Identifying Genetic Biomarkers Predicting Response to Anti-Vascular Endothelial Growth Factor Injections in Diabetic Macular Edema
Abstract
1. Introduction
2. Results
2.1. Cohort Characteristics
2.2. Association Analysis
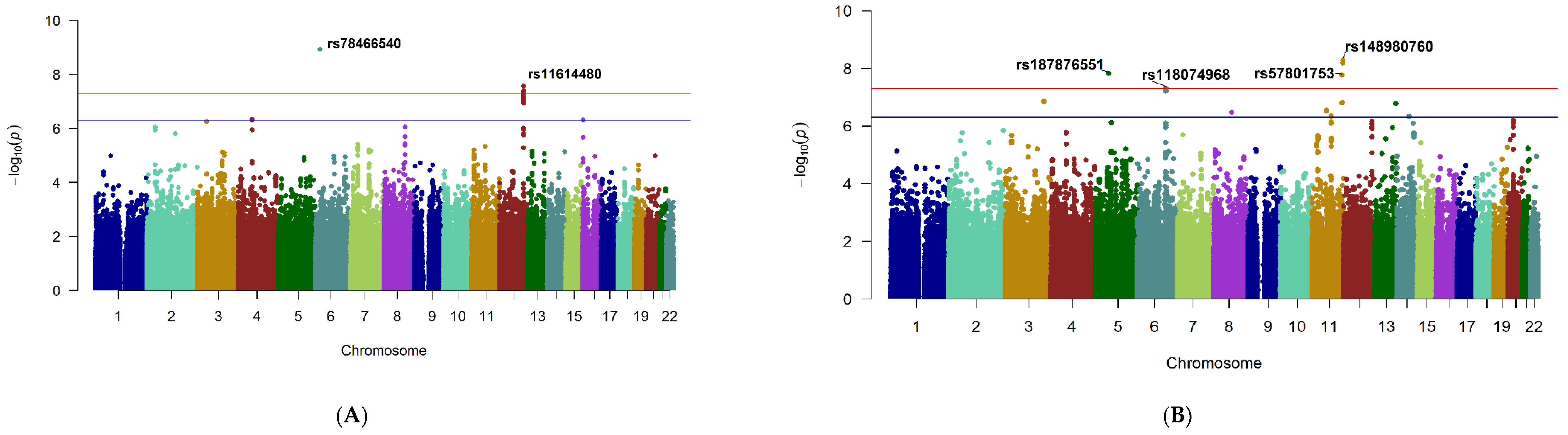
2.2.1. Genome-Wide Significant Loci
2.2.2. Suggestive Loci
2.2.3. Conditional Analysis
2.2.4. Logistic Regression Model
3. Functional Annotation
Genome-Wide Significant Loci
4. Ocular Tissue Database
5. Discussion
6. Conclusions
7. Materials and Methods
7.1. Study Design and Participants
7.2. Phenotyping
7.3. Outcome Measures
7.4. Genotyping and Quality Control
7.5. Imputation
7.6. GWAS Statistical Analysis
7.7. Demographic Data Statistical Analysis
7.8. Functional Annotation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lee, R.; Wong, T.Y.; Sabanayagam, C. Epidemiology of diabetic retinopathy, diabetic macular edema and related vision loss. Eye Vis. 2015, 2, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Bhatwadekar, A.; Shughoury, A.; Belamkar, A.; Ciulla, T. Genetics of Diabetic Retinopathy, a Leading Cause of Irreversible Blindness in the Industrialized World. Genes 2021, 12, 1200. [Google Scholar] [CrossRef] [PubMed]
- Arar, N.H.; Freedman, B.I.; Adler, S.G.; Iyengar, S.; Chew, E.Y.; Davis, M.D.; Satko, S.G.; Bowden, D.W.; Duggirala, R.; Elston, R.C.; et al. Heritability of the Severity of Diabetic Retinopathy: The FIND-Eye Study. Investig. Opthalmology Vis. Sci. 2008, 49, 3839–3845. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zeng, H.; Bao, S.; Wang, N.; Gillies, M.C. Diabetic macular edema: New concepts in patho-physiology and treatment. Cell Biosci. 2014, 4, 27. [Google Scholar] [CrossRef] [PubMed]
- Kent, D.; Vinores, S.A.; Campochiaro, P.A. Macular oedema: The role of soluble mediators. Br. J. Ophthalmol. 2000, 84, 542–545. [Google Scholar] [CrossRef][Green Version]
- Kim, L.; D’Amore, P.A. A Brief History of Anti-VEGF for the Treatment of Ocular Angiogenesis. Am. J. Pathol. 2012, 181, 376–379. [Google Scholar] [CrossRef]
- Ciulla, T.A.; Pollack, J.S.; Williams, D.F. Visual acuity outcomes and anti-VEGF therapy intensity in diabetic macular oedema: A real-world analysis of 28 658 patient eyes. Br. J. Ophthalmol. 2020, 105, 216–221. [Google Scholar] [CrossRef]
- Cai, S.; Bressler, N.M. Aflibercept, bevacizumab or ranibizumab for diabetic macular oedema: Recent clinically relevant findings from DRCR. net Protocol T. Curr. Opin. Ophthalmol. 2017, 28, 636–643. [Google Scholar] [CrossRef]
- Boyer, D.S.; Nguyen, Q.D.; Brown, D.M.; Basu, K.; Ehrlich, J.S.; Ride and Rise Research Group. Outcomes with as-needed ranibizumab after initial monthly therapy: Long-term outcomes of the phase III RIDE and RISE trials. Ophthalmology 2015, 122, 2504–2513.e1. [Google Scholar] [CrossRef]
- Brown, D.M.; Nguyen, Q.D.; Marcus, D.M.; Boyer, D.S.; Patel, S.; Feiner, L.; RIDE and RISE Research Group. Long-term outcomes of ranibizumab therapy for diabetic macular edema: The 36-month results from two phase III trials: RISE and RIDE. Ophthalmology 2013, 120, 2013–2022. [Google Scholar] [CrossRef]
