Abstract
Pathogenic bacteria causing human rickettsioses, transmitted in nature by arthropod vectors, primarily infect vascular endothelial cells lining the blood vessels, resulting in ‘endothelial activation’ and onset of innate immune responses. Nucleotide second messengers are long presumed to be the stimulators of type I interferons, of which bacterial cyclic-di-GMP (c-di-GMP) has been implicated in multiple signaling pathways governing communication with other bacteria and host cells, yet its importance in the context of rickettsial interactions with the host has not been investigated. Here, we report that all rickettsial genomes encode a putative diguanylate cyclase pleD, responsible for the synthesis of c-di-GMP. In silico analysis suggests that although the domain architecture of PleD is apparently well-conserved among different rickettsiae, the protein composition and sequences likely vary. Interestingly, cloning and sequencing of the pleD gene from virulent (Sheila Smith) and avirulent (Iowa) strains of R. rickettsii reveals a nonsynonymous substitution, resulting in an amino acid change (methionine to isoleucine) at position 236. Additionally, a previously reported 5-bp insertion in the genomic sequence coding for pleD (NCBI accession: NC_009882) was not present in the sequence of our cloned pleD from R. rickettsii strain Sheila Smith. In vitro infection of HMECs with R. rickettsii (Sheila Smith), but not R. rickettsii (Iowa), resulted in dynamic changes in the levels of pleD up to 24 h post-infection. These findings thus provide the first evidence for the potentially important role(s) of c-di-GMP in the determination of host-cell responses to pathogenic rickettsiae. Further studies into molecular mechanisms through which rickettsial c-di-GMP might regulate pathogen virulence and host responses should uncover the contributions of this versatile bacterial second messenger in disease pathogenesis and immunity to human rickettsioses.
1. Introduction
The genus Rickettsia, in the family Rickettsiaceae of the order Rickettsiales, now encompasses an expanding number of Gram-negative, intracellular α-proteobacteria distributed across the globe and transmitted to humans as accidental hosts by arthropod vectors, including fleas, lice, mites, and ticks. The most prominent pathogenic species causing vector-borne rickettsioses belong to the spotted-fever group represented by R. rickettsii and R. conorii, and typhus group, including R. prowazekii and R. typhi [1,2]. As endotheliotropic pathogens, rickettsiae preferentially infect endothelial cells lining the small- and medium-sized vessels, resulting in systemic inflammation and compromised barrier integrity as the major pathophysiological sequelae, leading to increased vascular permeability and tissue edema in vital organs such as the lungs and brain [3,4]. Consequently, host–pathogen interactions with a focus on endothelial responses and vascular dysfunction or damage constitute an important area of study to achieve a comprehensive mechanistic understanding of rickettsial pathogenesis and immunity. Not surprisingly, a majority of endothelial responses to disseminated infection with rickettsiae are mediated through the activation of cellular regulators, including nuclear transcription factor-kappa B (NF-kB) and interferon regulatory factors [5,6,7]. In this regard, our prior findings have documented induction of a type I interferon (IFN) response by cultured human microvascular endothelial cells (HMECs) infected with R. conorii and its involvement in the autocrine and/or paracrine activation of the Janus kinase-signal transducer and activator of the transcription (JAK-STAT) signaling pathway, and increased expression of a number of downstream IFN-stimulated innate-response genes [8,9].
Cyclic dinucleotides are now well-appreciated to manifest important roles in the determination of host innate immune responses to bacterial and viral infections. Bacteria produce cyclic diguanylate monophosphate (c-di-GMP), cyclic diadenylate monophosphate (c-di-AMP), and cyclic guanosine monophosphate-adenosine monophosphate (c-GAMP) as second messengers for the fine regulation of a variety of cellular processes [10,11]. Of these, c-di-GMP synthesized from two GTP molecules by diguanylate cyclases (DGCs) has been determined to be ubiquitously utilized by bacteria for the governance of numerous life-cycle activities, including biofilm formation, motility, pathogenesis, and virulence [11,12,13]. The catabolism of c-di-GMP into linear 5-phosphoguanylyl-(3-5)-guanosine (pGpG) or GMP occurs through phosphodiesterase activities (PDEs). Importantly, catalytic domains of DGCs and PDEs, named as GGDEF and EAL or HD-GYP, respectively, have been extensively characterized biochemically, structurally, and genetically to suggest a high degree of conservation with an almost ubiquitous presence in bacteria [14,15]. The active site of the GGDEF domain is involved in GTP binding, resulting in the synthesis of c-di-GMP, which is then recognized by the mammalian innate immune system to trigger host type I interferon responses [16,17]. The ubiquitous effects of c-di-GMP on the survival and virulence strategies of most bacterial species, in conjunction with the lack of both synthetic and degradative enzymes in all other domains of life, make this bacterial messenger a promising candidate for the development of immune-based adjuvants or therapeutics [14].
Despite the tendency for evolution by genomic reduction, the genomes of Rickettsia species carry pleD predicted to encode for a putative diguanylate cyclase responsible for the synthesis of c-di-GMP. However, the expression of this important gene in target host cells and its functional roles in rickettsial pathogenesis remain an open question. As the critical first step to address this important knowledge gap, the aims of this study focus upon the demonstration of the presence of pleD in the genomes of different Rickettsia species, characterization of its domain architecture in well-characterized strains of varying virulence, and determination of its expression profile during rickettsial infection of the host microvascular endothelium as the preferred target niche.
2. Results
2.1. PleD Is Present and Conserved among Rickettsia Species
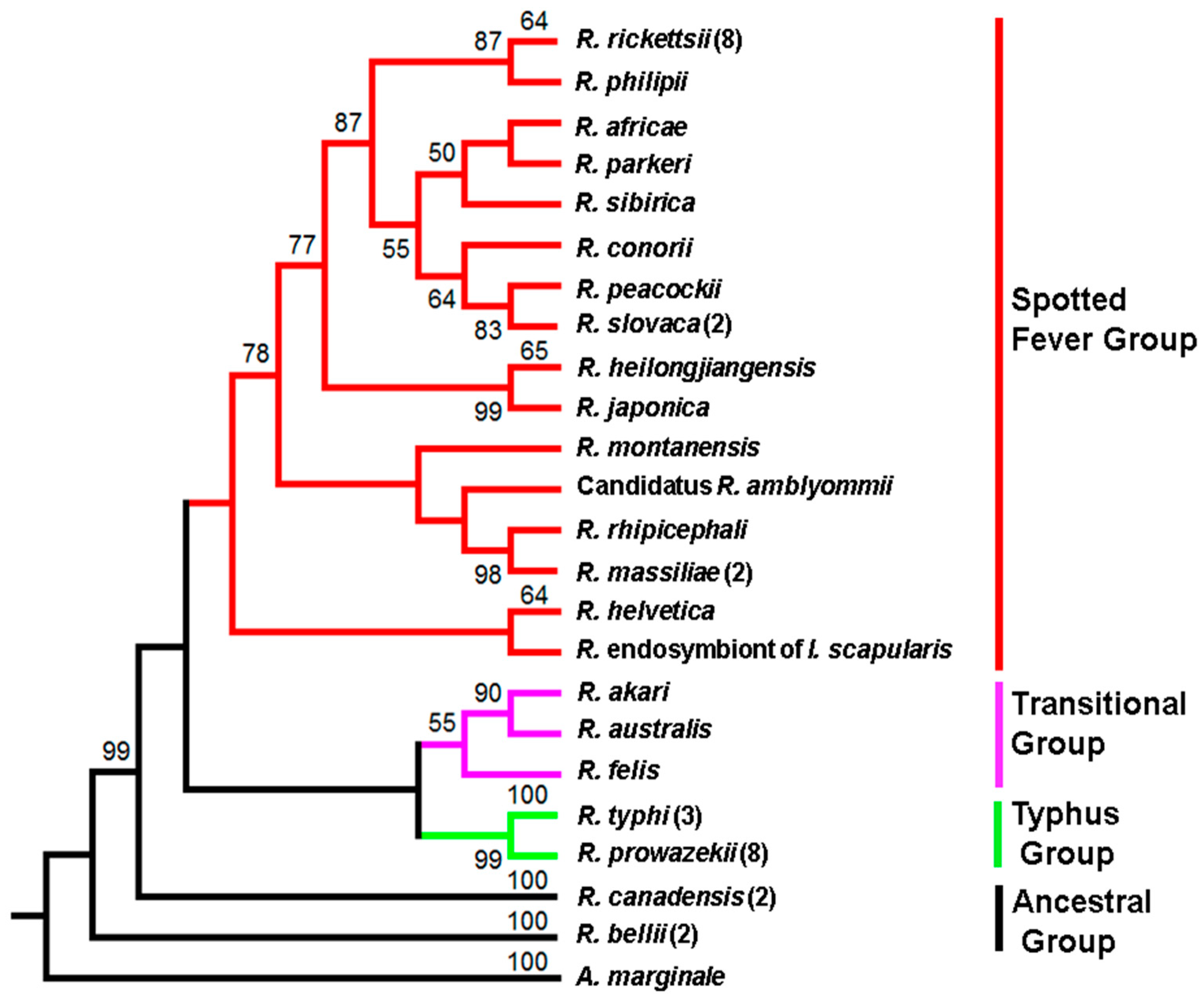
A phylogenetic tree based on amino-acid composition of PleD from representative species of all major subgroups, i.e., ancestral, spotted fever, typhus, and translational, illustrates its presence and conservation in the Genus Rickettsia. The protein with a predicted approximate molecular mass of ~52 kDa is encoded by a nucleotide sequence of 1353 bases and is comprised of 450 amino acids. As shown in Figure 1, the phylogeny is based upon protein sequences for 16 different species from the spotted fever group, 3 from the transitional group, and 2 each from the typhus and ancestral group. For spotted fever species, namely R. rickettsii, R. massiliae, and R. slovaca, both major prototypical representatives of the typhus group R. prowazekii and R. typhi, and R. bellii and R. canadensis of the ancestral group, different specific strains for which the genomes have been sequenced and annotated were also included.
