Identification of New QTLs for Dietary Fiber Content in Aegilops biuncialis
Abstract
1. Introduction
2. Results
2.1. Characterization of Quality Traits
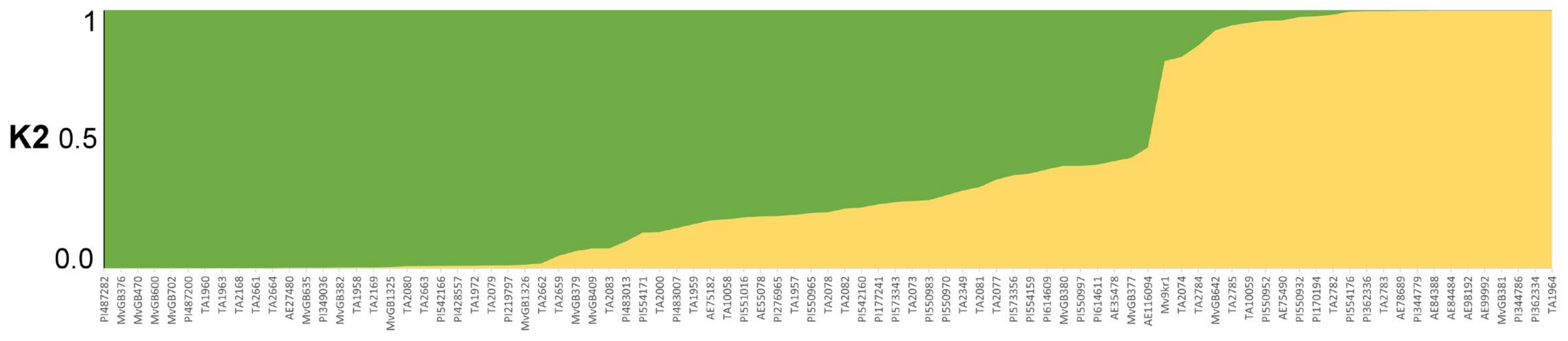
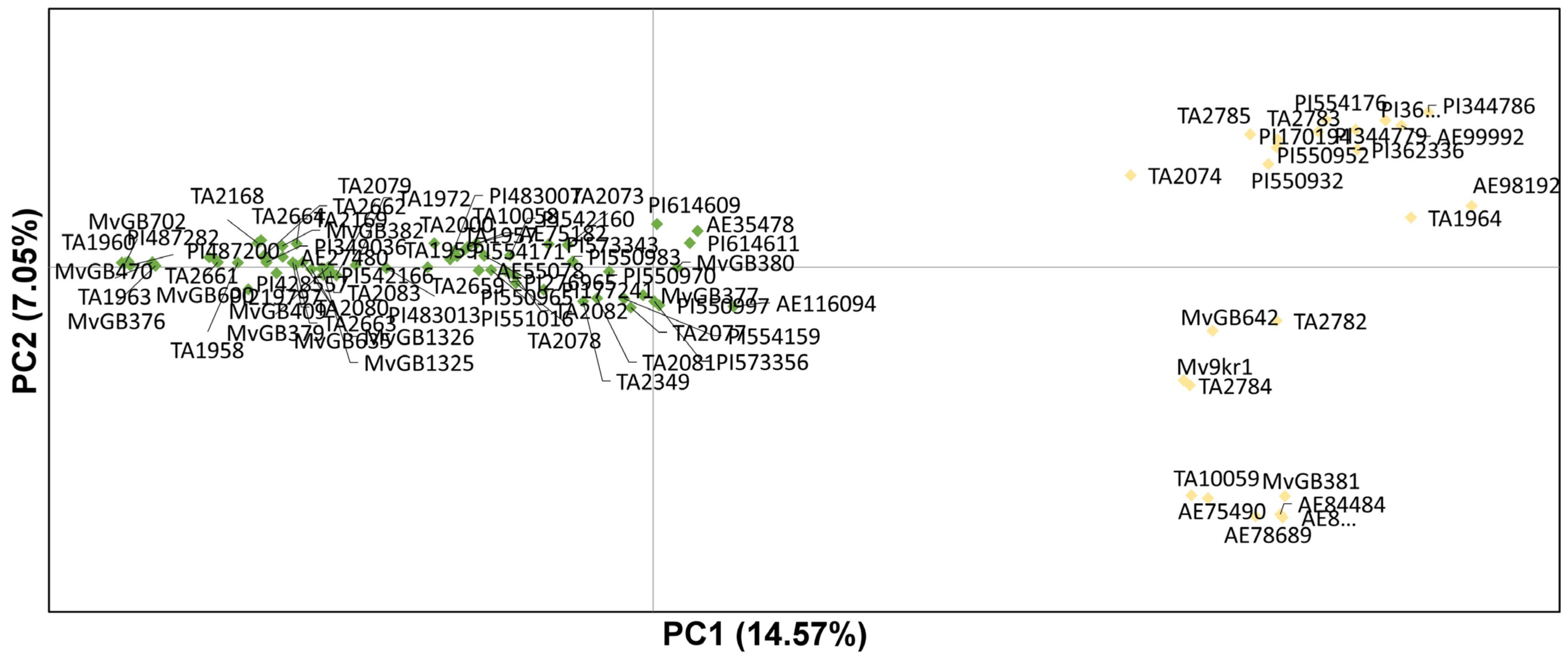
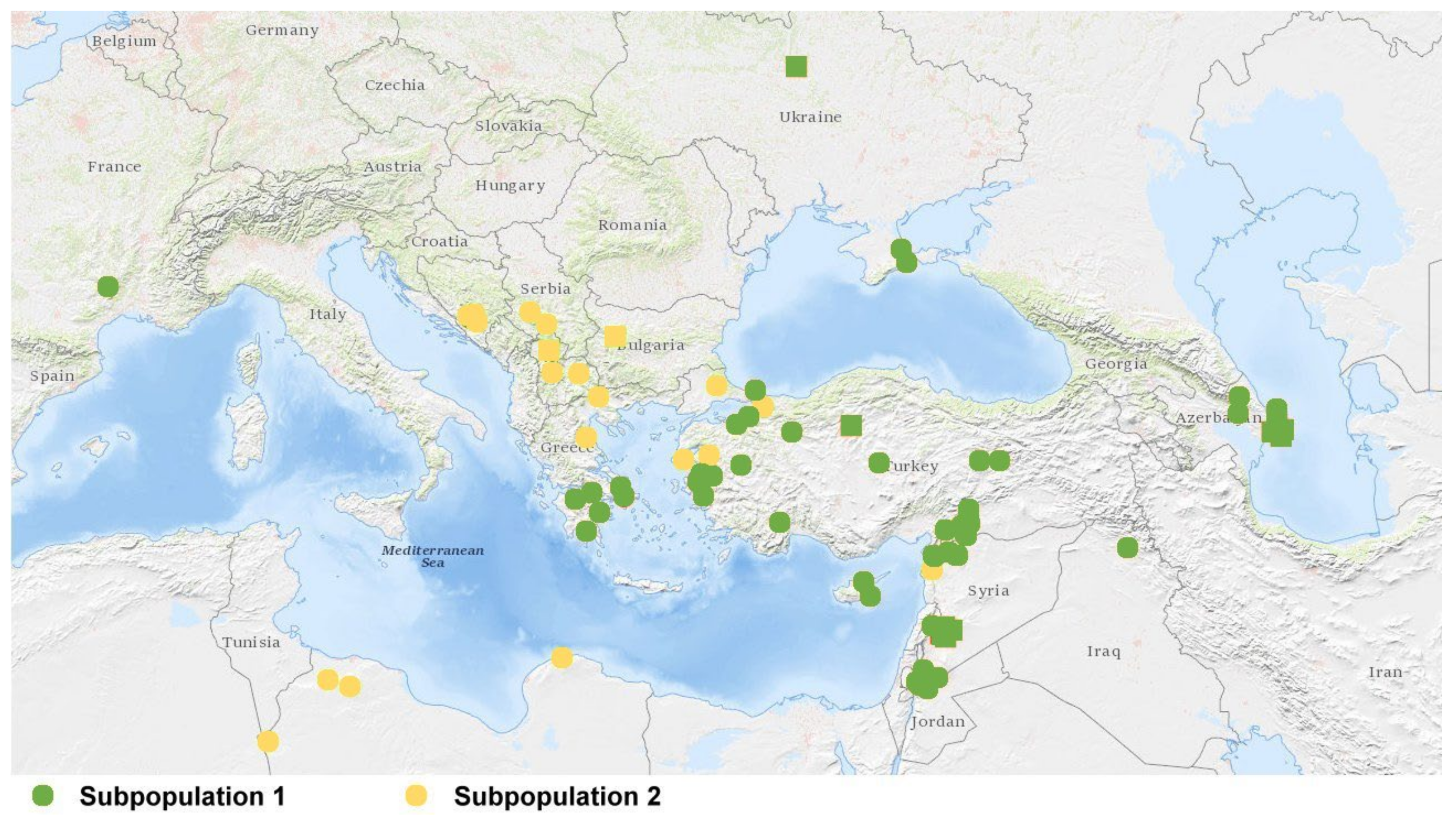
2.2. Population Structure
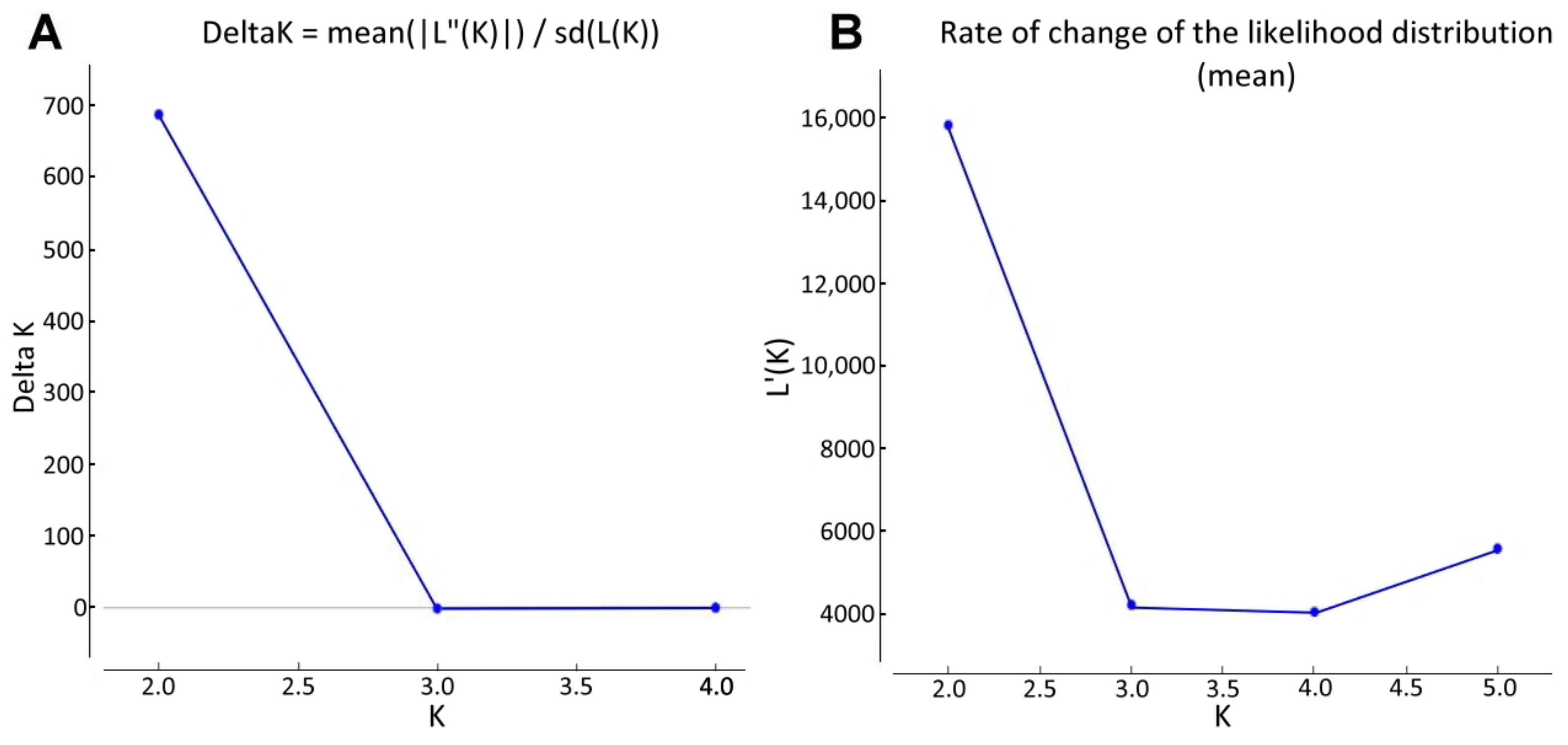
2.3. Determination of Marker-Trait Associations
2.4. Chromosomal Location and Functional Annotation of the QTLs
3. Discussion
3.1. Grain Composition of Ae. biuncialis
3.2. Genetic Diversity of the Ae. biuncialis Collection
3.3. GWAS and Candidate Gene Identification
4. Materials and Methods
4.1. Plant Material
4.2. Field Trial
4.3. Quantitative Determination of Grain Composition
4.4. QTL and Candidate Gene Detection
4.5. Sequence Similarity Analysis and Functional Annotation
4.6. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Collins, H.M.; Burton, R.A.; Topping, D.L.; Liao, M.-L.; Bacic, A.; Fincher, G.B. Variability in fine structures of noncellulosic cell wall polysaccharides from cereal grains: Potential importance in human health and nutrition. Cereal Chem. 2010, 87, 272–282. [Google Scholar] [CrossRef]
- Mares, D.J.; Stone, B.A. Studies on Lolium multiflorum endosperm in tissue culture II. Fine structure of cells and cell walls and the development of cell walls. Aust. J. Biol. Sci. 1973, 26, 135–150. [Google Scholar] [CrossRef]
- Courtin, C.M.; Gelders, G.G.; Delcour, J.A. Use of two endoxylanases with different substrate selectivity for understanding arabinoxylan functionality in wheat flour breadmaking. Cereal Chem. 2001, 78, 564–571. [Google Scholar] [CrossRef]
- Frederix, S.A.; van Hoeymissen, K.E.; Courtin, C.M.; Delcour, J.A. Water-extractable and water-unextractable arabinoxylans affect gluten agglomeration behavior during wheat flour gluten-starch separation. J. Agric. Food Chem. 2004, 52, 7950–7956. [Google Scholar] [CrossRef]
- Shewry, P.R.; Piironen, V.; Lampi, A.-M.; Edelmann, M.; Kariluoto, S.; Nurmi, T.; Fernandez-Orozco, R.; Ravel, C.; Charmet, G.; Andersson, A.A.M.; et al. The HEALTHGRAIN wheat diversity screen: Effects of genotype and environment on phytochemicals and dietary fiber components. J. Agric. Food Chem. 2010, 58, 9291–9298. [Google Scholar] [CrossRef]
- Saulnier, L.; Sado, P.-E.; Branlard, G.; Charmet, G.; Guillon, F. Wheat arabinoxylans: Exploiting variation in amount and composition to develop enhanced varieties. J. Cereal Sci. 2007, 46, 261–281. [Google Scholar] [CrossRef]
- Andersson, A.A.M.; Kamal-Eldin, A.; Fraś, A.; Boros, D.; Aman, P. Alkylresorcinols in wheat varieties in the HEALTHGRAIN Diversity Screen. J. Agric. Food Chem. 2008, 56, 9722–9725. [Google Scholar] [CrossRef]
- Gebruers, K.; Dornez, E.; Boros, D.; Fraś, A.; Dynkowska, W.; Bedo, Z.; Rakszegi, M.; Delcour, J.A.; Courtin, C.M. Variation in the content of dietary fiber and components thereof in wheats in the HEALTHGRAIN Diversity Screen. J. Agric. Food Chem. 2008, 56, 9740–9749. [Google Scholar] [CrossRef]
- Rakszegi, M.; Boros, D.; Kuti, C.; Láng, L.; Bedo, Z.; Shewry, P.R. Composition and end-use quality of 150 wheat lines selected for the HEALTHGRAIN Diversity Screen. J. Agric. Food Chem. 2008, 56, 9750–9757. [Google Scholar] [CrossRef]
- Shewry, P.R.; Piironen, V.; Lampi, A.-M.; Nyström, L.; Li, L.; Rakszegi, M.; Fraś, A.; Boros, D.; Gebruers, K.; Courtin, C.M.; et al. Phytochemical and fiber components in oat varieties in the HEALTHGRAIN Diversity Screen. J. Agric. Food Chem. 2008, 56, 9777–9784. [Google Scholar] [CrossRef]
- Ward, J.L.; Poutanen, K.; Gebruers, K.; Piironen, V.; Lampi, A.-M.; Nyström, L.; Andersson, A.A.M.; Aman, P.; Boros, D.; Rakszegi, M.; et al. The HEALTHGRAIN Cereal Diversity Screen: Concept, results, and prospects. J. Agric. Food Chem. 2008, 56, 9699–9709. [Google Scholar] [CrossRef] [PubMed]