- Pelosini, L.; Hull, C.C.; Boyce, J.F.; McHugh, D.; Stanford, M.R.; Marshall, J. Optical Coherence Tomography May Be Used to Predict Visual Acuity in Patients with Macular Edema. Investig. Opthalmology Vis. Sci. 2011, 52, 2741–2748. [Google Scholar] [CrossRef] [PubMed]
- Bressler, S.B.; Odia, I.; Maguire, M.G.; Dhoot, D.S.; Glassman, A.R.; Jampol, L.M.; Diabetic Retinopathy Clinical Research Network. Factors Associated with Visual Acuity and Central Subfield Thickness Changes When Treating Diabetic Macular Edema With Anti–Vascular Endothelial Growth Factor Therapy: An Exploratory Analysis of the Protocol T Randomized Clinical Trial. JAMA Ophthalmol. 2019, 137, 382–389. [Google Scholar] [CrossRef] [PubMed]
- Sophie, R.; Lu, N.; Campochiaro, P.A. Predictors of Functional and Anatomic Outcomes in Patients with Diabetic Macular Edema Treated with Ranibizumab. Ophthalmology 2015, 122, 1395–1401. [Google Scholar] [CrossRef]
- Toraih, E.A.; Abd Elghany, A.A.; Abd El Fadeal, N.M.; Al Ageeli, E.; Fawzy, M.S. Deciphering the role of circulating lncRNAs: RNCR2, NEAT2, CDKN2B-AS1, and PVT1 and the possible prediction of anti-VEGF treatment outcomes in diabetic retinopathy patients. Graefes Arch. Clin. Exp. Ophthalmol. 2019, 257, 1897–1913. [Google Scholar] [CrossRef]
- Tetikoğlu, M.; Yüksel, Z.; Aktas, S.; Sağdik, H.M.; Özcura, F. VEGF-A gene polymorphisms and responses to intravitreal ranibizumab treatment in patients with diabetic macular edema. Int. Ophthalmol. 2017, 38, 2381–2388. [Google Scholar] [CrossRef] [PubMed]
- El-Shazly, S.F.; El-Bradey, M.H.; Tameesh, M.K. Vascular endothelial growth factor gene polymorphism prevalence in patients with diabetic macular oedema and its correlation with anti-vascular endothelial growth factor treatment outcomes. Clin. Exp. Ophthalmol. 2013, 42, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Dabir, S.; Das, D.; Nallathambi, J.; Mangalesh, S.; Yadav, N.; Schouten, J.A.G. Differential systemic gene expression profile in patients with diabetic macular edema: Responders versus nonresponders to standard treatment. Indian J. Ophthalmol. 2014, 62, 66–73. [Google Scholar] [CrossRef]
- Abdelghany, A.A.; Toraih, E.A.; Mohamed, A.A.; Lashine, R.M.; Saleh, M.H.; Nafie, M.S.; Fawzy, M.S. Association of VEGF gene family variants with central macular thickness and visual acuity after Aflibercept short-term treatment in diabetic patients: A pilot study. Ophthalmic Res. 2020, 64, 261–272. [Google Scholar] [CrossRef]
- Mohammad, H.M.; Abdelghany, A.A.; Al Ageeli, E.; Kattan, S.W.; Hassan, R.; A Toraih, E.; Fawzy, M.S.; Mokhtar, N. Long Non-Coding RNAs Gene Variants as Molecular Markers for Diabetic Retinopathy Risk and Response to Anti-VEGF Therapy. Pharmacogenomics Pers. Med. 2021, 4, 997–1014. [Google Scholar] [CrossRef]