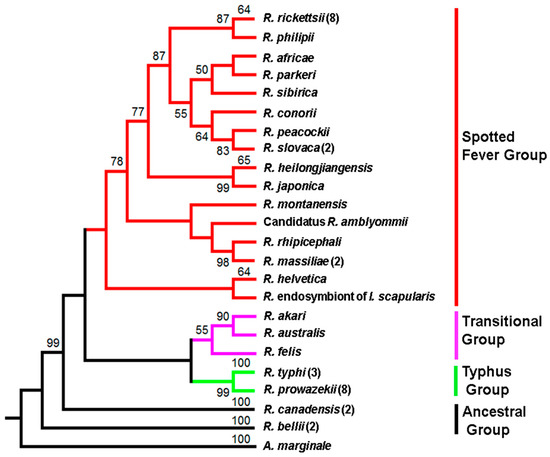
Figure 1.
Phylogeny of Rickettsia species inferred from PleD protein sequences. The numbers above the branches indicate bootstrap support values. The PleD sequence of A. marginale str. Florida was used as an outgroup. The number in parenthesis indicates the number of strains used in the analysis. Branch color represents Rickettsia species belonging to different groups. Red: spotted fever group, Purple: Transitional group, Green: Typhus group, and Black: Ancestral group.
2.2. Preservation of Domain Organization of PleD in Rickettsia Species
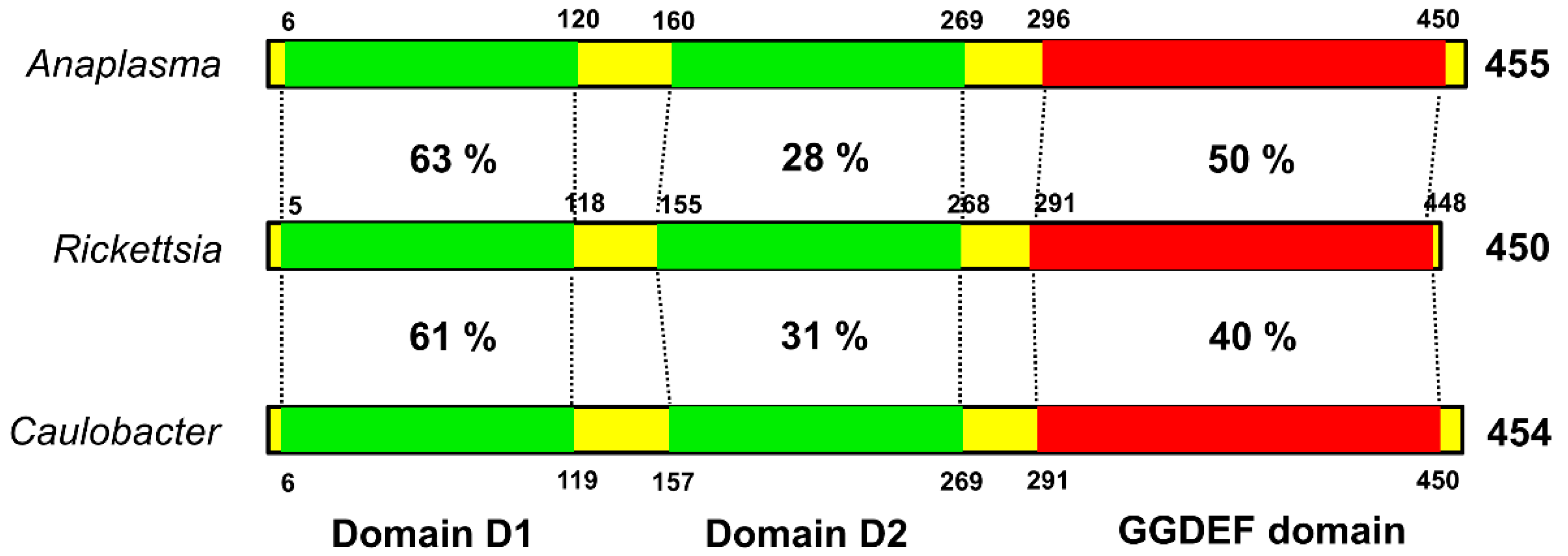
The biological importance of c-di-GMP as a bacterial second messenger is underscored by the omnipresence of the diguanylate cyclase (DGC) domain (also referred to as GGDEF or DUF1) in bacterial genomes. Structurally, PleD is composed of a CheY-like receiver domain (D1), a CheY-like adapter domain (D2), and a DGC domain. The D1 domain contains conserved residues, acts as a phosphoacceptor, and is involved in activation of the response regulator. The D2 domain is involved in dimerazation and binding to cyclic di-GMP at the DGC-D2 interface, leading to structural stabilization and prevention of catalysis [18]. Despite nearly ubiquitous distribution and documented regulatory relevance of DGC proteins in bacteria, structural and functional information about this class of regulators in Rickettsia species is largely missing. To address this, we performed the amino-acid sequence (blastp) comparison of the PleD domains of Rickettsia with the corresponding domains of a pathogenic α-proteobacterium Anaplasma phagocytophilum and a free-living, dimorphic α-proteobacterium Caulobacter crescentus. Such analysis revealed the highest level of conservation in D1 as suggested by a homology of 63 and 61%, respectively, with Anaplasma and Caulobacter. A similar comparison for the catalytic GGDEF domain suggested identity levels of 50% between rickettsial and Anaplasma PleD, and 40% between rickettsial and Caulobacter PleD, while adapter domain D2 displayed significant divergence among these bacteria (Figure 2).