- Rahman, S.; Islam, S.; Yu, Z.; She, M.; Nevo, E.; Ma, W. Current progress in understanding and recovering the wheat genes lost in evolution and domestication. Int. J. Mol. Sci. 2020, 21, 5836. [Google Scholar] [CrossRef] [PubMed]
- Van Slageren, M.W. Wild Wheats: A Monograph of Aegilops L. and Amblyopyrum (Jaub. & Spach) Eig (Poaceae); Agricultural University: Wageningen, The Netherlands, 1994. [Google Scholar]
- Schneider, A.; Molnár, I.; Molnár-Láng, M. Utilisation of Aegilops (goatgrass) species to widen the genetic diversity of cultivated wheat. Euphytica 2008, 163, 1–19. [Google Scholar] [CrossRef]
- Zhang, P.; Dundas, I.S.; McIntosh, R.A.; Xu, S.S.; Park, R.F.; Gill, B.S.; Friebe, B. Wheat–Aegilops introgressions. In Alien Introgression in Wheat: Cytogenetics, Molecular Biology, and Genomics; Molnár-Láng, M., Ceoloni, C., Doležel, J., Eds.; Springer Science + Business Media: Cham, Switzerland, 2015; pp. 221–243. ISBN 978-3-319-23494-6. [Google Scholar]
- Kishii, M. An update of recent use of Aegilops species in wheat breeding. Front. Plant Sci. 2019, 10, 585. [Google Scholar] [CrossRef] [PubMed]
- Kilian, B.; Mammen, K.; Millet, E.; Sharma, R.; Graner, A.; Salamini, F.; Hammer, K.; Özkan, H. Aegilops. Wild Crop Relatives: Genomic and Breeding Resources; Springer: Cham, Switzerland, 2011; pp. 1–76. [Google Scholar]
- Ivanizs, L.; Monostori, I.; Farkas, A.; Megyeri, M.; Mikó, P.; Türkösi, E.; Gaál, E.; Lenykó-Thegze, A.; Szőke-Pázsi, K.; Szakács, É.; et al. Unlocking the genetic diversity and population structure of a wild gene source of wheat, Aegilops biuncialis Vis., and its relationship with the heading time. Front. Plant Sci. 2019, 10, 1531. [Google Scholar] [CrossRef]
- Makkouk, K.M.; Ghulam, W.; Comeau, A. Resistance to barley yellow dwarf luteovirus in Aegilops species. Can. J. Plant. Sci. 1994, 74, 631–634. [Google Scholar] [CrossRef]
- Li, H.; Dong, Z.; Ma, C.; Tian, X.; Xiang, Z.; Xia, Q.; Ma, P.; Liu, W. Discovery of powdery mildew resistance gene candidates from Aegilops biuncialis chromosome 2Mb based on transcriptome sequencing. PLoS ONE 2019, 14, e0220089. [Google Scholar] [CrossRef]
- Damania, A.B.; Pecetti, L. Variability in a collection of Aegilops species and evaluation for yellow rust resistance at two locations in Northern Syria. J. Genet. Breed. 1990, 44, 97–102. [Google Scholar]
- Dimov, A.; Zaharieva, M.; Mihova, S. Rust and powdery mildew resistance in Aegilops accessions from Bulgaria. In Biodiversity and Wheat Improvement; Damania, A.B., Ed.; John Wiley & Sons: New York, NY, USA; Chichester, UK, 1993; pp. 165–169. [Google Scholar]
- Kwiatek, M.; Błaszczyk, L.; Wiśniewska, H.; Apolinarska, B. Aegilops-Secale amphiploids: Chromosome categorisation, pollen viability and identification of fungal disease resistance genes. J. Appl. Genet. 2012, 53, 37–40. [Google Scholar] [CrossRef][Green Version]
- Olivera, P.D.; Rouse, M.N.; Jin, Y. Identification of new sources of resistance to wheat stem rust in Aegilops spp. in the tertiary genepool of wheat. Front. Plant Sci. 2018, 9, 1719. [Google Scholar] [CrossRef]
- Ekmekci, Y.; Terzioglu, S. Changes in the electrophoretic pattern of soluble shoot proteins of wild and cultivated tetraploid wheats following cold acclimation and freezing. Isr. J. Plant Sci. 2002, 50, 95–102. [Google Scholar] [CrossRef]
- Colmer, T.D.; Flowers, T.J.; Munns, R. Use of wild relatives to improve salt tolerance in wheat. J. Exp. Bot. 2006, 57, 1059–1078. [Google Scholar] [CrossRef] [PubMed]
- Darkó, É.; Khalil, R.; Dobi, Z.; Kovács, V.; Szalai, G.; Janda, T.; Molnár, I. Addition of Aegilops biuncialis chromosomes 2M or 3M improves the salt tolerance of wheat in different way. Sci. Rep. 2020, 10, 22327. [Google Scholar] [CrossRef] [PubMed]