- Sun, J.; Xiong, Y.; Jiang, K.; Xin, B.; Jiang, T.; Wei, R.; Zou, K.; Tan, H.; Jiang, T.; Yang, A.; et al. Hypoxia-sensitive long noncoding RNA CASC15 promotes lung tumorigenesis by regulating the SOX4/β-catenin axis. J. Exp. Clin. Cancer Res. 2021, 40, 12. [Google Scholar] [CrossRef]
- Zhu, Z.; Zhang, F.; Hu, H.; Bakshi, A.; Robinson, M.R.; Powell, J.E.; Montgomery, G.W.; Goddard, M.E.; Wray, N.R.; Visscher, P.M.; et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 2016, 48, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Gusev, A.; Ko, A.; Shi, H.; Bhatia, G.; Chung, W.; Penninx, B.W.J.H.; Jansen, R.; de Geus, E.J.C.; Boomsma, D.I.; Wright, F.A.; et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 2016, 48, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Tweedie, S.; Braschi, B.; Gray, K.; Jones, T.E.M.; Seal, R.L.; Yates, B.; Bruford, E.A. Genenames.org: The HGNC and VGNC resources in 2021. Nucleic Acids Res. 2021, 49, D939–D946. [Google Scholar] [PubMed]
- Hormozdiari, F.; van de Bunt, M.; Segrè, A.V.; Li, X.; Joo, J.W.J.; Bilow, M.; Sul, J.H.; Sankararaman, S.; Pasaniuc, B.; Eskin, E. Colocalization of GWAS and eQTL Signals Detects Target Genes. Am. J. Hum. Genet. 2016, 99, 1245–1260. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Wang, K.-S.; Aragam, N. NTM and NR3C2 polymorphisms influencing intelligence: Family-based association studies. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2011, 35, 154–160. [Google Scholar] [CrossRef]
- Romero-Aroca, P.; Baget-Bernaldiz, M.; Pareja-Rios, A.; Lopez-Galvez, M.; Navarro-Gil, R.; Verges, R. Diabetic Macular Edema Pathophysiology: Vasogenic versus Inflammatory. J. Diabetes Res. 2016, 2016, 2156273. [Google Scholar] [CrossRef]
- Kerr, N.M.; Johnson, C.S.; de Souza, C.F.; Chee, K.-S.; Good, W.R.; Green, C.R.; Danesh-Meyer, H.V. Immunolocalization of Gap Junction Protein Connexin43 (GJA1) in the Human Retina and Optic Nerve. Investig. Opthalmology Vis. Sci. 2010, 51, 4028–4034. [Google Scholar] [CrossRef]
- Chen, Y.; Huang, H.; Zhou, J.; Doumatey, A.; Lashley, K.; Chen, G.; Agyenim-Boateng, K.; Eghan, B.A.; Acheampong, J.; Fasanmade, O.; et al. Polymorphism of the endothelial nitric oxide synthase gene is associated with diabetic retinopathy in a cohort of West Africans. Mol. Vis. 2007, 13, 2142–2147. [Google Scholar]
- Altman, D.G.; Royston, P. The cost of dichotomising continuous variables. BMJ 2006, 332, 1080. [Google Scholar] [CrossRef]
- Naggara, O.; Raymond, J.; Guilbert, F.; Roy, D.; Weill, A.; Altman, D. Analysis by Categorizing or Dichotomizing Continuous Variables Is Inadvisable: An Example from the Natural History of Unruptured Aneurysms. Am. J. Neuroradiol. 2011, 32, 437–440. [Google Scholar] [CrossRef]
- Michaelides, M.; Kaines, A.; Hamilton, R.D.; Fraser-Bell, S.; Rajendram, R.; Quhill, F.; Boos, C.J.; Xing, W.; Egan, C.; Peto, T.; et al. A Prospective Randomized Trial of Intravitreal Bevacizumab or Laser Therapy in the Management of Diabetic Macular Edema (BOLT Study): 12-Month Data: Report. Ophthalmology 2010, 117, 1078–1086.e2. [Google Scholar] [CrossRef]
