E.; Roux, P.; Shorte, S.; Charneau, P.; Diaz-Griffero, F.; et al. Human Nucleoporins Promote HIV-1 Docking at the Nuclear Pore, Nuclear Import and Integration. PLoS ONE 2012, 7, e46037. [Google Scholar] [CrossRef] [Green Version]
- Zhang, R.; Mehla, R.; Chauhan, A. Perturbation of Host Nuclear Membrane Component RanBP2 Impairs the Nuclear Import of Human Immunodeficiency Virus -1 Preintegration Complex (DNA). PLoS ONE 2010, 5, e15620. [Google Scholar] [CrossRef]
- Lee, K.; Ambrose, Z.; Martin, T.D.; Oztop, I.; Mulky, A.; Julias, J.G.; Vandegraaff, N.; Baumann, J.G.; Wang, R.; Yuen, W.; et al. Flexible Use of Nuclear Import Pathways by HIV-1. Cell Host Microbe 2010, 7, 221–233. [Google Scholar] [CrossRef] [Green Version]
- Rasaiyaah, J.; Tan, C.P.; Fletcher, A.J.; Price, A.J.; Blondeau, C.; Hilditch, L.; Jacques, D.A.; Selwood, D.L.; James, L.C.; Noursadeghi, M.; et al. HIV-1 Evades Innate Immune Recognition through Specific Cofactor Recruitment. Nature 2013, 503, 402–405. [Google Scholar] [CrossRef]
- Condit, R.C.; Moussatche, N.; Traktman, P. In a Nutshell: Structure and Assembly of the Vaccinia Virion. Adv. Virus Res. 2006, 66, 31–124. [Google Scholar] [CrossRef]
- Tolonen, N.; Doglio, L.; Schleich, S.; Krijnse Locker, J. Vaccinia Virus DNA Replication Occurs in Endoplasmic Reticulum-Enclosed Cytoplasmic Mini-Nuclei. Mol. Biol. Cell 2001, 12, 2031–2046. [Google Scholar] [CrossRef] [Green Version]
- Sivan, G.; Martin, S.E.; Myers, T.G.; Buehler, E.; Szymczyk, K.H.; Ormanoglu, P.; Moss, B. Human Genome-Wide RNAi Screen Reveals a Role for Nuclear Pore Proteins in Poxvirus Morphogenesis. Proc. Natl. Acad. Sci. USA 2013, 110, 3519–3524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doorbar, J.; Gallimore, P.H. Identification of Proteins Encoded by the L1 and L2 Open Reading Frames of Human Papillomavirus 1a. J. Virol. 1987, 61, 2793–2799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melendy, T.; Sedman, J.; Stenlund, A. Cellular Factors Required for Papillomavirus DNA Replication. J. Virol. 1995, 69, 7857–7867. [Google Scholar] [CrossRef] [Green Version]
- Wilson, V.G.; West, M.; Woytek, K.; Rangasamy, D. Papillomavirus E1 Proteins: Form, Function, and Features. Virus Genes 2002, 24, 275–290. [Google Scholar] [CrossRef] [PubMed]
- Cueille, N.; Nougarede, R.; Mechali, F.; Philippe, M.; Bonne-Andrea, C. Functional Interaction between the Bovine Papillomavirus Virus Type 1 Replicative Helicase E1 and Cyclin E-Cdk2. J. Virol. 1998, 72, 7255–7262. [Google Scholar] [CrossRef] [Green Version]