- Molnár, I.; Gáspár, L.; Sárvári, É.; Dulai, S.; Hoffmann, B.; Molnár-Láng, M.; Galiba, G. Physiological and morphological responses to water stress in Aegilops biuncialis and Triticum aestivum genotypes with differing tolerance to drought. Funct. Plant Biol. 2004, 31, 1149–1159. [Google Scholar] [CrossRef] [PubMed]
- Dulai, S.; Molnár, I.; Szopkó, D.; Darkó, É.; Vojtkó, A.; Sass-Gyarmati, A.; Molnár-Láng, M. Wheat-Aegilops biuncialis amphiploids have efficient photosynthesis and biomass production during osmotic stress. J. Plant Physiol. 2014, 171, 509–517. [Google Scholar] [CrossRef]
- Farkas, A.; Molnár, I.; Dulai, S.; Rapi, S.; Oldal, V.; Cseh, A.; Kruppa, K.; Molnár-Láng, M. Increased micronutrient content (Zn, Mn) in the 3M(b)(4B) wheat-Aegilops biuncialis substitution and 3M(b).4BS translocation identified by GISH and FISH. Genome 2014, 57, 61–67. [Google Scholar] [CrossRef]
- Rakszegi, M.; Molnár, I.; Lovegrove, A.; Darkó, É.; Farkas, A.; Láng, L.; Bedő, Z.; Doležel, J.; Molnár-Láng, M.; Shewry, P. Addition of Aegilops U and M chromosomes affects protein and dietary fiber content of wholemeal wheat flour. Front. Plant Sci. 2017, 8, 1529. [Google Scholar] [CrossRef]
- Marcotuli, I.; Colasuonno, P.; Cutillo, S.; Simeone, R.; Blanco, A.; Gadaleta, A. β-glucan content in a panel of Triticum and Aegilops genotypes. Genet. Resour. Crop Evol. 2019, 66, 897–907. [Google Scholar] [CrossRef]
- Rakszegi, M.; Darkó, É.; Lovegrove, A.; Molnár, I.; Láng, L.; Bedő, Z.; Molnár-Láng, M.; Shewry, P. Drought stress affects the protein and dietary fiber content of wholemeal wheat flour in wheat/Aegilops addition lines. PLoS ONE 2019, 14, e0211892. [Google Scholar] [CrossRef]
- Schneider, A.; Linc, G.; Molnár, I.; Molnár-Láng, M. Molecular cytogenetic characterization of Aegilops biuncialis and its use for the identification of 5 derived wheat-Aegilops biuncialis disomic addition lines. Genome 2005, 48, 1070–1082. [Google Scholar] [CrossRef]
- Molnár, I.; Benavente, E.; Molnár-Láng, M. Detection of intergenomic chromosome rearrangements in irradiated Triticum aestivum-Aegilops biuncialis amphiploids by multicolour genomic in situ hybridization. Genome 2009, 52, 156–165. [Google Scholar] [CrossRef] [PubMed]
- Schneider, A.; Molnár-Láng, M. Detection of various U and M chromosomes in wheat-Aegilops biuncialis hybrids and derivatives using fluorescence in situ hybridisation and molecular markers. Czech J. Genet. Plant Breed. 2012, 48, 169–177. [Google Scholar] [CrossRef]
- Zhou, J.P.; Yao, C.H.; Yang, E.N.; Yin, M.Q.; Liu, C.; Ren, Z.L. Characterization of a new wheat-Aegilops biuncialis addition line conferring quality-associated HMW glutenin subunits. Genet. Mol. Res. 2014, 13, 660–669. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.P.; Cheng, Y.; Zang, L.L.; Yang, E.N.; Liu, C.; Zheng, X.L.; Deng, K.J.; Zhu, Y.Q.; Zhang, Y. Characterization of a new wheat-Aegilops biuncialis 1Mb(1B) substitution line with good quality-associated HMW glutenin subunit. Cereal Res. Commun. 2016, 44, 198–205. [Google Scholar] [CrossRef][Green Version]
- Wenzl, P.; Carling, J.; Kudrna, D.; Jaccoud, D.; Huttner, E.; Kleinhofs, A.; Kilian, A. Diversity Arrays Technology (DArT) for whole-genome profiling of barley. Proc. Natl. Acad. Sci. USA 2004, 101, 9915–9920. [Google Scholar] [CrossRef] [PubMed]
- Sansaloni, C.; Petroli, C.; Jaccoud, D.; Carling, J.; Detering, F.; Grattapaglia, D.; Kilian, A. Diversity Arrays Technology (DArT) and next-generation sequencing combined: Genome-wide, high throughput, highly informative genotyping for molecular breeding of Eucalyptus. BMC Proc. 2011, 5, P54. [Google Scholar] [CrossRef]