- Massin, P.; Bandello, F.; Garweg, J.G.; Hansen, L.L.; Harding, S.P.; Larsen, M. Safety and efficacy of ranibizumab in diabetic macular edema (RESOLVE Study): A 12-month, randomized, controlled, double-masked, multicenter phase II study. Diabetes Care 2010, 33, 2399–2405. [Google Scholar] [CrossRef] [PubMed]
- Korobelnik, J.-F.; Daien, V.; Faure, C.; Tadayoni, R.; Giocanti-Auregan, A.; Dot, C.; Kodjikian, L.; Massin, P. Real-world outcomes following 12 months of intravitreal aflibercept monotherapy in patients with diabetic macular edema in France: Results from the APOLLON study. Graefe’s Arch. Clin. Exp. Ophthalmol. 2020, 258, 521–528. [Google Scholar] [CrossRef]
- Bhandari, S.; Nguyen, V.; Fraser-Bell, S.; Mehta, H.; Viola, F.; Baudin, F.; Gabrielle, P.-H.; Creuzot-Garcher, C.; Gillies, M.; Barthelmes,, D. Ranibizumab or Aflibercept for Diabetic Macular Edema: Comparison of One-Year Outcomes from the Fight Retinal Blindness! Registry. Ophthalmology 2019, 127, 608–615. [Google Scholar] [CrossRef] [PubMed]
- Banaee, T.; Ashraf, M.; Conti, F.F.; Singh, R.P. Switching Anti-VEGF Drugs in the Treatment of Diabetic Macular Edema. Ophthalmic Surg. Lasers Imaging Retin. 2017, 48, 748–754. [Google Scholar] [CrossRef] [PubMed]
- Hussain, R.M.; Ciulla, T. Treatment strategies for refractory diabetic macular edema: Switching anti-VEGF treatments, adopting corticosteroid-based treatments, and combination therapy. Expert Opin. Biol. Ther. 2016, 16, 365–374. [Google Scholar] [CrossRef]
- Gregori, N.Z.; Feuer, W.; Rosenfeld, P.J. Novel method for analyzing snellen visual acuity measurements. Retina 2010, 30, 1046–1050. [Google Scholar] [CrossRef]
- Elman, M.J.; Aiello, L.P.; Beck, R.W.; Bressler, N.M.; Bressler, S.B.; Edwards, A.R.; Ferris, F.L., 3rd; Friedman, S.M.; Glassman, A.R.; Miller, K.M.; et al. Randomized Trial Evaluating Ranibizumab Plus Prompt or Deferred Laser or Triamcinolone Plus Prompt Laser for Diabetic Macular Edema. Ophthalmology 2010, 117, 1064–1077.e35. [Google Scholar] [CrossRef]
- Boyer, D.S.; Hopkins, J.J.; Sorof, J.; Ehrlich, J.S. Anti-vascular endothelial growth factor therapy for diabetic macular edema. Ther. Adv. Endocrinol. Metab. 2013, 4, 151–169. [Google Scholar] [CrossRef]
- Holekamp, N.M.; Campbell, J.; Almony, A.; Ingraham, H.; Marks, S.; Chandwani, H.; Cole, A.L.; Kiss, S. Vision Outcomes Following Anti–Vascular Endothelial Growth Factor Treatment of Diabetic Macular Edema in Clinical Practice. Am. J. Ophthalmol. 2018, 191, 83–91. [Google Scholar] [CrossRef]
- Ashraf, M.; Souka, A.; Adelman, R. Predicting outcomes to anti-vascular endothelial growth factor (VEGF) therapy in diabetic macular oedema: A review of the literature. Br. J. Ophthalmol. 2016, 100, 1596–1604. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; He, J.; Zhao, S.; Wu, H.; Zhong, X.; Sheng, Q.; Samuels, D.; Shyr, Y.; Long, J. Illumina human exome genotyping array clustering and quality control. Nat. Protoc. 2014, 9, 2643–2662. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. GigaScience 2015, 4, 7. [Google Scholar] [CrossRef] [PubMed]
