Transcriptional Regulation of the Synaptic Vesicle Protein Synaptogyrin-3 (SYNGR3) Gene: The Effects of NURR1 on Its Expression
Abstract
1. Introduction
2. Results
2.1. In Silico Analysis of the 5′-Flanking Region of the Human SYNGR3 Gene
2.2. Characterization of the 5′-Flanking Region of the Human SYNGR3 Gene
2.3. In Silico and Promoter Analyses Showed XCPE1 Decreased SYNGR3 Promoter Activity in SH-SY5Y and HEK293 Cells
2.4. Site-Directed Mutagenesis Confirmed Functional XCPE1 in the 5′-Untranslated Region between TIS (+1) and ATG (+159) of the SYNGR3 Gene
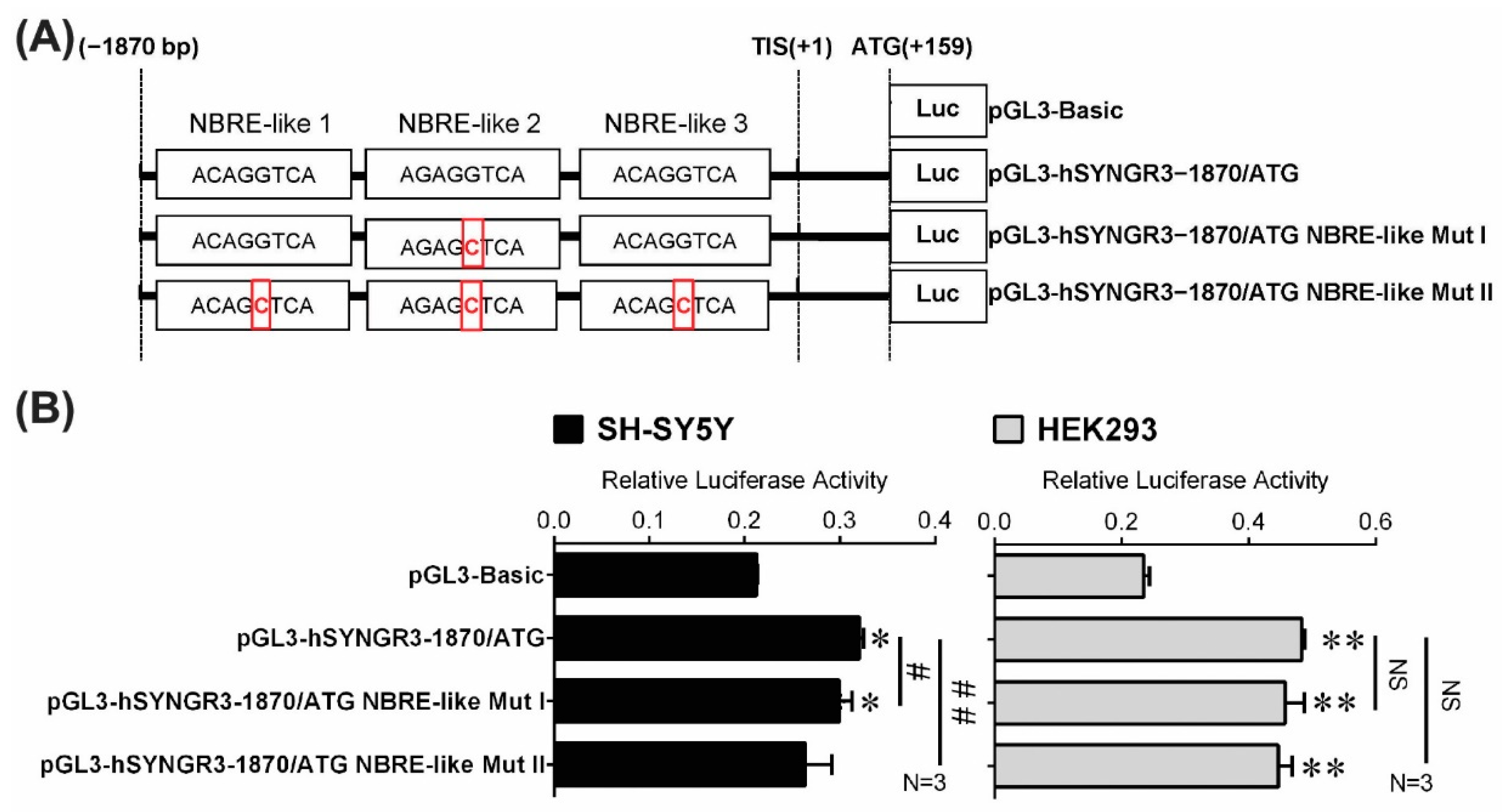
2.5. Site-Directed Mutagenesis Confirmed Functional Putative ncNBRE Sites in the 5′-Flanking Region of the Human SYNGR3 Gene
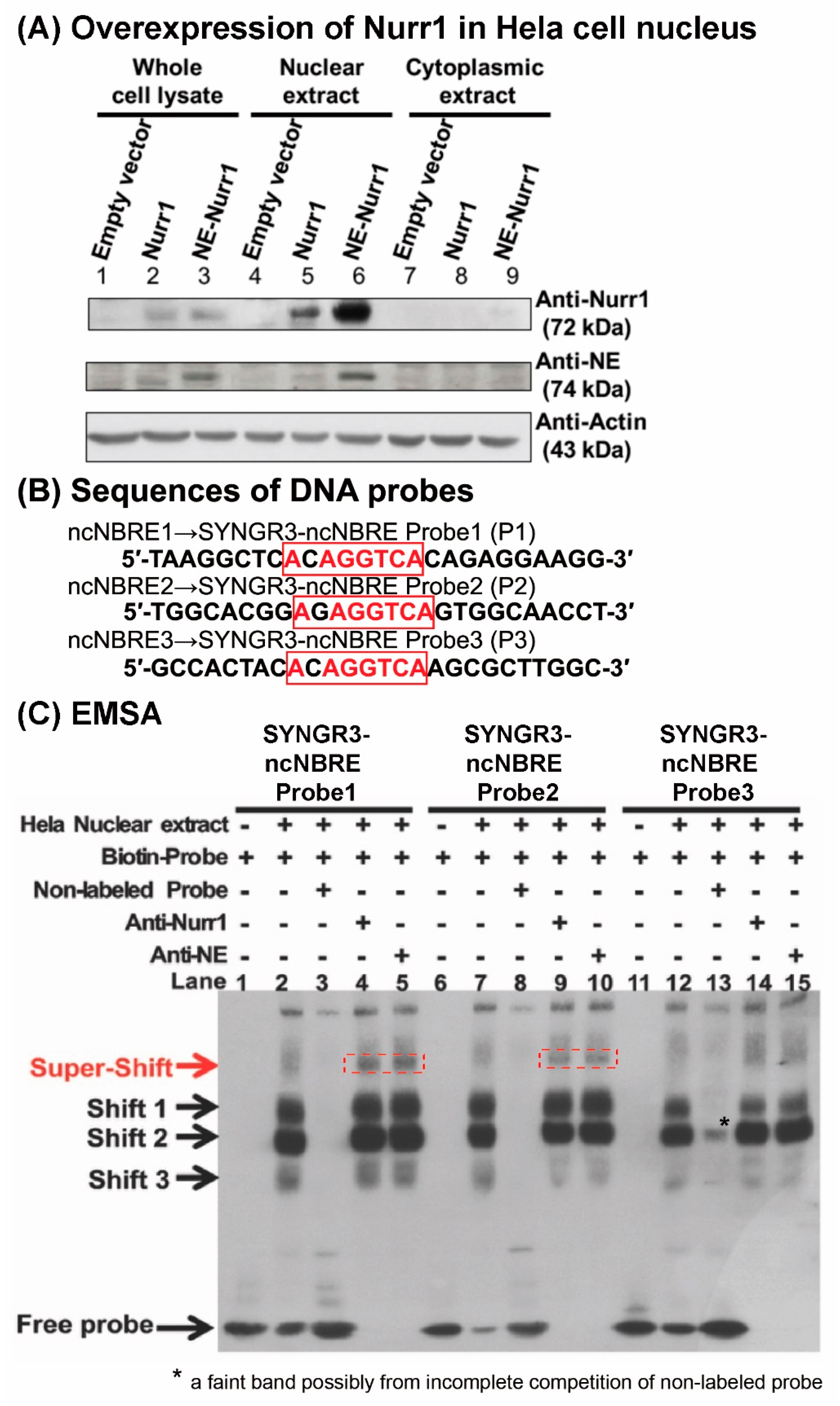
2.6. NURR1 Protein Specifically Bound to Three Putative ncNBRE Sites in the 5′-Flanking Region of the Human SYNGR3 Gene
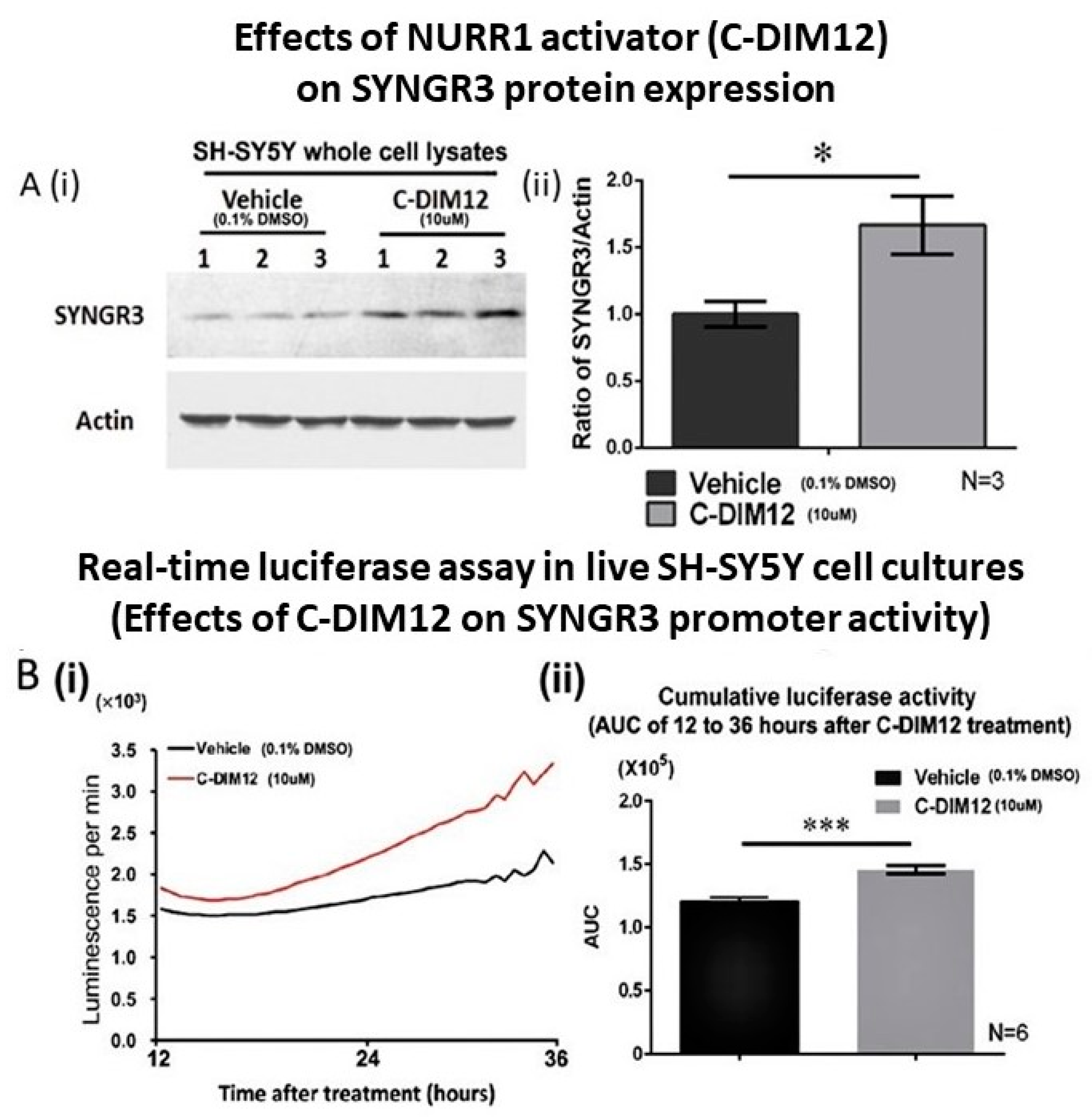
2.7. C-DIM12 (NURR1 Activator) Increased SYNGR3 Protein Expression in SH-SY5Y Cells and Its Promoter Activity
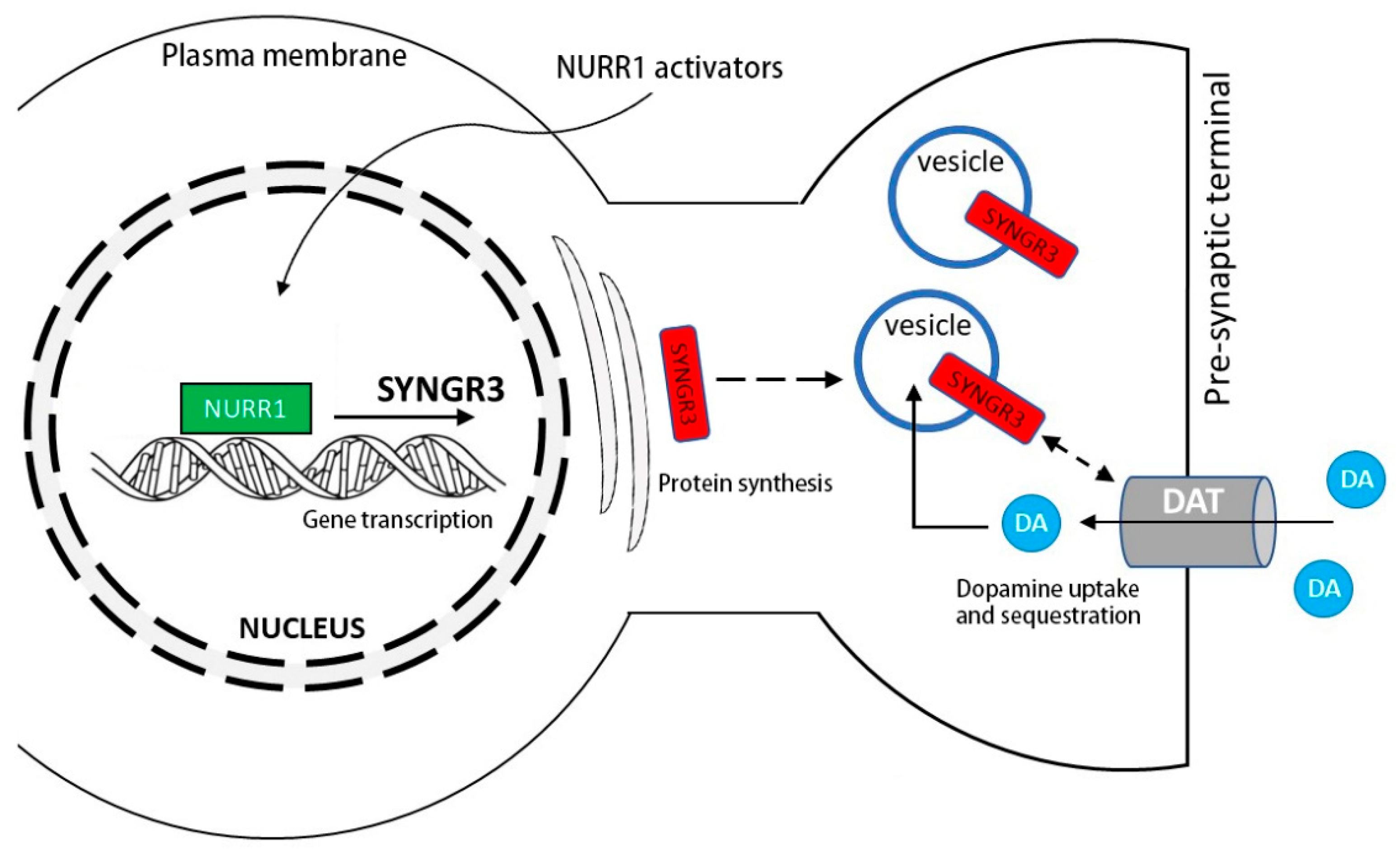
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Cell Cultures
5.2. In Silico Analysis of SYNGR3 5′-Flanking Sequence
5.3. Human SYNGR3 Promoter–Luciferase Fusion Constructs for Reporter Gene Assays
5.4. Human SYNGR3 Gene Promoter Activity Assay
5.5. Site-Directed Mutagenesis of Three Putative ncNBRE Sites in the SYNGR3 Promoter Region
5.6. Construction of the Expression Plasmids Encoding NURR1 (pcDNA3.1(+)-NURR1) and NE-NURR1 (pcDNA3.1(+)-NE-NURR1) Proteins
5.7. Overexpression of NURR1 and NE-NURR1 in HeLa Cells
5.8. Treatment of SH-SY5Y Cells with C-DIM12 (NURR1 Activator)
5.9. Western Blots
5.10. Statistical Analyses
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2016, 44, D7–D19. [Google Scholar] [CrossRef] [PubMed]
- Burgess, D.L.; Davis, C.F.; Gefrides, L.A.; Noebels, J.L. Identification of three novel Ca(2+) channel gamma subunit genes reveals molecular diversification by tandem and chromosome duplication. Genome Res. 1999, 9, 1204–1213. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cunningham, F.; Achuthan, P.; Akanni, W.; Allen, J.; Amode, M.R.; Armean, I.M.; Bennett, R.; Bhai, J.; Billis, K.; Boddu, S.; et al. Ensembl 2019. Nucleic Acids Res. 2019, 47, D745–D751. [Google Scholar] [CrossRef] [PubMed]
- Boutet, E.; Lieberherr, D.; Tognolli, M.; Schneider, M.; Bansal, P.; Bridge, A.J.; Poux, S.; Bougueleret, L.; Xenarios, I. UniProtKB/Swiss-Prot, the Manually Annotated Section of the UniProt KnowledgeBase: How to Use the Entry View. Plant Bioinform. 2016, 1374, 23–54. [Google Scholar]