- Kilian, A.; Wenzl, P.; Huttner, E.; Carling, J.; Xia, L.; Blois, H.; Caig, V.; Heller-Uszynska, K.; Jaccoud, D.; Hopper, C.; et al. Diversity Arrays Technology: A generic genome profiling technology on open platforms. In Data Production and Analysis in Population Genomics: Methods and Protocols; Pompanon, F., Bonin, A., Eds.; Humana Press: New York, NY, USA, 2012; pp. 67–89. ISBN 978-1-61779-870-2. [Google Scholar]
- Kumar, A.; Seetan, R.; Mergoum, M.; Tiwari, V.K.; Iqbal, M.J.; Wang, Y.; Al-Azzam, O.; Šimková, H.; Luo, M.-C.; Dvorak, J.; et al. Radiation hybrid maps of the D-genome of Aegilops tauschii and their application in sequence assembly of large and complex plant genomes. BMC Genom. 2015, 16, 800. [Google Scholar] [CrossRef]
- Olivera, P.D.; Kilian, A.; Wenzl, P.; Steffenson, B.J. Development of a genetic linkage map for Sharon goatgrass (Aegilops sharonensis) and mapping of a leaf rust resistance gene. Genome 2013, 56, 367–376. [Google Scholar] [CrossRef]
- Molnár, I.; Vrána, J.; Burešová, V.; Cápal, P.; Farkas, A.; Darkó, É.; Cseh, A.; Kubaláková, M.; Molnár-Láng, M.; Doležel, J. Dissecting the U, M, S and C genomes of wild relatives of bread wheat (Aegilops spp.) into chromosomes and exploring their synteny with wheat. Plant J. 2016, 88, 452–467. [Google Scholar] [CrossRef]
- Said, M.; Holušová, K.; Farkas, A.; Ivanizs, L.; Gaál, E.; Cápal, P.; Abrouk, M.; Martis-Thiele, M.M.; Kalapos, B.; Bartoš, J.; et al. Development of DNA markers from physically mapped loci in Aegilops comosa and Aegilops umbellulata using single-gene FISH and chromosome sequences. Front. Plant Sci. 2021, 12, 689031. [Google Scholar] [CrossRef]
- Li, S.; Morris, C.F.; Bettge, A.D. Genotype and environment variation for arabinoxylans in hard winter and spring wheats of the U.S. Pacific Northwest. Cereal Chem. 2009, 86, 88–95. [Google Scholar] [CrossRef]
- Tremmel-Bede, K.; Szentmiklóssy, M.; Tömösközi, S.; Török, K.; Lovegrove, A.; Shewry, P.R.; Láng, L.; Bedő, Z.; Vida, G.; Rakszegi, M. Stability analysis of wheat lines with increased level of arabinoxylan. PLoS ONE 2020, 15, e0232892. [Google Scholar] [CrossRef] [PubMed]
- Martinant, J.-P.; Billot, A.; Bouguennec, A.; Charmet, G.; Saulnier, L.; Branlard, G. Genetic and environmental variations in water-extractable arabinoxylans content and flour extract viscosity. J. Cereal Sci. 1999, 30, 45–48. [Google Scholar] [CrossRef]
- Dornez, E.; Gebruers, K.; Joye, I.J.; de Ketelaere, B.; Lenartz, J.; Massaux, C.; Bodson, B.; Delcour, J.A.; Courtin, C.M. Effects of genotype, harvest year and genotype-by-harvest year interactions on arabinoxylan, endoxylanase activity and endoxylanase inhibitor levels in wheat kernels. J. Cereal Sci. 2008, 47, 180–189. [Google Scholar] [CrossRef]
- Monte, J.V.; de Nova, P.J.; Soler, C. AFLP-based analysis to study genetic variability and relationships in the Spanish species of the genus Aegilops. Hereditas 2001, 135, 233–238. [Google Scholar] [CrossRef]
- Okuno, K.; Ebana, K.; Noov, B.; Yoshida, H. Genetic diversity of Central Asian and north Caucasian Aegilops species as revealed by RAPD markers. Genet. Resour. Crop Evol. 1998, 45, 389–394. [Google Scholar] [CrossRef]
- Arrigo, N.; Felber, F.; Parisod, C.; Buerki, S.; Alvarez, N.; David, J.; Guadagnuolo, R. Origin and expansion of the allotetraploid Aegilops geniculata, a wild relative of wheat. New Phytol. 2010, 187, 1170–1180. [Google Scholar] [CrossRef]
- Meimberg, H.; Rice, K.J.; Milan, N.F.; Njoku, C.C.; McKay, J.K. Multiple origins promote the ecological amplitude of allopolyploid Aegilops (Poaceae). Am. J. Bot. 2009, 96, 1262–1273. [Google Scholar] [CrossRef]
- Caldwell, K.S.; Russell, J.; Langridge, P.; Powell, W. Extreme population-dependent linkage disequilibrium detected in an inbreeding plant species, Hordeum vulgare. Genetics 2006, 172, 557–567. [Google Scholar] [CrossRef]
- Kim, S.; Plagnol, V.; Hu, T.T.; Toomajian, C.; Clark, R.M.; Ossowski, S.; Ecker, J.R.; Weigel, D.; Nordborg, M. Recombination and linkage disequilibrium in Arabidopsis thaliana. Nat. Genet. 2007, 39, 1151–1155. [Google Scholar] [CrossRef]