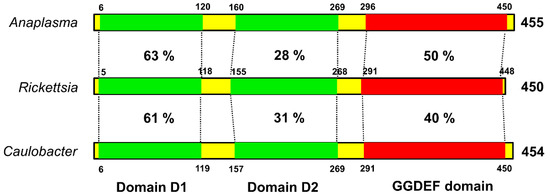
Figure 2.
The conserved domain architecture of rickettsial PleD. The rickettsial PleD domains (receiver domain 1, adapter domain 2, and GGDEF domain) were identified using blastp and compared with their counterparts in Anaplasma and Caulobacter. The receiver domain 1 and GGDEF domain of Rickettsia showed higher sequence identity with those present in Anaplasma and Caulobacter, whereas the adapter domain 2 is relatively less conserved. Sequence identity is presented as percentage. The numbers above/below the bars correspond to domain coordinates.
2.3. Sequencing and Comparison of pleD from Virulent and Avirulent Strains of R. rickettsii
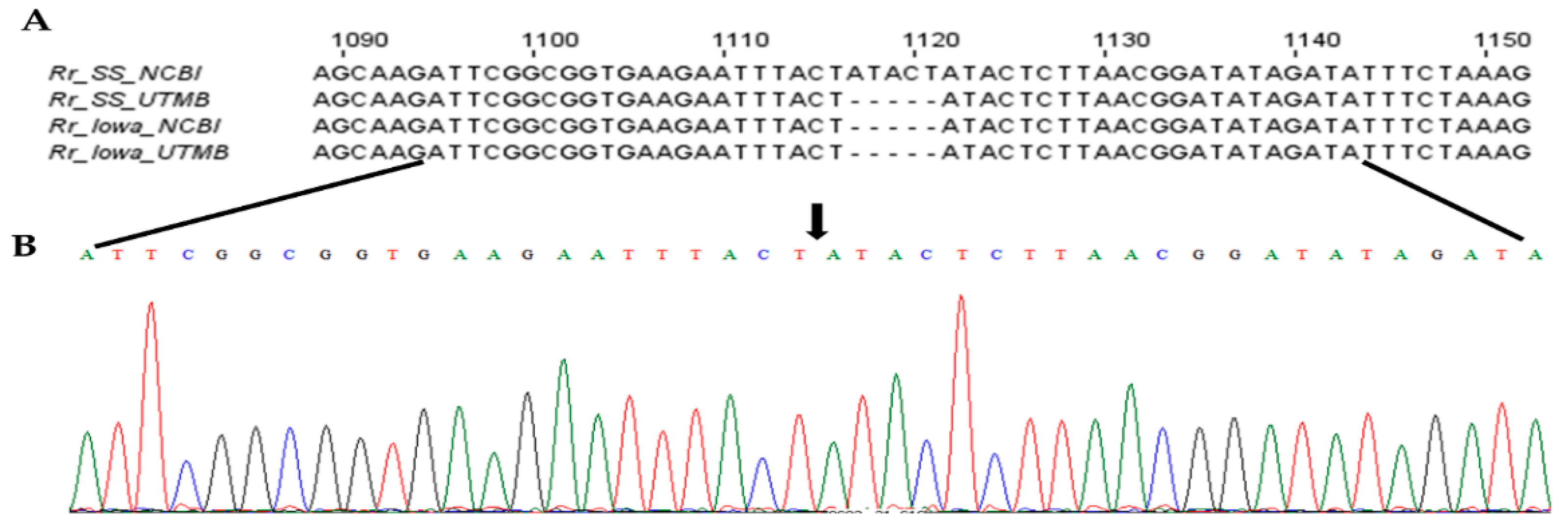
We next set out to investigate the possibility of PleD’s contributions to the host-cell responses to infection and a potential virulence factor of rickettsiae. Differences in the pathogenicity and virulence of different strains of spotted-fever group Rickettsia species, including R. rickettsii, are well established [3,19]. In a previously published study, application of bioinformatics-based strategies to define the biological basis of variations in virulence revealed a number of single-nucleotide polymorphisms, insertions, and deletions unique to specific strains and capable of distinguishing between them to allow for the distinction between virulent and avirulent R. rickettsii [20]. Intriguingly, our database search for the genomic sequences of pleD in a highly virulent Sheila Smith strain vis-à-vis an avirulent Iowa strain of R. rickettsii resulted in the identification of a fully annotated pleD in Iowa, but not in R. rickettsii Sheila Smith, and alignment of published sequences (NCBI Accession numbers NC_009882 (Sheila Smith) and NC_010263 (Iowa)) suggested a 5-bp insertion (ATACT) at positions 1117 to 1121, with respect to the start site, of the pleD gene in R. rickettsii Sheila Smith. To resolve this possible ambiguity, we cloned and sequenced pleD from both strains to demonstrate the absence of 5-bp insertion in R. rickettsii Sheila Smith and a 100% match between the sequences from Iowa and Sheila Smith strains of R. rickettsii (Figure 3; Supplementary File S1).
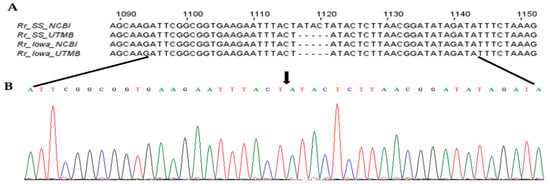
Figure 3.
The nucleotide sequence and chromatogram showing the absence of 5-base insertion in the pleD gene of R. rickettsii str. Sheila Smith. The full-length pleD from R. rickettsii strains Sheila Smith (Rr_SS_UTMB) and Iowa (Rr_Iowa_UTMB) were sequenced and aligned with sequences available in NCBI (Rr_SS_NCBI and Rr_Iowa_NCBI). The sequence alignment (A) and chromatogram (B) show the absence (shown by black arrow) of ‘ATACT’ indel in pleD sequence of R. rickettsii str. Sheila Smith.