- Lentz, M.R.; Pak, D.; Mohr, I.; Botchan, M.R. The E1 Replication Protein of Bovine Papillomavirus Type 1 Contains an Extended Nuclear Localization Signal That Includes a P34cdc2 Phosphorylation Site. J. Virol. 1993, 67, 1414–1423. [Google Scholar] [CrossRef] [Green Version]
- Ma, T.; Zou, N.; Lin, B.Y.; Chow, L.T.; Harper, J.W. Interaction between Cyclin-Dependent Kinases and Human Papillomavirus Replication-Initiation Protein E1 Is Required for Efficient Viral Replication. Proc. Natl. Acad. Sci. USA 1999, 96, 382–387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zanardi, T.A.; Stanley, C.M.; Saville, B.M.; Spacek, S.M.; Lentz, M.R. Modulation of Bovine Papillomavirus DNA Replication by Phosphorylation of the Viral E1 Protein. Virology 1997, 228, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Lentz, M.R. A Carboxyl-Terminal Serine of the Bovine Papillomavirus E1 Protein Is Phosphorylated in Vivo and in Vitro. Virus Res. 2002, 83, 213–219. [Google Scholar] [CrossRef]
- McShan, G.D.; Wilson, V.G. Casein Kinase II Phosphorylates Bovine Papillomavirus Type 1 E1 in Vitro at a Conserved Motif. J. Gen. Virol. 1997, 78 Pt 1, 171–177. [Google Scholar] [CrossRef] [Green Version]
- Rangasamy, D.; Woytek, K.; Khan, S.A.; Wilson, V.G. SUMO-1 Modification of Bovine Papillomavirus E1 Protein Is Required for Intranuclear Accumulation. J. Biol. Chem. 2000, 275, 37999–38004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rangasamy, D.; Wilson, V.G. Bovine Papillomavirus E1 Protein Is Sumoylated by the Host Cell Ubc9 Protein. J. Biol. Chem. 2000, 275, 30487–30495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The Species Severe Acute Respiratory Syndrome-Related Coronavirus: Classifying 2019-NCoV and Naming It SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar] [CrossRef] [Green Version]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A Pneumonia Outbreak Associated with a New Coronavirus of Probable Bat Origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L.; et al. A SARS-CoV-2 Protein Interaction Map Reveals Targets for Drug Repurposing. Nature 2020, 583, 459–468. [Google Scholar] [CrossRef]
- Miorin, L.; Kehrer, T.; Sanchez-Aparicio, M.T.; Zhang, K.; Cohen, P.; Patel, R.S.; Cupic, A.; Makio, T.; Mei, M.; Moreno, E.; et al. SARS-CoV-2 Orf6 Hijacks Nup98 to Block STAT Nuclear Import and Antagonize Interferon Signaling. Proc. Natl. Acad. Sci. USA 2020, 117, 28344–28354. [Google Scholar] [CrossRef]