- Kedra, D.; Pan, H.-Q.; Seroussi, E.; Fransson, I.; Guilbaud, C.; Collins, J.E.; Dunham, I.; Blennow, E.; Roe, B.A.; Piehl, F.; et al. Characterization of the human synaptogyrin gene family. Hum. Genet. 1998, 103, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.S.; Tachibana, I.; Pohl, U.; Lee, H.K.; Thanarajasingam, U.; Portier, B.P.; Ueki, K.; Ramaswamy, S.; Billings, S.J.; Mohrenweiser, H.W.; et al. A Transcript Map of the Chromosome 19q-Arm Glioma Tumor Suppressor Region. Genomics 2000, 64, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.G.; Gross, B.E.; Demir, E.; Rodchenkov, I.; Babur, O.; Anwar, N.; Schultz, N.; Bader, G.D.; Sander, C. Pathway Commons, a web resource for biological pathway data. Nucleic Acids Res. 2011, 39, D685–D690. [Google Scholar] [CrossRef]
- Hubner, K.; Windoffer, R.; Hutter, H.; Leube, R.E. Tetraspan vesicle membrane proteins: Synthesis, subcellular localization, and functional properties. Int. Rev. Cytol. 2002, 214, 103–159. [Google Scholar]
- Egaña, L.A.; Cuevas, R.A.; Baust, T.B.; Parra, L.A.; Leak, R.K.; Hochendoner, S.; Peña, K.; Quiroz, M.; Hong, W.C.; Dorostkar, M.M. Physical and functional interaction between the dopamine transporter and the synaptic vesicle protein synaptogyrin-3. J. Neurosci. 2009, 29, 4592–4604. [Google Scholar] [CrossRef]
- Sossi, V.; de la Fuente-Fernández, R.; Schulzer, M.; Troiano, A.R.; Ruth, T.J.; Stoessl, A.J. Dopamine transporter relation to dopamine turnover in Parkinson’s disease: A positron emission tomography study. Ann. Neurol. 2007, 62, 468–474. [Google Scholar] [CrossRef]
- Lachén-Montes, M.; Mendizuri, N.; Schvartz, D.; Fernández-Irigoyen, J.; Sánchez, J.C.; Santamaría, E. Proteomic Characterization of the Olfactory Molecular Imbalance in Dementia with Lewy Bodies. Int. J. Mol. Sci. 2020, 21, 6371. [Google Scholar] [CrossRef] [PubMed]
- Busche, M.A.; Konnerth, A. Neuronal hyperactivity—A key defect in Alzheimer’s disease? Bioessays 2015, 37, 624–632. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.B.; Cao, Q.; Yan, Z. Transcriptomic analysis of human brains with Alzheimer’s disease reveals the altered expression of synaptic genes linked to cognitive deficits. Brain Commun. 2021, 3, fcab12–fcab123. [Google Scholar] [CrossRef] [PubMed]
- Morton, A.J.; Faull, R.L.M.; Edwardson, J.M. Abnormalities in the synaptic vesicle fusion machinery in Huntington’s disease. Brain Res. Bull. 2001, 56, 111–117. [Google Scholar] [CrossRef]