2.4. A Nonsynonymous Substitution in the Nucleotide Sequence Results in an Amino-Acid Change at Position 236 of PleD in Different Strains of R. rickettsii
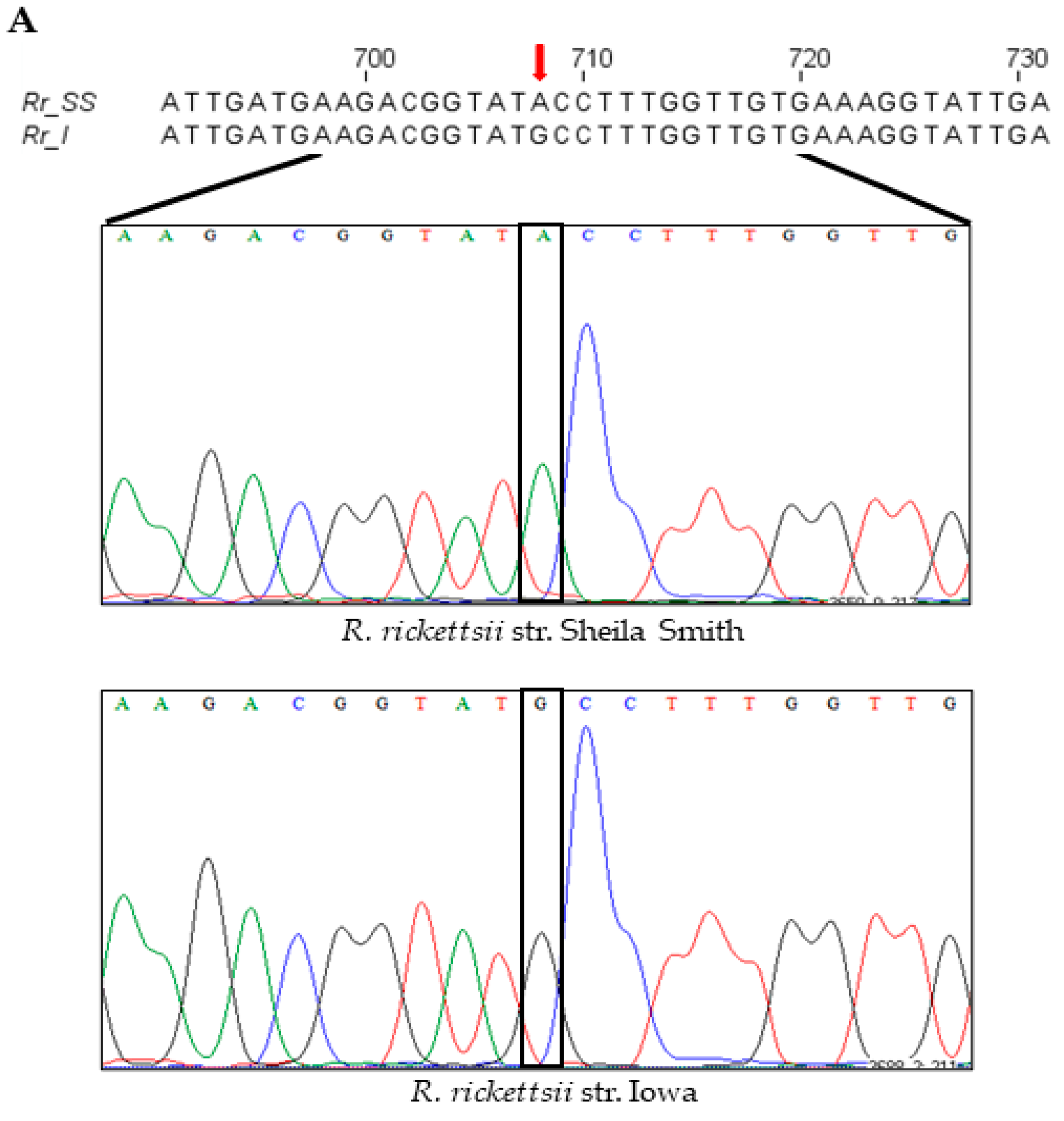
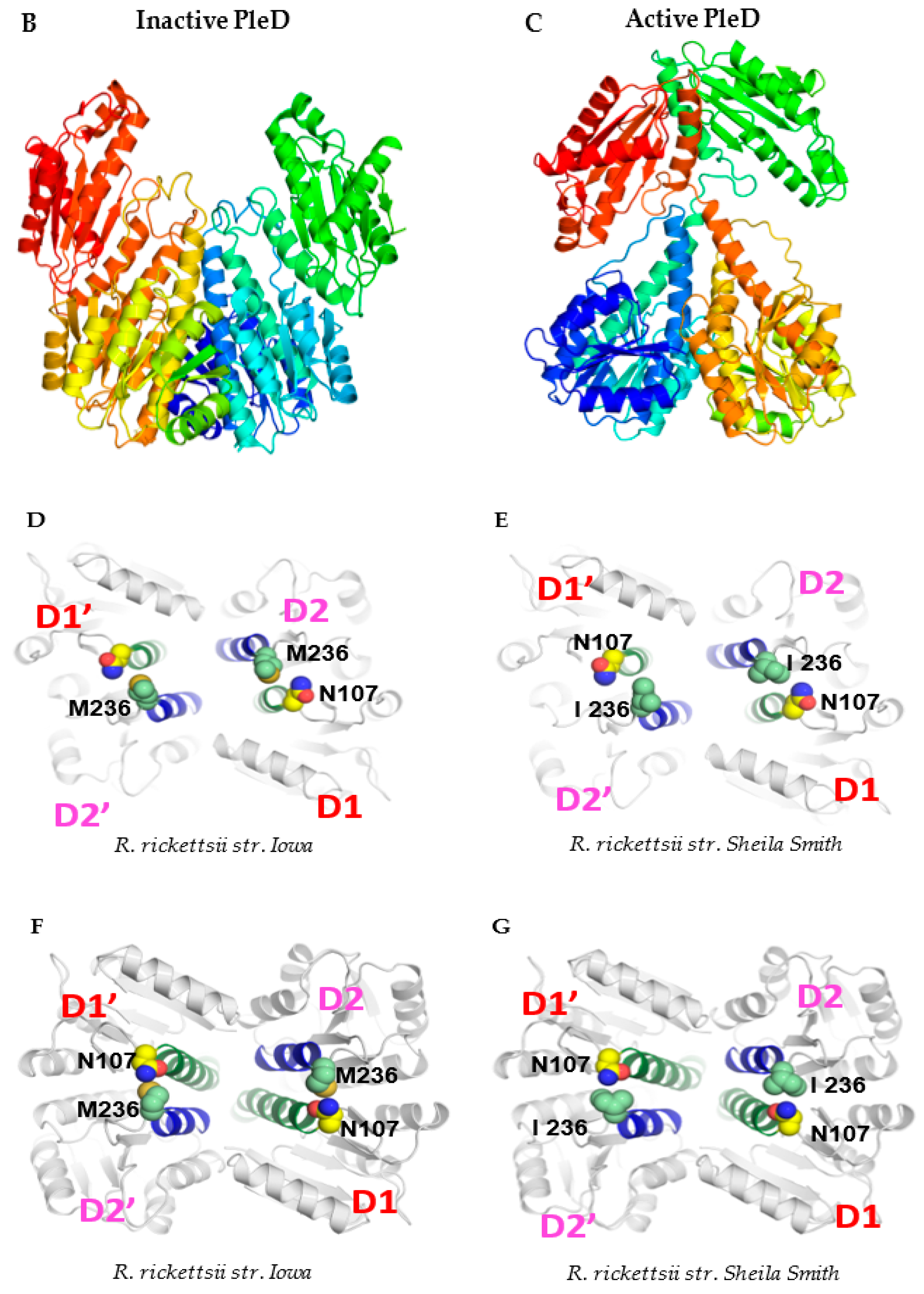
An intriguing and serendipitous finding emerging from the cloning, sequencing, and comparison of the composition of pleD from virulent Sheila Smith and avirulent Iowa strains of R. rickettsii was the identification of a single-nucleotide substitution (A→G; ‘A’ in Sheila Smith to ‘G’ in Iowa) at position 708 in the coding sequence (Figure 4A). This nucleotide change led to a nonsynonymous substitution of the amino acid at position 236 in the resultant protein, which accordingly was determined to be an isoleucine in strain Sheila Smith versus a methionine in Iowa (Supplementary File S1). In consideration of the structural and functional implications of nonsynonymous mutations due to variations in amino-acid sequences, we next performed a predictive analysis of tertiary structures using the MODELLER program [21] and generated the inactive and active models of encoded PleD proteins from both Iowa and Sheila Smith strains. As expected, the nonactivated PleD formed a weak dimer-interface interaction, while the activated PleD exhibited a strong dimer-interface interaction of the D1 and D2 domains (Figure 4B,C). Despite exhibiting mostly similar structural conformations in both active and inactive forms, the local interactions involving the receiver domain D1 and adaptor domain D2 domains were slightly different between the strains (Figure 4D–G). Interestingly, we identified that the amino acid at position 236 present in D2 is a predicted key residue which is involved in forming an interdomain bridge with asparagine at position 106 present in D1 domain. Importantly, the nature of the interactions between N106-M236 in the PleD of Iowa strain varies compared to the interaction between N106-I236 in the Sheila Smith strain potentially enhancing the stability of the active form of PleD in the virulent Sheila Smith strain.