- Addetia, A.; Lieberman, N.A.P.; Phung, Q.; Hsiang, T.-Y.; Xie, H.; Roychoudhury, P.; Shrestha, L.; Loprieno, M.A.; Huang, M.-L.; Gale, M.; et al. SARS-CoV-2 ORF6 Disrupts Bidirectional Nucleocytoplasmic Transport through Interactions with Rae1 and Nup98. mBio 2021, 12, e00065-21. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical Features of Patients Infected with 2019 Novel Coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Qin, C.; Zhou, L.; Hu, Z.; Zhang, S.; Yang, S.; Tao, Y.; Xie, C.; Ma, K.; Shang, K.; Wang, W.; et al. Dysregulation of Immune Response in Patients with Coronavirus 2019 (COVID-19) in Wuhan, China. Clin. Infect. Dis. 2020, 71, 762–768. [Google Scholar] [CrossRef]
- Tan, M.; Liu, Y.; Zhou, R.; Deng, X.; Li, F.; Liang, K.; Shi, Y. Immunopathological Characteristics of Coronavirus Disease 2019 Cases in Guangzhou, China. Immunology 2020, 160, 261–268. [Google Scholar] [CrossRef]
- Xu, Z.; Shi, L.; Wang, Y.; Zhang, J.; Huang, L.; Zhang, C.; Liu, S.; Zhao, P.; Liu, H.; Zhu, L.; et al. Pathological Findings of COVID-19 Associated with Acute Respiratory Distress Syndrome. Lancet Respir. Med. 2020, 8, 420–422. [Google Scholar] [CrossRef] [PubMed]
- Hirano, T.; Murakami, M. COVID-19: A New Virus, but a Familiar Receptor and Cytokine Release Syndrome. Immunity 2020, 52, 731–733. [Google Scholar] [CrossRef] [PubMed]
- Murakami, M.; Kamimura, D.; Hirano, T. Pleiotropy and Specificity: Insights from the Interleukin 6 Family of Cytokines. Immunity 2019, 50, 812–831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thakur, K.T.; Miller, E.H.; Glendinning, M.D.; Al-Dalahmah, O.; Banu, M.A.; Boehme, A.K.; Boubour, A.L.; Bruce, S.S.; Chong, A.M.; Claassen, J.; et al. COVID-19 Neuropathology at Columbia University Irving Medical Center/New York Presbyterian Hospital. Brain 2021, 144, 2696–2708. [Google Scholar] [CrossRef]
- Poyiadji, N.; Shahin, G.; Noujaim, D.; Stone, M.; Patel, S.; Griffith, B. COVID-19-Associated Acute Hemorrhagic Necrotizing Encephalopathy: Imaging Features. Radiology 2020, 296, E119–E120. [Google Scholar] [CrossRef] [Green Version]
- Moriguchi, T.; Harii, N.; Goto, J.; Harada, D.; Sugawara, H.; Takamino, J.; Ueno, M.; Sakata, H.; Kondo, K.; Myose, N.; et al. A First Case of Meningitis/Encephalitis Associated with SARS-Coronavirus-2. Int. J. Infect. Dis. 2020, 94, 55–58. [Google Scholar] [CrossRef]