- Li, Y.; Haseneyer, G.; Schön, C.-C.; Ankerst, D.; Korzun, V.; Wilde, P.; Bauer, E. High levels of nucleotide diversity and fast decline of linkage disequilibrium in rye (Secale cereale L.) genes involved in frost response. BMC Plant Biol. 2011, 11, 6. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, V.K.; Wang, S.; Danilova, T.; Koo, D.H.; Vrána, J.; Kubaláková, M.; Hřibová, E.; Rawat, N.; Kalia, B.; Singh, N.; et al. Exploring the tertiary gene pool of bread wheat: Sequence assembly and analysis of chromosome 5Mg of Aegilops geniculata. Plant J. 2015, 84, 733–746. [Google Scholar] [CrossRef] [PubMed]
- Molnár, I.; Šimková, H.; Leverington-Waite, M.; Goram, R.; Cseh, A.; Vrána, J.; Farkas, A.; Doležel, J.; Molnár-Láng, M.; Griffiths, S. Syntenic relationships between the U and M genomes of Aegilops, wheat and the model species Brachypodium and rice as revealed by COS markers. PLoS ONE 2013, 8, e70844. [Google Scholar] [CrossRef]
- Edae, E.A.; Olivera, P.D.; Jin, Y.; Poland, J.A.; Rouse, M.N. Genotype-by-sequencing facilitates genetic mapping of a stem rust resistance locus in Aegilops umbellulata, a wild relative of cultivated wheat. BMC Genom. 2016, 17, 1039. [Google Scholar] [CrossRef] [PubMed]
- Edae, E.A.; Olivera, P.D.; Jin, Y.; Rouse, M.N. Genotyping-by-sequencing facilitates a high-density consensus linkage map for Aegilops umbellulata, a wild relative of cultivated wheat. G3 Genes Genomes Genet. 2017, 7, 1551–1561. [Google Scholar] [CrossRef]
- Zhang, H.; Jia, J.; Gale, M.D.; Devos, K.M. Relationships between the chromosomes of Aegilops umbellulata and wheat. Theor. Appl. Genet. 1998, 96, 69–75. [Google Scholar] [CrossRef]
- Marcotuli, I.; Colasuonno, P.; Hsieh, Y.S.Y.; Fincher, G.B.; Gadaleta, A. Non-Starch polysaccharides in durum wheat: A review. Int. J. Mol. Sci. 2020, 21, 2933. [Google Scholar] [CrossRef]
- Schreiber, M.; Wright, F.; MacKenzie, K.; Hedley, P.E.; Schwerdt, J.G.; Little, A.; Burton, R.A.; Fincher, G.B.; Marshall, D.; Waugh, R.; et al. The barley genome sequence assembly reveals three additional members of the CslF (1,3;1,4)-β-glucan synthase gene family. PLoS ONE 2014, 9, e90888. [Google Scholar] [CrossRef]
- Burton, R.A.; Jobling, S.A.; Harvey, A.J.; Shirley, N.J.; Mather, D.E.; Bacic, A.; Fincher, G.B. The genetics and transcriptional profiles of the cellulose synthase-like HvCslF gene family in barley. Plant Physiol. 2008, 146, 1821–1833. [Google Scholar] [CrossRef]
- Islam, S.; Rahman, I.A.; Islam, T.; Ghosh, A. Genome-wide identification and expression analysis of glutathione S-transferase gene family in tomato: Gaining an insight to their physiological and stress-specific roles. PLoS ONE 2017, 12, e0187504. [Google Scholar] [CrossRef]
- Charmet, G.; Masood-Quraishi, U.; Ravel, C.; Romeuf, I.; Balfourier, F.; Perretant, M.R.; Joseph, J.L.; Rakszegi, M.; Guillon, F.; Sado, P.E.; et al. Genetics of dietary fibre in bread wheat. Euphytica 2009, 170, 155–168. [Google Scholar] [CrossRef]
- Quraishi, U.M.; Murat, F.; Abrouk, M.; Pont, C.; Confolent, C.; Oury, F.X.; Ward, J.; Boros, D.; Gebruers, K.; Delcour, J.A.; et al. Combined meta-genomics analyses unravel candidate genes for the grain dietary fiber content in bread wheat (Triticum aestivum L.). Funct. Integr. Genom. 2011, 11, 71–83. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, R.A.C.; Dupree, P.; Shewry, P.R. A novel bioinformatics approach identifies candidate genes for the synthesis and feruloylation of arabinoxylan. Plant Physiol. 2007, 144, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Lovegrove, A.; Wilkinson, M.D.; Freeman, J.; Pellny, T.K.; Tosi, P.; Saulnier, L.; Shewry, P.R.; Mitchell, R.A.C. RNA interference suppression of genes in glycosyl transferase families 43 and 47 in wheat starchy endosperm causes large decreases in arabinoxylan content. Plant Physiol. 2013, 163, 95–107. [Google Scholar] [CrossRef]