- Lepeta, K.; Lourenco, M.V.; Schweitzer, B.C.; Martino Adami, P.V.; Banerjee, P.; Catuara-Solarz, S.; La Fuente Revenga, M.; Guillem, A.M.; Haidar, M.; Ijomone, O.M.; et al. Synaptopathies: Synaptic dysfunction in neurological disorders—A review from students to students. J. Neurochem. 2016, 138, 785–805. [Google Scholar] [CrossRef]
- Miller, R.M.; Callahan, L.M.; Casaceli, C.; Chen, L.; Kiser, G.L.; Chui, B.; Kaysser-Kranich, T.M.; Sendera, T.J.; Palaniappan, C.; Federoff, H.J. Dysregulation of Gene Expression in the 1-Methyl-4-Phenyl-1,2,3,6-Tetrahydropyridine-Lesioned Mouse Substantia Nigra. J. Neurosci. 2004, 24, 7445–7454. [Google Scholar] [CrossRef]
- Palese, F.; Pontis, S.; Realini, N.; Piomelli, D. A protective role for N-acylphosphatidylethanolamine phospholipase D in 6-OHDA-induced neurodegeneration. Sci. Rep. 2019, 9, 15927. [Google Scholar] [CrossRef]
- Simunovic, F.; Yi, M.; Wang, Y.; Macey, L.; Brown, L.T.; Krichevsky, A.M.; Andersen, S.L.; Stephens, R.M.; Benes, F.M.; Sonntag, K.C. Gene expression profiling of substantia nigra dopamine neurons: Further insights into Parkinson’s disease pathology. Brain 2009, 132, 1795–1809. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, Y.; Wang, M.; Song, W.-M.; Shen, Q.; McKenzie, A.; Choi, I.; Zhou, X.; Pan, P.-Y.; Yue, Z.; et al. The landscape of multiscale transcriptomic networks and key regulators in Parkinson’s disease. Nat. Commun. 2019, 10, 5234. [Google Scholar] [CrossRef]
- Emilsson, L.; Saetre, P.; Jazin, E. Alzheimer’s disease: mRNA expression profiles of multiple patients show alterations of genes involved with calcium signaling. Neurobiol. Dis. 2006, 21, 618–625. [Google Scholar] [CrossRef]
- Saetre, P.; Jazin, E.; Emilsson, L. Age-related changes in gene expression are accelerated in Alzheimer’s disease. Synapse 2011, 65, 971–974. [Google Scholar] [CrossRef] [PubMed]
- McInnes, J.; Wierda, K.; Snellinx, A.; Bounti, L.; Wang, Y.-C.; Stancu, I.-C.; Apóstolo, N.; Gevaert, K.; Dewachter, I.; Spires-Jones, T.L.; et al. Synaptogyrin-3 Mediates Presynaptic Dysfunction Induced by Tau. Neuron 2018, 97, 823–835.e8. [Google Scholar] [CrossRef] [PubMed]
- Tokusumi, Y.; Ma, Y.; Song, X.; Jacobson, R.H.; Takada, S. The New Core Promoter Element XCPE1 (X Core Promoter Element 1) Directs Activator-, Mediator-, and TATA-Binding Protein-Dependent but TFIID-Independent RNA Polymerase II Transcription from TATA-Less Promoters. Mol. Cell Biol. 2007, 27, 1844–1858. [Google Scholar] [CrossRef] [PubMed]
- Saxonov, S.; Berg, P.; Brutlag, D.L. A Genome-Wide Analysis of CpG Dinucleotides in the Human Genome Distinguishes Two Distinct Classes of Promoters. Proc. Natl. Acad. Sci. USA 2006, 103, 1412–1417. [Google Scholar] [CrossRef] [PubMed]