- Lazarte-Rantes, C.; Guevara-Castañón, J.; Romero, L.; Guillén-Pinto, D. Acute Necrotizing Encephalopathy Associated With SARS-CoV-2 Exposure in a Pediatric Patient. Cureus 2021, 13, e15018. [Google Scholar] [CrossRef]
- Virhammar, J.; Kumlien, E.; Fällmar, D.; Frithiof, R.; Jackmann, S.; Sköld, M.K.; Kadir, M.; Frick, J.; Lindeberg, J.; Olivero-Reinius, H.; et al. Acute Necrotizing Encephalopathy with SARS-CoV-2 RNA Confirmed in Cerebrospinal Fluid. Neurology 2020, 95, 445–449. [Google Scholar] [CrossRef]
- Ghosh, R.; Dubey, S.; Finsterer, J.; Chatterjee, S.; Ray, B.K. SARS-CoV-2-Associated Acute Hemorrhagic, Necrotizing Encephalitis (AHNE) Presenting with Cognitive Impairment in a 44-Year-Old Woman without Comorbidities: A Case Report. Am. J. Case Rep. 2020, 21, e925641. [Google Scholar] [CrossRef]
- Walker, E.J.; Younessi, P.; Fulcher, A.J.; McCuaig, R.; Thomas, B.J.; Bardin, P.G.; Jans, D.A.; Ghildyal, R. Rhinovirus 3C Protease Facilitates Specific Nucleoporin Cleavage and Mislocalisation of Nuclear Proteins in Infected Host Cells. PLoS ONE 2013, 8, e71316. [Google Scholar] [CrossRef] [Green Version]
- Johnston, S.L.; Pattemore, P.K.; Sanderson, G.; Smith, S.; Lampe, F.; Josephs, L.; Symington, P.; O’Toole, S.; Myint, S.H.; Tyrrell, D.A. Community Study of Role of Viral Infections in Exacerbations of Asthma in 9-11 Year Old Children. BMJ 1995, 310, 1225–1229. [Google Scholar] [CrossRef] [Green Version]
- Kaur, B.; Anderson, H.R.; Austin, J.; Burr, M.; Harkins, L.S.; Strachan, D.P.; Warner, J.O. Prevalence of Asthma Symptoms, Diagnosis, and Treatment in 12-14 Year Old Children across Great Britain (International Study of Asthma and Allergies in Childhood, ISAAC UK). BMJ 1998, 316, 118–124. [Google Scholar] [CrossRef] [Green Version]
- Meng, W.; Wang, X.-J.; Wang, H.-C.R. Targeting Nuclear Proteins for Control of Viral Replication. Crit. Rev. Microbiol. 2019, 45, 495–513. [Google Scholar] [CrossRef]
- Bouvier, N.M.; Palese, P. The Biology of Influenza Viruses. Vaccine 2008, 26 (Suppl. S4), D49–D53. [Google Scholar] [CrossRef] [Green Version]
- Smith, J.A.; Daniel, R. Following the Path of the Virus: The Exploitation of Host DNA Repair Mechanisms by Retroviruses. ACS Chem. Biol. 2006, 1, 217–226. [Google Scholar] [CrossRef]