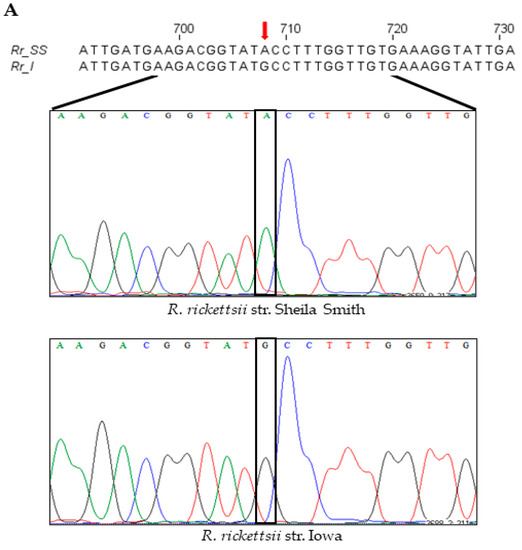
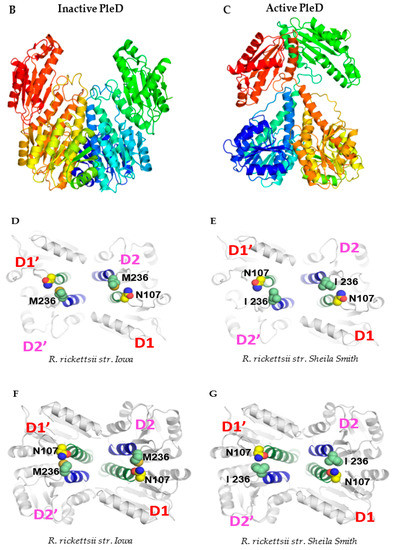
Figure 4.
A single-nucleotide polymorphism (SNP) at position 708, with respect to the gene start site, in the pleD of R. rickettsii str. Sheila Smith and Iowa results in an amino-acid change at a key residue of the protein. (A) Nucleotide sequence and chromatograms of R. rickettsii str. Sheila Smith and Iowa showing SNP (red arrow) at position 708 with respect to the gene start site. Ribbon representation of the dimeric model structures (perpendicular to the two-fold axis of the stem) of the inactive (B) and active (C) PleD. In panels (D–G), the views of the PleD inactive and active dimeric models are rotated by 90° around a horizontal axis with respect to the top panels (B,C), and shown is the bottom view of the (D1/D2) dimer stem with the diguanylate cyclase (DGC) domain in the rear hide for clarity. The residue N107 (yellow) on receiver domain D1 and M236/I236 (light green) on adaptor domain D2 are highlighted. The α-helix5 on receiver domain and α-helix4 on adaptor domain are colored in green and blue, respectively.
2.5. Differences in pleD Expression Pattern during Infection of Host Endothelial Cells with Virulent and Avirulent Strains of R. rickettsii
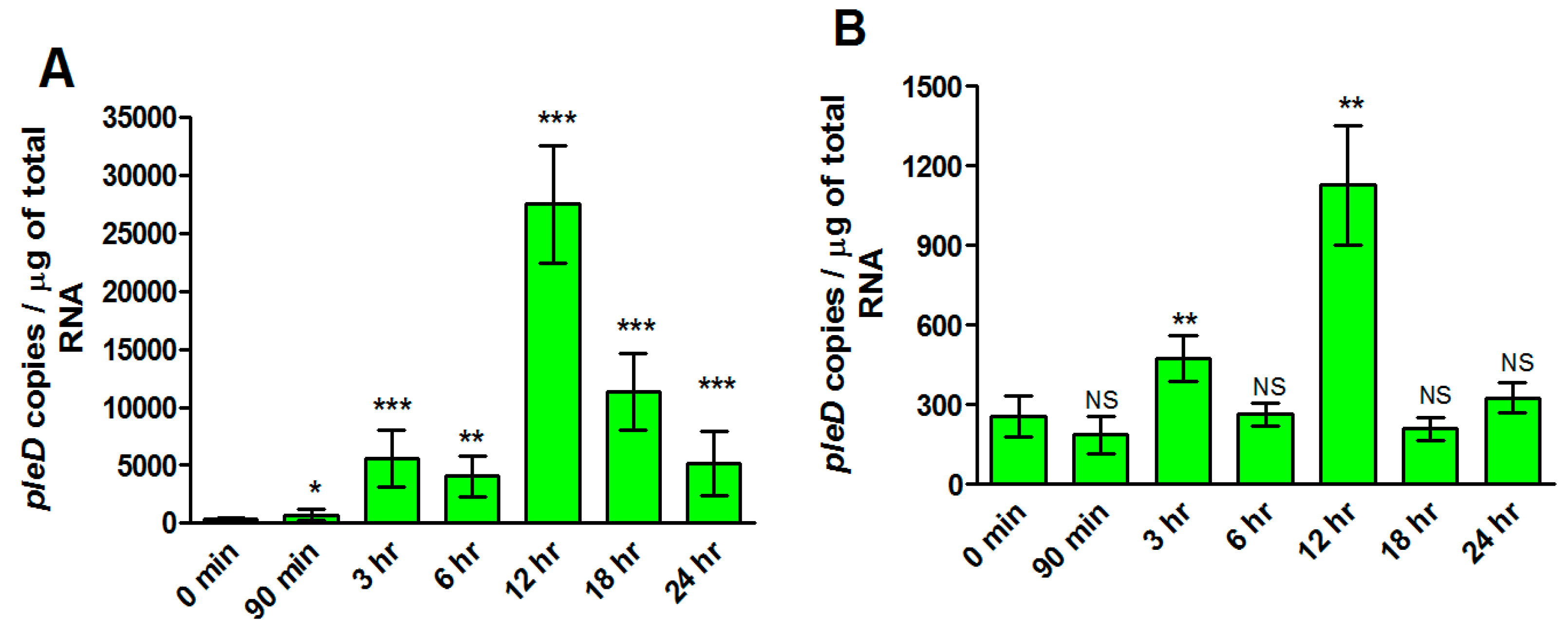
We finally determined expression profiles of rickettsial pleD in human microvascular endothelial cells (ECs) infected with R. rickettsii strains (MOI = 20) at opposite ends of virulence spectrum. We quantified the bacterial load using citrate synthase-based qPCR assay to ensure that both strains had similar levels of infection at all time points. No significant difference in rickettsial copy number was observed between virulent Sheila Smith and avirulent Iowa strains (Supplementary File S2). Interestingly, pleD expression was clearly evident and significantly higher as compared to the basal levels (at 0 h) at all studied timepoints up to 24 h post-infection in ECs infected with the virulent Sheila Smith strain. In contrast, avirulent Iowa triggered such a response of much lower magnitude with significant changes compared to the baseline only at 3 and 12 h post-infection (Figure 5). Absolute quantitation of pleD copy numbers further suggested a robust expression of the transcript during infection with virulent R. rickettsii, as evidenced by an increase of greater than 10- and 25-fold at 3 and 12 h post-infection when compared to host cells concurrently infected with the avirulent strain.
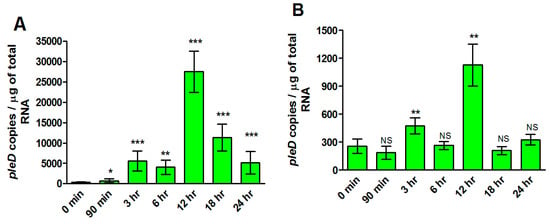
Figure 5.
Absolute quantification of pleD expression in R. rickettsii str. Sheila Smith and Iowa during the infection of host endothelial cells, in vitro. Significant differences in pleD expression levels are observed in host cells infected with R. rickettsii str. Sheila Smith (A) versus R. rickettsii str. Iowa (B) strain. Highest expression of pleD is observed in both R. rickettsii str. Sheila Smith and R. rickettsii str. Iowa at 12 h post-infection when compared to the control (0 min). * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001, not significant (NS) p > 0.05.