- Goncalves, I.; Duret, L.; Mouchiroud, D. Nature and structure of human genes that generate retropseudogenes. Genome Res. 2000, 10, 672–678. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rivenbark, A.G.; Jones, W.D.; Risher, J.D.; Coleman, W.B. DNA methylation-dependent epigenetic regulation of gene expression in MCF-7 breast cancer cells. Epigenetics 2006, 1, 32–44. [Google Scholar] [CrossRef]
- Wade, D.P.; Lindahl, G.E.; Lawn, R.M. Apolipoprotein(a) gene transcription is regulated by liver-enriched trans-acting factor hepatocyte nuclear factor 1 alpha. J. Biol. Chem. 1994, 269, 19757–19765. [Google Scholar] [CrossRef]
- Yang, C.; Bolotin, E.; Jiang, T.; Sladek, F.M.; Martinez, E. Prevalence of the initiator over the TATA box in human and yeast genes and identification of DNA motifs enriched in human TATA-less core promoters. Gene 2007, 389, 52–65. [Google Scholar] [CrossRef]
- Anish, R.; Hossain, M.B.; Jacobson, R.H.; Takada, S. Characterization of Transcription from TATA-Less Promoters: Identification of a New Core Promoter Element XCPE2 and Analysis of Factor Requirements. PLoS ONE 2009, 4, e5103. [Google Scholar] [CrossRef]
- García-Yagüe, Á.J.; Rada, P.; Rojo, A.I.; Lastres-Becker, I.; Cuadrado, A. Nuclear Import and Export Signals Control the Subcellular Localization of Nurr1 Protein in Response to Oxidative Stress. J. Biol. Chem. 2013, 288, 5506–5517. [Google Scholar] [CrossRef]
- Loppi, S.; Kolosowska, N.; Kärkkäinen, O.; Korhonen, P.; Huuskonen, M.; Grubman, A.; Dhungana, H.; Wojciechowski, S.; Pomeshchik, Y.; Giordano, M.; et al. HX600, a synthetic agonist for RXR-Nurr1 heterodimer complex, prevents ischemia-induced neuronal damage. Brain Behav. Immun. 2018, 73, 670–681. [Google Scholar] [CrossRef] [PubMed]
- Rajan, S.; Jang, Y.; Kim, C.-H.; Kim, W.; Toh, H.T.; Jeon, J.; Song, B.; Serra, A.; Lescar, J.; Yoo, J.Y.; et al. PGE1 and PGA1 bind to Nurr1 and activate its transcriptional function. Nat. Chem. Biol. 2020, 16, 876–886. [Google Scholar] [CrossRef] [PubMed]
- Katsuki, H. Nuclear receptors of NR1 and NR4 subfamilies in the regulation of microglial functions and pathology. Pharmacol. Res. Perspect. 2021, 9, e00766. [Google Scholar] [CrossRef] [PubMed]
- Jakaria, M.; Haque, M.E.; Cho, D.-Y.; Azam, S.; Kim, I.-S.; Choi, D.-K. Molecular Insights into NR4A2(Nurr1): An Emerging Target for Neuroprotective Therapy Against Neuroinflammation and Neuronal Cell Death. Mol. Neurobiol. 2019, 56, 5799–5814. [Google Scholar] [CrossRef] [PubMed]