- Jayappa, K.D.; Ao, Z.; Yao, X. The HIV-1 Passage from Cytoplasm to Nucleus: The Process Involving a Complex Exchange between the Components of HIV-1 and Cellular Machinery to Access Nucleus and Successful Integration. Int. J. Biochem. Mol. Biol. 2012, 3, 70–85. [Google Scholar]
- Siliciano, R.F.; Greene, W.C. HIV Latency. Cold Spring Harb. Perspect. Med. 2011, 1, a007096. [Google Scholar] [CrossRef] [Green Version]
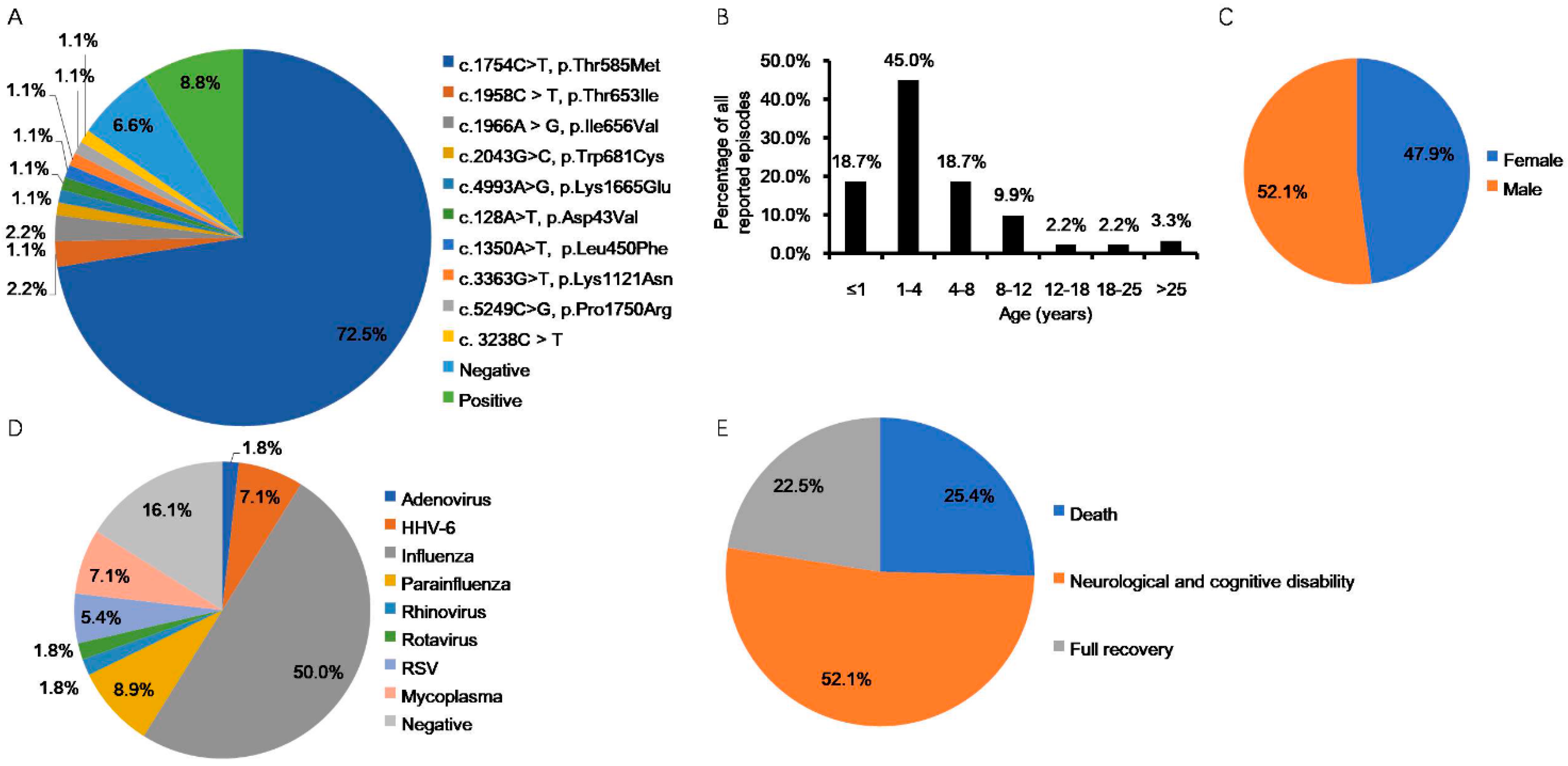
| Study, (n Cases) | Country of Origin | Age | Gender (F:M) | Infectious Agents | Recurrent Cases (n, %) | RanBP2 Mutation (n, %) | Outcome (n, %) | References |
|---|---|---|---|---|---|---|---|---|
| Neilson et al., 2009 (32) a | USA, Australia, UK, Switzerland, Denmark, Greece, Germany | Range b (n, %) <1 y: (1/32, 3%) 1~3 y: (3/32, 9%) 8~12 y: (1/32, 3%) 13~18 y: (1/32, 3%) >25 y: (1/32, 3%) N/A: (25/32, 78%) | 15:17 | Influenza (8 epi), Parainfluenza (1 epi), Mycoplasma (1 epi) | 14/32, 44% | c.1880C>T, p.Thr585Met (30/32, 94%); c.2085C>T, p.Thr653Ile (1/32, 3%); c.2094A>G, p.Ile656Val (1/32, 3%) | D (2/32, 6%), NCD (5/32, 16%), FR (4/32, 13%), N/A (21/32, 65%) | [37,46,55,56] |
| Loh et al., 2010 (1) | UK | 18 m (1st epi) 3 y 11 m (2nd epi) | 0:1 | Adenovirus (1st epi), Influenza (2nd epi) | 1/1, 100% | 3238C>T (1/1, 100%) | FR (1/1, 100%) | [57] |
| Marco et al., 2010 (3) | USA (Eastern Europeandescent) | C1:11m C2:23m C3:18 m (1st epi), 8 y (2nd~4th epi) | 0:3 | C1: HHV-6 C2: Influenza C3: N/A | 1/3, 33% | C1-3: negative (3/3, 100%) c | D (3/3, 100%) | [58] |
| Gilson et al., 2011 (1) | UK | 5 y | 1:0 | N/A | 0 | c.1754C>T; p.Thr585Met (1/1, 100%) | NCD (1/1, 100%) | [59] |
| Howayyer et al., 2011 (1) | Canada (Canadian Aboriginal Cree descent) | 34 m (1st epi), 5 y (2nd epi) | 0:1 | Negative (1st epi), Influenza (2nd epi) | 1/1, 100% | g.33868A>G p.Ile656Val (1/1, 100%) | NCD (1/1, 100%) | [60] |
| Lönnqvist et al., 2011 (6) | Finland | C1:12 y C2:9 m (1st epi), 6 y (2nd epi) C3-6:7 m to 6 y | 3:3 | N/A | 2/6, 33% d | c.1880C>T, p.Thr585Met (6/6, 100%) | FR (1/6, 17%) NCD (5/6, 83%) | [61] |
| Bergaminoet al., 2012 (1) | Italy | 5m(1st epi), 18 m (2nd epi), 26 m (3rd epi) | 0:1 | Rotavirus (1st epi), RSV (2nd epi) | 1/1, 100% | c.1880C>T, p.Thr585Met (1/1, 100%) | FR (1/1, 100%) | [62] |
| Lee et al., 2012 (1) | Korea | 12 m (1st epi), 22 m (2nd epi) | 0:1 | Influenza (1st and 2nd epi) | 1/1, 100% | Negative c (1/1, 100%) | NCD (1/1, 100%) | [63] |