- Zeng, W.; Jiang, N.; Nadella, R.; Killen, T.L.; Nadella, V.; Faik, A. A glucurono(arabino)xylan synthase complex from wheat contains members of the GT43, GT47, and GT75 families and functions cooperatively. Plant Physiol. 2010, 154, 78–97. [Google Scholar] [CrossRef]
- Anders, N.; Wilkinson, M.D.; Lovegrove, A.; Freeman, J.; Tryfona, T.; Pellny, T.K.; Weimar, T.; Mortimer, J.C.; Stott, K.; Baker, J.M.; et al. Glycosyl transferases in family 61 mediate arabinofuranosyl transfer onto xylan in grasses. Proc. Natl. Acad. Sci. USA 2012, 109, 989–993. [Google Scholar] [CrossRef]
- Hatfield, R.D.; Nevins, D.J. Hydrolytic activity and substrate specificity of an endoglucanase from Zea mays seedling cell walls. Plant Physiol. 1987, 83, 203–207. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Y.; Zhang, H.; Zhai, H.; Liu, Q.; He, S. A soluble starch synthase I gene, IbSSI, alters the content, composition, granule size and structure of starch in transgenic sweet potato. Sci. Rep. 2017, 7, 2315. [Google Scholar] [CrossRef]
- Naumoff, D.G. Hierarchical classification of glycoside hydrolases. Biochemestry 2011, 76, 622–635. [Google Scholar] [CrossRef]
- McArthur, J.B.; Chen, X. Glycosyltransferase engineering for carbohydrate synthesis. Biochem. Soc. Trans. 2016, 44, 129–142. [Google Scholar] [CrossRef]
- Molnár-Láng, M.; Linc, G.; Sutka, J. Transfer of the recessive crossability allele kr1 from Chinese Spring into the winter wheat variety Martonvásári 9. Euphytica 1996, 90, 301–305. [Google Scholar] [CrossRef]
- Molnár-Láng, M.; Molnár, I.; Szakács, É.; Linc, G.; Bedö, Z. Production and molecular cytogenetic identification of wheat-alien hybrids and introgression lines. In Genomics of Plant Genetic Resources; Tuberosa, R., Graner, A., Frison, E.A., Eds.; Springer: New York, NY, USA, 2014; pp. 255–283. ISBN 978-94-007-7572-5. [Google Scholar]
- ICC Standard Method No. 167. Grain and Grain Products for Food and Feed. Determination of Crude Protein in Grain and Grain Products for Food and Feed by the Dumas Combustion Principle; International Association for Cereal Science and Technology: Vienna, Austria, 2000. [Google Scholar]
- Douglas, S. A rapid method for the determination of pentosans in wheat flour. Food Chem. 1981, 7, 139–145. [Google Scholar] [CrossRef]
- Finnie, S.M.; Bettge, A.D.; Morris, C.F. Influence of cultivar and environment on water-soluble and water-insoluble arabinoxylans in soft wheat. Cereal Chem. 2006, 83, 617–623. [Google Scholar] [CrossRef]
- ICC Standard Method No. 166. Determination of β-Glucan in Barley, Oat and Rye; International Association for Cereal Science and Technology: Vienna, Austria, 1998. [Google Scholar]
- AACC Method No. 32-23.01. β-Glucan Content of Barley and Oats, Rapid Enzymatic Procedure. In AACC Approved Methods of Analysis, 11th ed.; American Association of Cereal Chemists: St Paul, MN, USA, 2009. [Google Scholar]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Dominant markers and null alleles. Mol. Ecol. Notes 2007, 7, 574–578. [Google Scholar] [CrossRef]
- Hubisz, M.J.; Falush, D.; Stephens, M.; Pritchard, J.K. Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Resour. 2009, 9, 1322–1332. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Dabney, A.; Storey, J.D.; Warnes, G.R. Qvalue: Q-Value Estimation for False Discovery Rate Control. R Package Version 1.16.0. 2008. Available online: http://bioconductor.org/packages/2.4/bioc/html/qvalue.html (accessed on 21 May 2021).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018; Available online: https://www.R-project.org/ (accessed on 21 May 2021).