- Murphy, E.P.; Dobson, A.D.; Keller, C.; Conneely, O.M. Differential regulation of transcription by the NURR1/NUR77 subfamily of nuclear transcription factors. Gene Expr. 1996, 5, 169–179. [Google Scholar] [PubMed]
- Wallen-Mackenzie, A.; Mata de Urquiza, A.; Petersson, S.; Rodriguez, F.J.; Friling, S.; Wagner, J.; Ordentlich, P.; Lengqvist, J.; Heyman, R.A.; Arenas, E.; et al. Nurr1-RXR heterodimers mediate RXR ligand-induced signaling in neuronal cells. Genes Dev. 2003, 17, 3036–3047. [Google Scholar] [CrossRef] [PubMed]
- Ramsden, D.B.; Parsons, R.B.; Ho, S.L.; Waring, R.H. The aetiology of idiopathic Parkinson’s disease. J. Clin. Pathol. 2001, 54, 369–380. [Google Scholar]
- Kim, S.M.S.M.; Cho, S.Y.S.Y.; Kim, M.W.M.W.; Roh, S.R.S.R.; Shin, H.S.H.S.; Suh, Y.H.Y.H.; Geum, D.D.; Lee, M.A.M.A. Genome-Wide Analysis Identifies NURR1-Controlled Network of New Synapse Formation and Cell Cycle Arrest in Human Neural Stem Cells. Mol. Cells 2020, 43, 551–571. [Google Scholar]
- Gao, H.; Wang, D.; Jiang, S.; Mao, J.; Yang, X. NF-κB is negatively associated with Nurr1 to reduce the inflammatory response in Parkinson’s disease. Mol. Med. Rep. 2021, 23, 396. [Google Scholar] [CrossRef]
- Itokazu, Y.; Fuchigami, T.; Morgan, J.C.; Yu, R.K. Intranasal infusion of GD3 and GM1 gangliosides downregulates alpha-synuclein and controls tyrosine hydroxylase gene in a PD model mouse. Mol. Ther. 2021, 29, 3059–3071. [Google Scholar] [CrossRef]
- Le, W.; Pan, T.; Huang, M.; Xu, P.; Xie, W.; Zhu, W.; Zhang, X.; Deng, H.; Jankovic, J. Decreased NURR1 gene expression in patients with Parkinson’s disease. J. Neurol. Sci. 2008, 273, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Moran, L.B.; Croisier, E.; Duke, D.C.; Kalaitzakis, M.E.; Roncaroli, F.; Deprez, M.; Dexter, D.T.; Pearce, R.K.B.; Graeber, M.B. Analysis of alpha-synuclein, dopamine and parkin pathways in neuropathologically confirmed parkinsonian nigra. Acta Neuropathol. 2007, 113, 253–263. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.-H.; Wong, C.F.; Tan, H.L.; Yang, X.J.; Ditlev, J.; Matsuda, D.; Khoo, S.K.; Sugimura, J.; Fujioka, T.; Furge, K.A.; et al. Genomic expression and single-nucleotide polymorphism profiling discriminates chromophobe renal cell carcinoma and oncocytoma. BMC Cancer 2010, 10, 196. [Google Scholar] [CrossRef] [PubMed]