| Wolf et al., 2013 (1) | Switzerland | 36 y | 1:0 | Negative | 0 | c.1880C>T, p.Thr585Met (1/1, 100%) | FR (1/1, 100%) | [64] |
| Denier et al., 2014 (3) | France | C1: 1 y (1st epi), 2 y (2nd epi), 9 y (3rd epi) C2:6 m (1st epi), 2 y (2nd epi), 6 y (3rd epi), 23 y (4th epi) C3: 5 y | 2:1 | C1: N/A C2: Influenza (2nd and 4th epi) C3: N/A | 2/3 (67%) | C1: N/A (1/3, 33%) C2–3:c.1754C>T, p.Thr585Met (2/3, 67%) | C1: D (1/3, 33%) C2–3: NCD (2/3, 67%) | [65] |
| McSwiney et al., 2014 (1) | Australia | 3 y | 1:0 | Influenza | 0 | Positive (1/1, 100%) | NCD (1/1, 100%) | [66] |
| Di Meglio et al., 2014 (2) | France (Tunisian descent) | C1: 9 m (1st epi), 9 y (2nd epi) C2: 9 m | 1:1 | N/A | 1/2, 50% | C1-2:c.1880C>T, p.Thr585Met (2/2, 100%) | C1: D (1/2, 50%), C2: NCD (1/2, 50%) | [67] |
| Anand et al., 2015 (1) | UK | 28 m | 1:0 | Influenza | 0 | c.2085C>T, p.Thr653Ile (1/1, 100%) | FR (1/1, 100%) | [68] |
| Bloch et al., 2015 (2) | Switzerland | C1:10 y C2:40 y | 1:1 | C1-2:Influenza | 0 | C1-2:c. 1754 C>Tp.Thr585Met (2/2, 100%) | C1-2: NCD (2/2, 100%) | [69] |
| Singhet al., 2015 (2) | UK | C1:2 y 7 m C2:1 y 4 m | 2:0 | C1: N/A C2: negative | 0 | C1-2:c.1880C>T: p.Thr585Met (2/2, 100%) | C1: NCD (1/2, 50%) C2: FR (1/2, 50%) | [55] |
| Sell et al., 2016 (2) | Germany | C1:10 m C2:19 m (1st epi), 22 m (2nd epi), 36 m (3rd epi) | 0:2 | C1: HHV-6 C2: N/A | 1/2, 50% | C1: c. 1754 C>T, p.Thr585Met (1/2, 50%) C2: c.2043G>C, p.Trp681Cys (1/2, 50%) | C1: NCD (1/2, 50%) C2: D (1/2, 50%) | [38] |
| Sondhi et al., 2016 (1) | India | 3.5 y (1st epi), 3 y 11 m (2nd epi), | 1:0 | Negative (1st and 2nd epi) | 1/1, 100% | c.1754 C>T, p.Thr585Met (1/1, 100%) | NCD (1/1, 100%) | [41] |
| Nishimura et al., 2016 (2) | Japan | C1:3 y 5 m C2:4 y 8 m | 0:2 | C1: N/A C2: Influenza | 0 | Negative (2/2, 100%) c | C1: D (1/2, 50%) C2: N/A (1/2, 50%) | [70] |
| Lee et al., 2017 (2) | South Korea | C1:2 y C2:12 m | 2:0 | C1: Negative C2: Mycoplasma | 0 | C1: N/A (1/2, 50%), C2:c.1754C>T, p.Thr585Met (1/2, 50%) | C1: D (1/2, 50%) C2: NCD (1/2, 50%) | [71] |
| Alawadhi et al., 2018 (1) | Canada | 6 y | 1:0 | Negative | 0 | c.4993A>G, p.Lys1665Glu (1/1, 100%) | NCD (1/1, 100%) | [45] |
| Howard et al., 2018 (2) | Mexico | C1:5 y C2:17 m | 1:1 | C1-2: Influenza | 0 | C1-2:c.1754C>T, p.Thr585Met (2/2, 100%) | C1: FR (1/2, 50%) C2: D (1/2, 50%) | [72] |
| Isikay et al., 2018 (1) | Turkey | 12 y (1st epi), 14 y (2nd epi) | 0:1 | N/A | 1/1, 100% | c.1754C>T, p.Thr585Met (1/1, 100%) | FR (1/1, 100%) | [73] |