- International Wheat Genome Sequencing Consortium (IWGSC); Appels, R.; Eversole, K.; Feuillet, C.; Keller, B.; Rogers, J.; Stein, N.; Pozniak, C.J.; Choulet, F.; Distelfeld, A.; et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar] [CrossRef] [PubMed]
- Eddy, S.R. A new generation of homology search tools based on probabilistic inference. Genome Inform. 2009, 23, 205–211. [Google Scholar] [PubMed]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef] [PubMed]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef]
- Virk, D.S.; Pandit, D.B.; Sufian, M.A.; Ahmed, F.; Siddique, M.A.; Samad, M.A.; Rahman, M.M.; Islam, M.M.; Ortiz-Ferrara, G.; Joshi, K.D.; et al. REML is an effective analysis for mixed modelling of unbalanced on-farm varietal trials. Exp. Agric. 2009, 45, 77–91. [Google Scholar] [CrossRef]


| 2016 | 2017 | |||||||
|---|---|---|---|---|---|---|---|---|
| Mv9kr1 | Ae. biuncialis Collection | Mv9kr1 | Ae. biuncialis Collection | |||||
| Mean * | Min | Max | Mean * | Min | Max | |||
| BG (mg/g) | 9.44 | 38.10 | 22.70 | 54.90 | 8.62 | 35.30 | 19.50 | 51.20 |
| TP (mg/g) | 40.17 | 40.11 | 29.60 | 50.77 | 40.24 | 40.40 | 31.90 | 50.71 |
| WEP (mg/g) | 10.79 | 10.82 | 6.83 | 15.42 | 10.63 | 10.68 | 7.25 | 15.46 |
| Protein (%) | 12.91 | 26.61 | 19.61 | 33.49 | 13.01 | 27.20 | 22.80 | 33.00 |
| Mean Squares | h2 | |||
|---|---|---|---|---|
| G | E | G × E | ||
| BG | 244.6 *** | 1366.5 *** | 17.7 *** | 0.93 |
| TP | 131.7 *** | 13.4 n.s. | 61.7 *** | 0.54 |
| WEP | 24.8 *** | 3.04 ** | 3.7 *** | 0.61 |
| Protein | 27.7 *** | 59.3 *** | 3.8 *** | 0.27 |
| Trait | QTL | Marker | Effect | Chr (UM) a | Chr (ABD) b | R2 | p | LOD | Candidate Annotation |
|---|---|---|---|---|---|---|---|---|---|
| BG | 1 | 100022501_F_0 | −0.88 | 4M/6U | 4ABD | 0.128 | 6.87 × 10−4 | 4.542 | glutathione S-transferase 3-like † |
| 2 | 100013840_F_1 | −0.78 | 5M | 5ABD | 0.125 | 7.72 × 10−4 | 3.113 | - | |
| 3 | 100079925_F_0 | −1.62 | 1M **/1U | 1ABD | 0.149 | 2.26 × 10−4 | 3.647 | - | |
| Protein | 4 | 100001630_F_1 | −3.64 | 6M | - | 0.124 | 8.39 × 10−4 | 3.076 | putative ripening-related protein 6 † |
| 1 | 100022501_F_2 | −5.76 | 4M/6U | 4ABD | 0.159 | 1.30 × 10−4 | 3.886 | glutathione S-transferase 3-like † | |
| 5 | 100011893_F_1 | 4.67 | 2M/2U | 2ABD | 0.139 | 3.68 × 10−4 | 3.435 | - | |
| 6 | 100074730_F_0 | −1.67 | 1M **/1U | 1ABD | 0.140 | 3.54 × 10−4 | 3.451 | - | |
| 7 | 100016211_F_2 | −8.13 | 2M/6U | 2ABD | 0.189 | 2.53 × 10−5 | 4.597 | - | |
| 8 | 100030135_F_1 | −2.43 | 5M | 5ABD | 0.125 | 7.84 × 10−4 | 3.106 | DNA-binding transcription factor activity * | |
| 9 | 100054424_F_0 | 1.33 | 2M/2U | 2ABD | 0.126 | 7.44 × 10−4 | 3.128 | - | |
| 10 | 100033763_F_0 | −2.13 | 4M/6U | 2A 6B 4D | 0.122 | 9.34 × 10−4 | 3.030 | - | |
| 11 | 100016524_F_0 | −3.64 | 7M/7U | 7ABD | 0.145 | 2.78 × 10−4 | 3.556 | - | |
| 12 | 100024379_F_1 | 3.85 | 3M/3U | 3ABD | 0.138 | 3.92 × 10−4 | 3.407 | - | |
| 2 | 100013840_F_0 | −3.10 | 5M | 5ABD | 0.147 | 2.41 × 10−4 | 3.618 | - | |
| 13 | 100013808_F_0 | 5.39 | 5M/5U | 5ABD | 0.123 | 8.56 × 10−4 | 3.068 | DNA-binding transcription factor activity * | |
| TP | 5 | 100011893_F_2 | 0.10 | 2M/2U | 2ABD | 0.125 | 8.01 × 10−4 | 3.096 | - |
| 7 | 100016211_F_0 | −12.70 | 2M/6U | 2ABD | 0.184 | 3.44 × 10−5 | 4.464 | - | |
| 8 | 100030135_F_0 | −6.40 | 5M | 5ABD | 0.144 | 2.96 × 10−4 | 3.529 | DNA-binding transcription factor activity * | |
| WEP | 4 | 100001630_F_0 | −1.67 | 6M | - | 0.145 | 2.80 × 10−4 | 3.554 | putative ripening-related protein 6 † |
| 14 | 100009067_F_0 | −3.16 | 1M/1U | 1ABD | 0.131 | 5.85 × 10−4 | 3.233 | 1-deoxy-D-xylulose-5-phosphate synthase activity * | |