- Pyeon, D.; Newton, N.A.; Lambert, P.F.; den Boon, J.A.; Sengupta, S.; Marsit, C.J.; Woodworth, C.D.; Connor, J.P.; Haugen, T.H.; Smith, E.M.; et al. Fundamental differences in cell cycle deregulation in human papillomavirus-positive and human papillomavirus-negative head/neck and cervical cancers. Cacner Res. 2007, 67, 4605–4619. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Ren, C.-X.; Zhang, J.-M.; Xin, X.-Y.; Hua, T.; Wang, H.-B.; Wang, H.-B. The Effects of miR-195-5p/MMP14 on Proliferation and Invasion of Cervical Carcinoma Cells Through TNF Signaling Pathway Based on Bioinformatics Analysis of Microarray Profiling. Cell Physiol. Biochem. 2018, 50, 1398–1413. [Google Scholar] [CrossRef] [PubMed]
- Hammond, S.L.; Popichak, K.A.; Li, X.; Hunt, L.G.; Richman, E.H.; Damale, P.U.; Chong, E.K.P.; Backos, D.S.; Safe, S.; Tjalkens, R.B. The Nurr1 Ligand,1,1-bis(3′-Indolyl)-1-(p-Chlorophenyl)Methane, Modulates Glial Reactivity and Is Neuroprotective in MPTP-Induced Parkinsonism. J. Pharmacol. Exp. Ther. 2018, 365, 636–651. [Google Scholar] [CrossRef]
- Inamoto, T.; Papineni, S.; Chintharlapalli, S.; Cho, S.-D.; Safe, S.; Kamat, A.M. 1,1-Bis(3′-indolyl)-1-(p-chlorophenyl)methane activates the orphan nuclear receptor Nurr1 and inhibits bladder cancer growth. Mol. Cancer Ther. 2008, 7, 3825–3833. [Google Scholar] [CrossRef]
- Li, X.; Lee, S.-O.; Safe, S. Structure-dependent activation of NR4A2 (Nurr1) by 1,1-bis(3′-indolyl)-1-(aromatic)methane analogs in pancreatic cancer cells. Biochem. Pharmacol. 2012, 83, 1445–1455. [Google Scholar] [CrossRef]
- De Miranda, B.R.; Popichak, K.A.; Hammond, S.L.; Jorgensen, B.A.; Phillips, A.T.; Safe, S.; Tjalkens, R.B. The Nurr1 Activator 1,1-Bis(3 ‘-Indolyl)-1-(p-Chlorophenyl) Methane Blocks Inflammatory Gene Expression in BV-2 Microglial Cells by Inhibiting Nuclear Factor kappa B. Mol. Pharmacol. 2015, 87, 1021–1034. [Google Scholar] [CrossRef]
- Carli, M.; Evenden, J.L.; Robbins, T.W. Depletion of unilateral striatal dopamine impairs initiation of contralateral actions and not sensory attention. Nature 1985, 313, 679–682. [Google Scholar] [CrossRef]
- Tepper, J.M.; Creese, I.; Schwartz, D.H. Stimulus-evoked changes in neostriatal dopamine levels in awake and anesthetized rats as measured by microdialysis. Brain Res. 1991, 559, 283–292. [Google Scholar] [CrossRef]
- Nguyen, M.; Wong, Y.C.; Ysselstein, D.; Severino, A.; Krainc, D. Synaptic, Mitochondrial, and Lysosomal Dysfunction in Parkinson’s Disease. Trends Neurosci. 2019, 42, 140–149. [Google Scholar] [CrossRef] [PubMed]
- Adams, J.R.; van Netten, H.; Schulzer, M.; Mak, E.; McKenzie, J.; Strongosky, A.; Sossi, V.; Ruth, T.J.; Lee, C.S.; Farrer, M.; et al. PET in LRRK2 mutations: Comparison to sporadic Parkinson’s disease and evidence for presymptomatic compensation. Brain 2005, 128, 2777–2785. [Google Scholar] [CrossRef]