| Soriano-Ramos et al., 2018 (1) | Spain | 7 m (1st epi), 19 m (2nd epi), 24 m (3rd epi), 10 y (4th epi) | 0:1 | N/A (1st~3rd epi), Influenza (4th epi) | 1/1, 100% | c.1754C>T, p.Thr585Met (1/1, 100%) | NCD (1/1, 100%) | [74] |
| Kelly et al., 2019 (1) | Australia | 15 m (1st epi), 27 m (2nd epi), 5 y (3rd epi), 22 y (4th epi) | 1:0 | Influenza (3rd epi) | 1/1, 100% | c.1754C>T, p.Thr585Met (1/1, 100%) | NCD (1/1, 100%) | [75] |
| Bashiri et al., 2020 (2) e | Saudi Arabia | N/A | N/A | N/A | N/A | c.3363G>T, p.Lys1121Asn (1/2, 50%), c.128A>T, p.Asp43Val (1/2, 50%) | N/A (2/2, 100%) | [43] |
| Chew et al., 2020 (3) | Malaysia | C1:11y C2:4y6m C3:2y6m | 3:0 | C1: Negative C2: Parainfluenza C3: Parainfluenza, Mycoplasma | 0 | C1-3: c.1754C>T, p.Thr585Met (3/3, 100%) | C1-2: NCD (2/3, 67%) C3: FR (1/3, 33%) | [76] |
| Chowet al., 2020 (2) | China | C4:15 m C5:9 m (1st epi), 22 m (2nd epi) | 1:1 | C4: N/A C5: Rhinovirus(1st epi), Parainfluenza(2nd epi) | 1/2, 50% | C4-5: c.1754C>T, p.Thr585Met (2/2, 100%) | C4-5: D (2/2, 100%) | [77] |
| Huang et al., 2020 (1) | China | 11 m | 0:1 | HHV-6 | 0 | c.1754C>T, p.Thr585Met (1/1, 100%) | N/A (1/1, 100%) | [78] |
| Iyer et al., 2020 (3) | India | C1:11m(1st epi), 15 m (2nd epi), 20 m (3rd epi) C2:9m C3:13m(1st epi), 24 m (2nd epi) | 1:2 | C1-3: N/A | 2/3, 67% | C1:c.5249C>G, p.Pro1750Arg f (1/3, 33%), C2–3: N/A (2/3, 67%) | C1–3: D (3/3, 100%) | [39] |
| Xavier et al., 2020 (1) | Portugal | 5y | 0:1 | Influenza | 0 | c.1754C>T, p.Thr585Met (1/1, 100%) | NCD (1/1, 100%) | [79] |
| Hartley et al., 2021 (2) | USA | C1: 9 m (1st epi), 2 y 9 m (2nd epi) C2: 6 y | 2:0 | C1: HHV-6 (1st epi) C2: negative | 1/2, 50% | C1: c.1754C>T; p.Thr585Met (1/2, 50%) C2: c.1350A>T, p.Leu450Phe (1/2, 50%) | C1–2: NCD (2/2, 100%) | [44] |
| Ohashi et al., 2021 (1) | Japan | 1 y 7 m (1st epi), 1 y 9 m (2nd epi) | 0:1 | Parainfluenza (1st epi), RSV (2nd epi) | 1/1, 100% | c.1754C>T, p.Thr585Met g (1/1, 100%) | NCD (1/1, 100%) | [80] |
| Paktinat et al., 2021 (3) | Iran | C1: 7 y (1st epi), 9 y (2nd epi) C2: 4 y C3: 4 y (1st epi), 6 y (2nd epi) | 1:2 | C1-3: N/A | 2/3, 67% | C1–2: c.1754C>T, p.Thr585Met (2/3, 67%) C3: N/A (1/3, 33%) | C1: NCD (1/3, 33%) C2: FR (1/3, 33%) C3: D (1/3, 33%) | [81] |
| Chatur et al., 2022 (7) | Canada | Range, 4 m–10 y | 3:4 | Influenza (3), Mycoplasma (1), RSV (1) N/A (2) | 3/7, 43% | Positive (7/7, 100%) | D (1/7, 14%), NCD (4/7, 57%), FR (2/7, 29) | [82] |