- Gogarten, S.M.; Bhangale, T.; Conomos, M.P.; Laurie, C.A.; McHugh, C.P.; Painter, I.; Zheng, X.; Crosslin, D.R.; Levine, D.; Lumley, T.; et al. GWASTools: An R/Bioconductor package for quality control and analysis of genome-wide association studies. Bioinformatics 2012, 28, 3329–3331. [Google Scholar] [CrossRef] [PubMed]
- Turner, S.D. qqman: An R package for visualizing GWAS results using Q-Q and manhattan plots. Biorxiv 2014. Biorxiv:005165. [Google Scholar]
- Watanabe, K.; Taskesen, E.; van Bochoven, A.; Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 2017, 8, 1–11. [Google Scholar] [CrossRef]
- Kircher, M.; Witten, D.M.; Jain, P.; O’roak, B.J.; Cooper, G.M.; Shendure, J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014, 46, 310–315. [Google Scholar] [CrossRef]
- Boyle, A.P.; Hong, E.L.; Hariharan, M.; Cheng, Y.; Schaub, M.A.; Kasowski, M.; Karczewski, K.J.; Park, J.; Hitz, B.C.; Weng, S.; et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012, 22, 1790–1797. [Google Scholar] [CrossRef]
- Ratnapriya, R.; Sosina, O.A.; Starostik, M.R.; Kwicklis, M.; Kapphahn, R.J.; Fritsche, L.G. Retinal transcriptome and eQTL analyses identify genes associated with age-related macular degeneration. Nat. Genet. 2019, 51, 606–610. [Google Scholar] [CrossRef]
- Wagner, A.; Anand, V.N.; Wang, W.-H.; Chatterton, J.E.; Sun, D.; Shepard, A.R.; Jacobson, N.; Pang, I.-H.; DeLuca, A.; Casavant, T.L.; et al. Exon-level expression profiling of ocular tissues. Exp. Eye Res. 2013, 111, 105–111. [Google Scholar] [CrossRef][Green Version]
| Variable | N = 220 |
|---|---|
| Patient related | Parameter |
| Age (years) | 66.35 (12.16) |
| Gender:Male, N (%) | 151 (68.6) |
| BMI (kg/m2) | 33.76 (8.00) |
| DM: T2, N (%) | 181 (82.3) |
| DM duration (years) | 22.74 (10.15) |
| HbA1c (mg/dl) | 8.38 (1.63) |
| HTN: Yes, N (%) | 188 (85.5) |
| Nephropathy: Yes, N (%) | 120 (54.5) |
| Hyperlipidemia: Yes, N (%) | 198 (90) |
| Smoker: Yes, N (%) | 111 (50.5) |
| Eye related | |
| Baseline BCVA (ETDRS letters) | 64.25 (14.52) |
| Final BCVA (ETDRS letters) | 67.66 (15.54) |
| Change in BCVA (ETDRS letters) | 3.41 (12.05) * |
| Baseline CMT (microns) | 380.57 (104.08) |
| Final CMT (microns) | 325.40 (75.38) |
| Change in CMT (microns) | −55.17 (99.16) * |
| Laterality: RE, N (%) | 113 (51.4) |
| Lens status: Phakic, N (%) | 141 (64.1) |
| DR duration (years) | 7.95 (4.30) |
| DR severity N (%) | |
| Mild DR | 45 (20.5) |
| Moderate DR | 67 (30.5) |
| Severe DR | 35 (15.9) |
| PDR | 72 (32.7) |
| Injection number | 8.02 (3.07) |
| Injection type: N (%) | |
| Bevacizumab | 126 (57.3) |
| Aflibercept | 25 (11.4) |
| Ranibizumab | 37 (16.8) |
| Mixed | 32 (14.5) |
| Locus * | Chr | Position † | Lead SNP | Ref | Alt | AAF | Beta ‡ | p Value |
|---|---|---|---|---|---|---|---|---|
| Change in CMT (microns) | ||||||||