3. Discussion
The diguanylate cyclase and phosphodiesterase groups of enzymes are important signal-transduction proteins involved in the regulation of some critically important attributes of bacterial pathogens via the effector molecule cyclic-di-GMP. Although contributions of c-di-GMP in the governance of processes determining the pathogenesis of many bacteria, such as biofilm formation, motility, and virulence gene expression are now well-established and documented, its biosynthesis and functional roles in Rickettsiales are generally poorly characterized and understood. It is important to acknowledge, however, a seminal investigation in this context by Lai et. al. [22], documenting the presence of a functional PleC histidine kinase and PleD diguanylate cyclase two-component system in Anaplasma phagocytophilum, another obligate intracellular bacterium which belongs to the family Anaplasmataceae of the order Rickettsiales and causes granulocytic anaplasmosis in humans. A rather intriguing finding of this study is the inhibitory effect of a hydrophobic c-di-GMP derivative, namely 2′-O-di(tert-butyldimethylsilyl)-c-di-GMP, on the infection of HL60 promyelocytic cells by A. phagocytophilum, demonstrating the potential involvement of a c-di-GMP-receptor complex in the regulation of intracellular A. phagocytophilum infection [22]. Based upon the conservation of PleD as a single GGDEF domain containing protein among different genera in the family Anaplasmataceae, analysis of cyclic-di-GMP signaling in Ehrlichia chaffeensis further suggests vital roles in its proliferation, maturation, and release from host cells with particular emphasis on the dispersion from morula and intracellular movement of bacteria [23]. The present study demonstrates the presence and conservation of PleD among all major subgroups and species of Rickettsiaceae, including well-known pathogens in the spotted fever and typhus groups, which are unique among bacteria due to their propensity to free themselves in the cytoplasm, in contrast to Anaplasma and Ehrlichia, which remain confined to membrane-bound vacuolar compartments in the host cell.
PleD is a multidomain protein with two N-terminal receiver domains arranged in tandem and a C-terminal GGDEF domain, which is highly conserved in many bacterial species. Due to its association with a variety of domains, it is involved in the sensing of signals in the periplasm, membrane, or cytoplasm. Another remarkable feature of proteins carrying a GGDEF domain is their presence in most bacteria, but absence in archaea and eukaryotes. Thus, it is not surprising that rickettsial GGDEF is seemingly relatively conserved when compared to that of A. phagocytophilum and C. crescentus, a free-living unicellular bacterium that transitions between an adhesive, sessile form and a motile, planktonic stage as part of its life cycle [24]. Although a similar trend of conservation for N-terminal receiver domain 1 is also evident, domain 2 of rickettsial PleD seems to diverge from that of Anaplasma and Caulobacter. Whether or not variations in host–pathogen interplay and life-style features might explain the biological basis of these similarities and differences remains an open area of inquiry to be pursued in further detail.
PleD is known to be active in a dimeric form. For instance, dimerization of PleD is required for optimal diguanylate cyclase activity and for sequestering the active form to the pole during differentiation in Caulobacter [25]. Based on structural analysis, we have predicted dimerization of PleD protein present in both R. rickettsii strains. Additionally, three key interactions between residues E100-R263; N106-M236; R114-D249 have been identified between the receiver domain D1 and adaptor domain D2. These interactions are known to act like a pivot for the transition of the PleD protein from inactive to active form. The nonsynonymous mutation resulting in an amino-acid change at position 236 (isoleucine in strain Sheila Smith versus a methionine in Iowa) could influence the equilibrium of active–inactive forms, as the hydrophobicity index of isoleucine (4.5) is very high compared to methionine (1.9) [26]. This mutation at the key inter-domain interaction can lead to a stable active form of PleD and result in increased production of cyclic di-guanosine monophosphate (c-di-GMP) in the Sheila Smith strain. However, ongoing functional studies aimed at determining the activity of diguanylate cyclase from both virulent and avirulent Rickettsia strains will shed light on the role of this mutation on the dimerization, stability, and catalytic activity of the enzyme.
In conclusion, we report the presence of PleD with conserved domain architecture in all Rickettsia species. Sequencing of the pleD gene from R. rickettsii str. Sheila Smith revealed the absence of a 5 bp indel and established the presence of a full-length gene encoding for a functional protein. Interestingly, a nonsynonymous mutation resulting in an amino-acid change at position 236 was identified in the PleD protein encoded by R. rickettsii strs. Sheila Smith (virulent) and Iowa (avirulent), and we postulate that this mutation at a key residue is likely to play a vital role in the stability and activity of the dimeric protein. Additionally, the expression of pleD transcripts was significantly different between virulent and avirulent Rickettsia strains, and further studies are currently ongoing to define the role for this protein in virulence and immune activation.
4. Materials and Methods
4.1. Rickettsia Species, Strains, and Sequences
The gene and/or protein sequences used in this study were obtained from a web-based information system named Pathosystems Resource Integration Center (PATRIC) [27]. As a genomics-centric relational database and bioinformatics resource, PATRIC provides consistency in the integration and annotation of all genomic and related data for a number of bacterial pathogens, including different species and strains of all major organisms classified into ancestral, spotted fever, typhus, and transitional subgroups of rickettsiae.
4.2. Cell Culture
An immortalized line of human dermal microvascular endothelial cells (CDC/EU.HMEC-1) displaying all major morphologic, phenotypic, and functional features of microvascular endothelium was obtained from the Centers for Disease Control and Prevention (Atlanta, GA, USA). HMECs were maintained in continuous cultures using MCDB131 growth medium (Caisson’s Laboratories) supplemented with fetal bovine serum (10% v/v; Aleken Biologicals), epidermal growth factor (10 ng/mL, Thermo Fisher Scientific, Waltham, MA, USA), L-glutamine (10mM, Thermo Fisher Scientific, Waltham, MA, USA), and hydrocortisone (1 μg/mL, Sigma) [28,29]. All experiments were performed at the passage level of ≤35.
4.3. Infection
A highly virulent strain (Sheila Smith) and an avirulent strain (Iowa) of R. rickettsii were grown intracellularly in cultured Vero cells and purified by differential centrifugation, as described previously [30]. Rickettsial stocks were subjected to slow freezing and thawing and stored at −80 °C as aliquots of ≤500 µL to minimize repeated freeze–thaw cycles. Infectivity titers of purified stocks were estimated by citrate synthase (gltA)-based quantitative PCR and/or plaque-formation assay [30,31]. HMECs were infected with R. rickettsii at an MOI (multiplicity of infection) of approximately 20 intracellular rickettsiae per cell following our standard laboratory protocols [30,32]. The viability of both mock-infected (controls) and R. rickettsii-infected HMECs were monitored microscopically.
4.4. Phylogenetic Analysis
The phylogenetic tree and evolutionary analyses of the PleD sequences belonging to different Rickettsia species were performed using MEGA [33]. The PleD protein sequences were downloaded from the PATRIC database and aligned using Clustal Omega. The bootstrap consensus tree inferred from 1000 replicates was taken to represent the evolutionary history of the taxa analyzed [34]. The branches corresponding to partitions reproduced in less than 90% bootstrap replicates are collapsed. The evolutionary history was inferred using the Maximum Likelihood method based on the JTT matrix-based model [35]. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log-likelihood value. The analysis involved 43 amino-acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 446 positions in the final dataset. The accession numbers of PleD sequences from different Rickettsia species used in the analysis are provided in Table S1.
4.5. Protein-Structure Prediction and Modeling
The active and inactive-conformation homology models of PleD protein were constructed in MODELLER [21]. This platform builds protein models from query sequences using the solved crystal structures contained in the RCSB Protein Data Bank (PDB) as templates. The modeled structures were subjected to constrained energy minimization to allow the global energy minimization and structural analysis using the AMBER 12 suite and VADAR [36,37]. Assessment of the stereochemistry of 3D models was performed using a Ramachandran plot (PROCHECK) [38], and Protein Structure Analysis (ProSA) [39]. The active and inactive forms of PleD protein were analyzed using PyMol [40].