| Virus Group (Baltimore Classification) | Virus Family | Virus | Consequence(s) (References) |
|---|---|---|---|
| I (dsDNA viruses) | Herpesviridae | HSV-1 | Reducing the levels of O-glycosylated RanBP2 [111] Facilitating HSV-1 capsid attachment to the nuclear surface [112,113] |
| Adenoviridae | Adenoviruses | Disrupting the nuclear envelope and facilitating the transport of viral DNA into the nucleus [114,115] | |
| Poxviridae | VACV | Maintaining the size and number of viral factories and facilitating viral yield of VACV [116] | |
| Papillomaviridae | BPV | Contributing to the import of viral protein E1 to the nucleus in bovines [117] | |
| IV ((+) ssRNA viruses) | Coronaviridae | SARS-CoV-2 | Downregulating the expression level of RanBP2and might facilitate the development of “cytokine storms” in most severe patients of COVID-19 [106,107,118,119] |
| Picornaviridae | HRV | Degrading RanBP2 and disrupting nuclear envelope permeability and nucleocytoplasmic trafficking [120] | |
| Flaviviridae | HCV | Increasing mRNA and protein levels of RanBP2, and might contribute to HCV replication, assembly, and viral immune evasion [121,122] | |
| JEV | Increasing RanBP2 expression and the knockdown of RanBP2 can increase JEV replication [123] | ||
| V ((−) ssRNA viruses) | Orthomyxoviridae | IAV | Unknown |
| VI (ssRNA-RT viruses) | Retroviridae | HIV-1 | Facilitating the rapid import of HIV-1 pre-integration complex into nucleus to evade innate immune sensors and facilitating viral infection [124,125,126,127,128,129] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, J.; Wang, Y.E.; Palazzo, A.F.; Shen, Q. Roles of Nucleoporin RanBP2/Nup358 in Acute Necrotizing Encephalopathy Type 1 (ANE1) and Viral Infection. Int. J. Mol. Sci. 2022, 23, 3548. https://doi.org/10.3390/ijms23073548
Jiang J, Wang YE, Palazzo AF, Shen Q. Roles of Nucleoporin RanBP2/Nup358 in Acute Necrotizing Encephalopathy Type 1 (ANE1) and Viral Infection. International Journal of Molecular Sciences. 2022; 23(7):3548. https://doi.org/10.3390/ijms23073548
Chicago/Turabian StyleJiang, Jing, Yifan E. Wang, Alexander F. Palazzo, and Qingtang Shen. 2022. "Roles of Nucleoporin RanBP2/Nup358 in Acute Necrotizing Encephalopathy Type 1 (ANE1) and Viral Infection" International Journal of Molecular Sciences 23, no. 7: 3548. https://doi.org/10.3390/ijms23073548