- Liu, H.F.; Lu, S.; Ho, P.W.L.; Tse, H.M.; Pang, S.Y.Y.; Kung, M.H.W.; Ho, J.W.M.; Ramsden, D.B.; Zhou, Z.J.; Ho, S.L. LRRK2 R1441G mice are more liable to dopamine depletion and locomotor inactivity. Ann. Clin. Transl. Neurol. 2014, 1, 199–208. [Google Scholar] [CrossRef]
- Ho, P.W.-L.; Leung, C.T.; Liu, H.; Pang, S.Y.; Lam, C.S.; Xian, J.; Li, L.; Kung, M.H.-W.; Ramsden, D.B.; Ho, S.-L. Age-dependent accumulation of oligomeric SNCA/a-synuclein from impaired degradation in mutant LRRK2 knockin mouse model of Parkinson disease: Role for therapeutic activation of chaperone-mediated autophagy (CMA). Autophagy 2020, 16, 347–370. [Google Scholar] [CrossRef] [PubMed]
- Li, L.C.; Dahiya, R. MethPrimer: Designing primers for methylation PCRs. Bioinformatics 2002, 18, 1427–1431. [Google Scholar] [CrossRef] [PubMed]
- Heinemeyer, T.; Wingender, E.; Reuter, I.; Hermjakob, H.; Kel, A.E.; Kel, O.V.; Ignatieva, E.V.; Ananko, E.A.; Podkolodnaya, O.A.; Kolpakov, F.A.; et al. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res. 1998, 26, 362–367. [Google Scholar] [CrossRef]
- Cartharius, K.; Frech, K.; Grote, K.; Klocke, B.; Haltmeier, M.; Klingenhoff, A.; Frisch, M.; Bayerlein, M.; Werner, T. MatInspector and beyond: Promoter analysis based on transcription factor binding sites. Bioinformatics 2005, 21, 2933–2942. [Google Scholar] [CrossRef]
- Ho, P.W.-L.; Tse, Z.H.-M.; Liu, H.-F.; Lu, S.; Ho, J.W.-M.; Kung, M.H.-W.; Ramsden, D.B.; Ho, S.-L. Assessment of Cellular Estrogenic Activity Based on Estrogen Receptor-Mediated Reduction of Soluble-Form Catechol-O-Methyltransferase (COMT) Expression in an ELISA-Based System. PLoS ONE 2013, 8, e74065. [Google Scholar]
- Barde, M.P.; Barde, P.J. What to use to express the variability of data: Standard deviation or standard error of mean? Perspect. Clin. Res. 2012, 3, 113–116. [Google Scholar] [CrossRef]




| Gene Name | Interact with SYNGR3 | Regulate SYNGR3 Transcription |
|---|---|---|
| ACSF2 | √ | |
| ATF2 | √ | |
| ATF3 | √ | |
| ATF4 | √ | |
| ATP1B4 | √ | |
| CREB1 | √ | |
| E4F1 | √ | |
| EHHADH | √ | |
| ESR2 | √ | |
| GAD2 | √ | |
| GLP1R | √ | |
| HSBP1L1 | √ | |
| IRF3 | √ | |
| JUN | √ | |
| MAPT | √ | |
| MIDN | √ | |
| MPP1 | √ | |
| NDRG4 | √ | |
| NOG | √ | |
| PLIN3 | √ | |
| PNKP | √ | |
| SH3GLB1 | √ | |
| SLC39A9 | √ | |
| SPG21 | √ | |
| TTPA | √ |
| Constructs | Primer | Sequence (5′→3′) |
|---|---|---|
| pCR4-TOPO-SYNGR3–1870/+224 | Forward | TTTGAAGGCATCCAGGCCAGTGGAACTCTAGTGCAAGGAAAAGTT |
| Reverse | CCGCGCAAAGCTCACGGGGTCCAG | |
| pGL3-SYNGR3–1870/ATG | Forward | TTTGAAGGCATCCAGGCCAGTGGAACTCTAGTGCAAGGAAAAGTT |