4.6. Cloning and Sequencing of Rickettsial pleD
Total genomic DNA from R. rickettsii strains Sheila Smith and Iowa was extracted from purified stocks using Qiagen Blood & Tissue kit (Qiagen, Germantown, MD, USA) following manufacturer’s instructions. Full-length pleD gene was amplified from the genomic DNA using Phusion high-fidelity taq polymerase (New England Biolabs, Ipswich, MA, USA) and using primers pleD-F (5′ CCTTTATCGCTGCCTATTTCTT 3′) and pleD-R (5′ GTGTTTTGCTGACAACCCATA 3′). The amplicon was verified by resolving on a 1.5% agarose gel and cloned into pGEM-T Easy vector (Promega, Madison, WI, USA) following manufacturer instructions. Plasmid was isolated from a positive clone carrying the pleD insert and was sequenced using T7 and SP6 primers at the institutional molecular genomics core facility. The resulting sequences were aligned with published sequences (NCBI Accession numbers NC_009882 (Sheila Smith) and NC_010263 (Iowa)) using Clustal Omega with default settings to identify indels and single-nucleotide polymorphisms.
4.7. Quantitative RT-PCR (RT-qPCR)
Total RNA was extracted from cells infected with R. rickettsii for different time durations and corresponding mock-infected controls was extracted using our optimized Tri Reagent ® protocol [30,41]. The RNA samples were subjected to DNase I (New England Biolabs, Ipswich, MA, USA) treatment to eliminate genomic DNA contamination, precipitated using 3M sodium acetate pH 5.5 and quantified using a MultiScan™ Go Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). The RNA samples were then enriched for bacterial coding transcripts using MicrobEnrich™ kit for removal of eukaryotic host RNA, and MicrobExpress™ kit for removal of bacterial rRNA. The enriched samples were then assessed for purity by qRT-PCR using human gapdh primers (GAPDH-qPCR-F: 5′ CCACTCCTCCACCTTTGAC 3′ and GAPDH-qPCR-R: 5′ ACCCTGTTGCTGTAGCCA 3′) and only samples which exhibited a CT >35 (indicating efficient removal of host transcriptome) were used for the study. Complementary DNA (cDNA) synthesis was carried out with random primers using High-Capacity cDNA Reverse Transcription kit (Applied Biosystems, Waltham, MA, USA) and absolute quantification of pleD expression was performed by real-time quantitative PCR. Briefly, a pleD amplicon was amplified using Rickettsia-specific primers pleD-qPCR-F (5′ TGTTATGATGCCGGAAATTGATGGA 3′) and pleD-qPCR-R (5′ AGCAGTATCGTTAATCGGCTTTGTT 3′), and cloned into a pGEM T-easy vector (Promega). The plasmid from a positive clone was then purified, linearized by restriction digestion, and used for generating the standard curve. SYBR green-assay-based absolute quantification of pleD expression was performed to test samples using the primers listed above.
Total DNA was extracted following our standard protocol and bacterial load in HMECs infected with R. rickettsii strains Sheila Smith and Iowa was estimated by citrate synthase (gltA)-based quantitative PCR assay, as described [30].
4.8. Statistical Analysis
A minimum of three independent biological replicates with two technical replicates were performed for each experiment. Statistical analysis was performed by unpaired t-test or Mann–Whitney test using GraphPad Prism software (GraphPad Software Inc., San Diego, CA, USA) and p value ≤ 0.05 was considered statistically significant.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms23073853/s1,
Author Contributions
Conceptualization, H.P.N. and S.K.S.; methodology, H.P.N., A.S., J.A. and K.M.S.; formal analysis, H.P.N., A.S., J.A., K.M.S. and S.K.S.; investigation, H.P.N. and S.K.S.; resources, H.P.N., A.S. and S.K.S.; data curation, H.P.N. and K.M.S.; writing—original draft preparation, H.P.N., K.M.S. and S.K.S.; writing—review and editing, H.P.N., K.M.S. and S.K.S.; visualization, H.P.N. and S.K.S.; supervision, H.P.N. and S.K.S.; project administration, H.P.N. and S.K.S.; funding acquisition, H.P.N. and S.K.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded, in part, by the National Institute of Allergy and Infectious Diseases at the National Institutes of Health, Bethesda, MD, USA, grant number “5R01 AI127899-04” and by the institutional funds from the Institute for Human Infections and Immunity at the University of Texas Medical Branch, Galveston, TX, USA.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The nucleotide sequence of full-length pleD gene from R. rickettsii strs. Sheila Smith and Iowa sequenced in this study are deposited in the NCBI GenBank and are available under accession numbers OM714535 (R. rickettsii str. Sheila Smith) and OM714536 (R. rickettsii str. Iowa).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sahni, S.K.; Narra, H.P.; Sahni, A.; Walker, D.H. Recent molecular insights into rickettsial pathogenesis and immunity. Future Microbiol. 2013, 8, 1265–1288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Narra, H.P.; Sahni, A.; Walker, D.H.; Sahni, S.K. Recent research milestones in the pathogenesis of human rickettsioses and opportunities ahead. Future Microbiol. 2020, 15, 753–765. [Google Scholar] [CrossRef] [PubMed]
- Sahni, A.; Fang, R.; Sahni, S.K.; Walker, D.H. Pathogenesis of Rickettsial Diseases: Pathogenic and Immune Mechanisms of an Endotheliotropic Infection. Annu. Rev. Pathol. Mech. Dis. 2019, 14, 127–152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walker, D.H. Changing Dynamics of Human–Rickettsial Interactions. Am. J. Trop. Med. Hyg. 2016, 94, 3–4. [Google Scholar] [CrossRef] [PubMed]
- Joshi, S.G.; Francis, C.W.; Silverman, D.J.; Sahni, S.K. Nuclear factor kappa B protects against host cell apoptosis during Rickettsia rickettsii infection by inhibiting activation of apical and effector caspases and maintaining mitochondrial integrity. Infect. Immun. 2003, 71, 4127–4136. [Google Scholar] [CrossRef] [Green Version]
- Clifton, D.R.; Rydkina, E.; Freeman, R.S.; Sahni, S.K. NF-kappaB activation during Rickettsia rickettsii infection of endothelial cells involves the activation of catalytic IkappaB kinases IKKalpha and IKKbeta and phosphorylation-proteolysis of the inhibitor protein IkappaBalpha. Infect. Immun. 2005, 73, 155–165. [Google Scholar] [CrossRef] [Green Version]
- Colonne, P.M.; Sahni, A.; Sahni, S.K. Rickettsia conorii infection stimulates the expression of ISG15 and ISG15 protease UBP43 in human microvascular endothelial cells. Biochem. Biophys. Res. Commun. 2011, 416, 153–158. [Google Scholar] [CrossRef] [Green Version]
- Colonne, P.M.; Eremeeva, M.E.; Sahni, S.K. Beta Interferon-Mediated Activation of Signal Transducer and Activator of Transcription Protein 1 Interferes with Rickettsia conorii Replication in Human Endothelial Cells. Infect. Immun. 2011, 79, 3733–3743. [Google Scholar] [CrossRef] [Green Version]