| 1 | 100022501_F_1 | −2.54 | 4M/6U | 4ABD | 0.221 | 4.29 × 10−6 | 5.367 | glutathione S-transferase 3-like † | |
| 15 | 100027188_F_0 | −3.55 | 7M/7U | 7ABD | 0.140 | 3.58 × 10−4 | 3.446 | - | |
| 5 | 100011893_F_0 | −0.33 | 2M/2U | 2ABD | 0.125 | 7.55 × 10−4 | 3.122 | - | |
| 16 | 100015451_F_0 | 4.12 | 1M ** | 1ABD | 0.183 | 3.50 × 10−5 | 4.456 | - | |
| 7 | 100016211_F_1 | −3.60 | 2M/6U | 2ABD | 0.198 | 1.54 × 10−5 | 4.812 | - | |
| 17 | 100013669_F_0 | 3.51 | 3M/3U | 3ABD | 0.149 | 2.19 × 10−4 | 3.660 | - | |
| 18 | 100033114_F_0 | −2.29 | 7M7U | 7ABD | 0.126 | 7.37 × 10−4 | 3.133 | DNA-binding transcription factor activity * | |
| 19 | 100006546_F_0 | −0.76 | 6U | 6ABD | 0.155 | 1.63 × 10−4 | 3.787 | - | |
| 20 | 100030958_F_0 | 4.47 | 6M | 6ABD | 0.123 | 8.59 × 10−4 | 3.066 | glycosyltransferase At5g20260 * | |
| 21 | 100041833_F_0 | −2.45 | - | - | 0.132 | 5.55 × 10−4 | 3.256 | - | |
| 22 | 100001948_F_0 | 2.88 | 3M/3U | 3ABD | 0.131 | 5.84 × 10−4 | 3.234 | soluble starch synthase * | |
| 23 | 100010676_F_0 | 3.93 | 1M **/4U | 6A 1B 1D | 0.126 | 7.34 × 10−4 | 3.135 | glycoside hydrolase/deacetylase superfamily * | |
| 24 | 100001383_F_0 | −2.25 | 3M/3U | 3ABD | 0.135 | 4.68 × 10−4 | 3.330 | O-acetyltransferase activity * | |
| 25 | 100070301_F_0 | −1.83 | 2M/2U | 2ABD | 0.120 | 9.91 × 10−4 | 3.004 | protein serine/threonine phosphatase activity * | |
| 12 | 100024379_F_0 | 3.15 | 3M/3U | 3ABD | 0.161 | 1.17 × 10−4 | 3.934 | - | |
| 26 | 100036161_F_0 | 2.05 | 3M/3U | 3ABD | 0.123 | 8.75 × 10−4 | 3.058 | - | |
| 27 | 100009019_F_0 | 3.06 | 6M/4U | 6ABD | 0.144 | 2.95 × 10−4 | 3.530 | glutamate receptor 2.8-like † | |
| 28 | 100024849_F_0 | 1.93 | 6M/6U | 6ABD | 0.127 | 6.97 × 10−4 | 3.157 | endoglucanase 5-like † | |
| 29 | 100010471_F_0 | 4.48 | 6M | 6ABD | 0.123 | 8.79 × 10−4 | 3.056 | - | |
| 30 | 100005191_F_0 | −3.51 | 4M/1U | 5A 4B 4D | 0.148 | 2.36 × 10-4 | 3.626 | flavonol synthase/flavanone 3-hydroxylase-like † | |
| 31 | 100006760_F_0 | −2.46 | 3M | 3ABD | 0.129 | 6.28 × 10−4 | 3.202 | - | |
| 32 | 100014148_F_0 | −2.45 | 3M/3U | 3ABD | 0.161 | 1.21 × 10−4 | 3.919 | - | |
| 33 | 100066878_F_0 | 3.09 | 4M/6U | 4ABD | 0.142 | 3.21 × 10−4 | 3.493 | - | |
| 34 | 100069132_F_0 | −2.00 | 1M **/6U | 1ABD | 0.166 | 9.09 × 10−5 | 4.041 | zinc ion binding * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ivanizs, L.; Marcotuli, I.; Rakszegi, M.; Kalapos, B.; Szőke-Pázsi, K.; Farkas, A.; Türkösi, E.; Gaál, E.; Kruppa, K.; Kovács, P.; et al. Identification of New QTLs for Dietary Fiber Content in Aegilops biuncialis. Int. J. Mol. Sci. 2022, 23, 3821. https://doi.org/10.3390/ijms23073821
Ivanizs L, Marcotuli I, Rakszegi M, Kalapos B, Szőke-Pázsi K, Farkas A, Türkösi E, Gaál E, Kruppa K, Kovács P, et al. Identification of New QTLs for Dietary Fiber Content in Aegilops biuncialis. International Journal of Molecular Sciences. 2022; 23(7):3821. https://doi.org/10.3390/ijms23073821
Chicago/Turabian StyleIvanizs, László, Ilaria Marcotuli, Marianna Rakszegi, Balázs Kalapos, Kitti Szőke-Pázsi, András Farkas, Edina Türkösi, Eszter Gaál, Klaudia Kruppa, Péter Kovács, and et al. 2022. "Identification of New QTLs for Dietary Fiber Content in Aegilops biuncialis" International Journal of Molecular Sciences 23, no. 7: 3821. https://doi.org/10.3390/ijms23073821
APA StyleIvanizs, L., Marcotuli, I., Rakszegi, M., Kalapos, B., Szőke-Pázsi, K., Farkas, A., Türkösi, E., Gaál, E., Kruppa, K., Kovács, P., Darkó, É., Szakács, É., Said, M., Cápal, P., Doležel, J., Gadaleta, A., & Molnár, I. (2022). Identification of New QTLs for Dietary Fiber Content in Aegilops biuncialis. International Journal of Molecular Sciences, 23(7), 3821. https://doi.org/10.3390/ijms23073821