| Reverse | CATGCCATGGCCGGCCCGGGCGGGACG | |
| pGL3-SYNGR3–1870/TIS | Forward | TTTGAAGGCATCCAGGCCAGTGGAACTCTAGTGCAAGGAAAAGTT |
| Reverse | CCGCTCGAGCGGGGGACGCGCGGGACGGGGCG | |
| pcDNA3.1(+)-NE-NURR1 | Forward | CCCAAGCTTATGACCAAAGAAAACCCGCGTAGCAACCAGGAAGAAAGCTATGATGATAACGAAAGCCCTTGTGTTCAGGCGC |
| Reverse | CGCGGATCCTTAGAAAGGTAAAGTGTCCAGG | |
| pcDNA3.1(+)-NURR1 | Forward | CCCAAGCTTATGCCTTGTGTTCAGGCGCAG |
| Reverse | CGCGGATCCTTAGAAAGGTAAAGTGTCCAGG |
| Constructs | Target Site to Be Mutated | Primer | Sequence |
|---|---|---|---|
| pGL3-SYNGR3–XCPE1-Mut | XCPE1 | Forward | GCGCGCGTCCAGCCCGGGCCG |
| Reverse | CGGCCCGGGCTGGACGCGCGC | ||
| pGL3-hSYNGR3–1870/ATG ncNBRE Mut I | ncNBRE1 | Forward | GATTGGCACGGAGAGCTCAGTGGCAACCTTG |
| Reverse | CAAGGTTGCCACTGAGCTCTCCGTGCCAATC | ||
| pGL3-hSYNGR3–1870/ATG ncNBRE Mut II | ncNBRE2 | Forward | TCCTAAGGCTCACAGCTCACAGAGGAAGGAC |
| Reverse | GTCCTTCCTCTGTGAGCTGTGAGCCTTAGGA | ||
| ncNBRE3 | Forward | CGCCACTACACAGCTCAAGCGCTTGGC | |
| Reverse | GCCAAGCGCTTGAGCTGTGTAGTGGCG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, L.; Ho, P.W.-L.; Liu, H.; Pang, S.Y.-Y.; Chang, E.E.-S.; Choi, Z.Y.-K.; Malki, Y.; Kung, M.H.-W.; Ramsden, D.B.; Ho, S.-L. Transcriptional Regulation of the Synaptic Vesicle Protein Synaptogyrin-3 (SYNGR3) Gene: The Effects of NURR1 on Its Expression. Int. J. Mol. Sci. 2022, 23, 3646. https://doi.org/10.3390/ijms23073646
Li L, Ho PW-L, Liu H, Pang SY-Y, Chang EE-S, Choi ZY-K, Malki Y, Kung MH-W, Ramsden DB, Ho S-L. Transcriptional Regulation of the Synaptic Vesicle Protein Synaptogyrin-3 (SYNGR3) Gene: The Effects of NURR1 on Its Expression. International Journal of Molecular Sciences. 2022; 23(7):3646. https://doi.org/10.3390/ijms23073646
Chicago/Turabian StyleLi, Lingfei, Philip Wing-Lok Ho, Huifang Liu, Shirley Yin-Yu Pang, Eunice Eun-Seo Chang, Zoe Yuen-Kiu Choi, Yasine Malki, Michelle Hiu-Wai Kung, David Boyer Ramsden, and Shu-Leong Ho. 2022. "Transcriptional Regulation of the Synaptic Vesicle Protein Synaptogyrin-3 (SYNGR3) Gene: The Effects of NURR1 on Its Expression" International Journal of Molecular Sciences 23, no. 7: 3646. https://doi.org/10.3390/ijms23073646
APA StyleLi, L., Ho, P. W.-L., Liu, H., Pang, S. Y.-Y., Chang, E. E.-S., Choi, Z. Y.-K., Malki, Y., Kung, M. H.-W., Ramsden, D. B., & Ho, S.-L. (2022). Transcriptional Regulation of the Synaptic Vesicle Protein Synaptogyrin-3 (SYNGR3) Gene: The Effects of NURR1 on Its Expression. International Journal of Molecular Sciences, 23(7), 3646. https://doi.org/10.3390/ijms23073646