- Colonne, P.M.; Sahni, A.; Sahni, S.K. Suppressor of cytokine signalling protein SOCS1 and UBP43 regulate the expression of type I interferon-stimulated genes in human microvascular endothelial cells infected with Rickettsia conorii. J. Med. Microbiol. 2013, 62, 968–979. [Google Scholar] [CrossRef]
- Gomelsky, M. cAMP, c-di-GMP, c-di-AMP and now cGMP: Bacteria use them all! Mol. Microbiol. 2011, 79, 562–565. [Google Scholar] [CrossRef] [Green Version]
- Yoon, S.H.; Waters, C.M. The ever-expanding world of bacterial cyclic oligonucleotide second messengers. Curr. Opin. Microbiol. 2021, 60, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Jenal, U.; Malone, J. Mechanisms of Cyclic-di-GMP Signaling in Bacteria. Annu. Rev. Genet. 2006, 40, 385–407. [Google Scholar] [CrossRef] [PubMed]
- Krol, E.; Schäper, S.; Becker, A. Cyclic di-GMP signaling controlling the free-living lifestyle of alpha-proteobacterial rhizobia. Biol. Chem. 2020, 401, 1335–1348. [Google Scholar] [CrossRef]
- Römling, U.; Galperin, M.Y.; Gomelsky, M. Cyclic di-GMP: The First 25 Years of a Universal Bacterial Second Messenger. Microbiol. Mol. Biol. Rev. 2013, 77, 1–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schirmer, T.; Jenal, U. Structural and mechanistic determinants of c-di-GMP signalling. Nat. Rev. Genet. 2009, 7, 724–735. [Google Scholar] [CrossRef] [PubMed]
- Mantoni, F.; Rossi, C.S.; Paiardini, A.; Di Matteo, A.; Cappellacci, L.; Petrelli, R.; Ricciutelli, M.; Paone, A.; Cutruzzolà, F.; Giardina, G.; et al. Studying GGDEF Domain in the Act: Minimize Conformational Frustration to Prevent Artefacts. Life 2021, 11, 31. [Google Scholar] [CrossRef]
- McWhirter, S.M.; Barbalat, R.; Monroe, K.M.; Fontana, M.F.; Hyodo, M.; Joncker, N.T.; Ishii, K.; Akira, S.; Colonna, M.; Chen, Z.; et al. A host type I interferon response is induced by cytosolic sensing of the bacterial second messenger cyclic-di-GMP. J. Exp. Med. 2009, 206, 1899–1911. [Google Scholar] [CrossRef]
- Christen, B.; Christen, M.; Paul, R.; Schmid, F.; Folcher, M.; Jenoe, P.; Meuwly, M.; Jenal, U. Allosteric control of cyclic di-GMP signaling. J. Biol. Chem. 2006, 281, 32015–32024. [Google Scholar] [CrossRef]
- Curto, P.; Simões, I.; Riley, S.P.; Martinez, J.J. Differences in Intracellular Fate of Two Spotted Fever Group Rickettsia in Macrophage-Like Cells. Front. Cell. Infect. Microbiol. 2016, 6, 80. [Google Scholar] [CrossRef] [Green Version]
- Clark, T.R.; Noriea, N.F.; Bublitz, D.C.; Ellison, D.W.; Martens, C.; Lutter, E.; Hackstadt, T. Comparative Genome Sequencing of Rickettsia rickettsii Strains That Differ in Virulence. Infect. Immun. 2015, 83, 1568–1576. [Google Scholar] [CrossRef] [Green Version]
- Webb, B.; Sali, A. Comparative Protein Structure Modeling Using MODELLER. Curr. Protoc. Protein Sci. 2016, 86, 5.6.1–5.6.27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, T.-H.; Kumagai, Y.; Hyodo, M.; Hayakawa, Y.; Rikihisa, Y. The Anaplasma phagocytophilum PleC Histidine Kinase and PleD Diguanylate Cyclase Two-Component System and Role of Cyclic Di-GMP in Host Cell Infection. J. Bacteriol. 2009, 191, 693–700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumagai, Y.; Matsuo, J.; Hayakawa, Y.; Rikihisa, Y. Cyclic di-GMP Signaling Regulates Invasion by Ehrlichia chaffeensis of Human Monocytes. J. Bacteriol. 2010, 192, 4122–4133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paul, R.; Weiser, S.; Amiot, N.C.; Chan, C.; Schirmer, T.; Giese, B.; Jenal, U. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 2004, 18, 715–727. [Google Scholar] [CrossRef] [Green Version]
- Paul, R.; Abel, S.; Wassmann, P.; Beck, A.; Heerklotz, H.; Jenal, U. Activation of the Diguanylate Cyclase PleD by Phosphorylation-mediated Dimerization. J. Biol. Chem. 2007, 282, 29170–29177. [Google Scholar] [CrossRef] [Green Version]
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef] [Green Version]
- Davis, J.J.; Wattam, A.R.; Aziz, R.K.; Brettin, T.; Butler, R.; Butler, R.M.; Chlenski, P.; Conrad, N.; Dickerman, A.; Dietrich, E.M.; et al. The PATRIC Bioinformatics Resource Center: Expanding data and analysis capabilities. Nucleic Acids Res. 2020, 48, D606–D612. [Google Scholar] [CrossRef] [Green Version]
- Sahni, A.; Patel, J.; Narra, H.P.; Schroeder, C.L.C.; Walker, D.H.; Sahni, S.K. Fibroblast growth factor receptor-1 mediates internalization of pathogenic spotted fever rickettsiae into host endothelium. PLoS ONE 2017, 12, e0183181. [Google Scholar] [CrossRef]
- Narra, H.P.; Sahni, A.; Alsing, J.; Schroeder, C.L.C.; Golovko, G.; Nia, A.M.; Fofanov, Y.; Khanipov, K.; Sahni, S.K. Comparative transcriptomic analysis of Rickettsia conorii during in vitro infection of human and tick host cells. BMC Genom. 2020, 21, 1–21. [Google Scholar] [CrossRef]
- Narra, H.P.; Schroeder, C.L.C.; Sahni, A.; Rojas, M.; Khanipov, K.; Fofanov, Y.; Sahni, S.K. Small Regulatory RNAs of Rickettsia conorii. Sci. Rep. 2016, 6, 36728. [Google Scholar] [CrossRef] [Green Version]
- Alhassan, A.; Liu, H.; McGill, J.; Cerezo, A.; Jakkula, L.U.M.R.; Nair, A.D.S.; Winkley, E.; Olson, S.; Marlow, D.; Sahni, A.; et al. Rickettsia rickettsii Whole-Cell Antigens Offer Protection against Rocky Mountain Spotted Fever in the Canine Host. Infect. Immun. 2019, 87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, J.G.; Narra, H.P.; Sepuru, K.M.; Sahni, A.; Golla, S.R.; Sahni, A.; Singh, A.; Schroeder, C.L.; Chowdhury, I.H.; Popov, V.L.; et al. Evolution, purification, and characterization of RC0497: A peptidoglycan amidase from the prototypical spotted fever species Rickettsia conorii. Biol. Chem. 2020, 401, 249–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992, 8, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Willard, L.; Ranjan, A.; Zhang, H.; Monzavi, H.; Boyko, R.F.; Sykes, B.D.; Wishart, D.S. VADAR: A web server for quantitative evaluation of protein structure quality. Nucleic Acids Res. 2003, 31, 3316–3319. [Google Scholar] [CrossRef]
- Case, D.A.; Darden, T.A.; Cheatham, T.E.; Simmerling, C.L.; Wang, J.; Duke, R.E.; Luo, R.; Walker, R.C.; Zhang, W.; Merz, K.M. Amber 12; University of California: San Francisco, CA, USA, 2012. [Google Scholar]
- Lovell, S.C.; Davis, I.W.; Arendall, W.B., 3rd; de Bakker, P.I.; Word, J.M.; Prisant, M.G.; Richardson, J.S.; Richardson, D.C. Structure validation by Calpha geometry: Phi,psi and Cbeta deviation. Proteins 2003, 50, 437–450. [Google Scholar] [CrossRef]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef] [Green Version]
- Schrödinger, L.L.C. The Pymol Molecular Graphics System; Version 2.5.2.; Schrödinger, LLC.: New York, NY, USA, 2021. [Google Scholar]
- Narra, H.P.; Sahni, A.; Khanipov, K.; Fofanov, Y.; Sahni, S.K. Global Transcriptomic Profiling of Pulmonary Gene Expression in an Experimental Murine Model of Rickettsia conorii Infection. Genes 2019, 10, 204. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).