| CASC15 | 6 | 21755718 | rs78466540 | A | G | 0.03 | 115.80 | 1.16 × 10−09 |
| RP11-116D17.1 | 12 | 115772072 | rs11614480 | T | C | 0.07 | 71.08 | 2.69 × 10−08 |
| 12 | 115772088 | rs11615848 | G | T | 0.07 | 71.08 | 2.69 × 10−08 | |
| 12 | 115772313 | rs11614887 | T | C | 0.07 | 71.02 | 2.73 × 10−08 | |
| 12 | 115772214 | rs11615870 | G | T | 0.07 | 71.02 | 2.73 × 10−08 | |
| 12 | 115772032 | rs11615833 | G | A | 0.07 | 70.84 | 4.04 × 10−08 | |
| Change in BCVA (ETDRS letters) | ||||||||
| NTM | 11 | 132228056 | rs148980760 | A | C | 0.03 | −17.98 | 5.30 × 10−09 |
| 11 | 132237087 | rs117744949 | G | A | 0.03 | −17.50 | 6.57 × 10−09 | |
| PGAM1P1 | 5 | 57535905 | rs187876551 | G | A | 0.02 | −21.80 | 1.52 × 10−08 |
| RP11-744N12.3 | 11 | 128524088 | rs57801753 | T | C | 0.02 | −20.19 | 1.71 × 10−08 |
| TBC1D32 | 6 | 121280029 | rs118074968 | A | G | 0.03 | −16.40 | 4.94 × 10−08 |
| Locus * | Chr | Position † | Lead SNP | CADD Score | RegulomeDB Score (v1.1) | eQTL (GTEx v8) | Tissue | eGENE ‡ | eQTL (EyeGEx) | eGENE ║ |
|---|---|---|---|---|---|---|---|---|---|---|
| Change in CMT (microns) | ||||||||||
| RP11-116D17.1 | 12 | 115772072 | rs11614480 | 18.42 | 5 | TRUE | Multiple tissues # | RP11-116D17.3 | FALSE | NA |
| CASC15 | 6 | 21755718 | rs78466540 | 1.37 | 7 | FALSE | NA | NA | FALSE | NA |
| Change in BCVA (ETDRS letters) | ||||||||||
| NTM | 11 | 132228056 | rs148980760 | 2.39 | 4 | FALSE | NA | NA | FALSE | NA |
| RP11-744N12.3 | 11 | 128524088 | rs57801753 | 3.91 | 4 | FALSE | NA | NA | FALSE | NA |
| PGAM1P1 | 5 | 57535905 | rs187876551 | 0.40 | 4 | FALSE | NA | NA | FALSE | NA |
| TBC1D32 | 6 | 121280029 | rs118074968 | 7.59 | 6 | TRUE | Skin-Not Sun Exposed | GJA1 | FALSE | NA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gurung, R.L.; FitzGerald, L.M.; Liu, E.; McComish, B.J.; Kaidonis, G.; Ridge, B.; Hewitt, A.W.; Vote, B.J.; Verma, N.; Craig, J.E.; et al. Identifying Genetic Biomarkers Predicting Response to Anti-Vascular Endothelial Growth Factor Injections in Diabetic Macular Edema. Int. J. Mol. Sci. 2022, 23, 4042. https://doi.org/10.3390/ijms23074042
Gurung RL, FitzGerald LM, Liu E, McComish BJ, Kaidonis G, Ridge B, Hewitt AW, Vote BJ, Verma N, Craig JE, et al. Identifying Genetic Biomarkers Predicting Response to Anti-Vascular Endothelial Growth Factor Injections in Diabetic Macular Edema. International Journal of Molecular Sciences. 2022; 23(7):4042. https://doi.org/10.3390/ijms23074042
Chicago/Turabian StyleGurung, Rajya L., Liesel M. FitzGerald, Ebony Liu, Bennet J. McComish, Georgia Kaidonis, Bronwyn Ridge, Alex W. Hewitt, Brendan J. Vote, Nitin Verma, Jamie E. Craig, and et al. 2022. "Identifying Genetic Biomarkers Predicting Response to Anti-Vascular Endothelial Growth Factor Injections in Diabetic Macular Edema" International Journal of Molecular Sciences 23, no. 7: 4042. https://doi.org/10.3390/ijms23074042
APA StyleGurung, R. L., FitzGerald, L. M., Liu, E., McComish, B. J., Kaidonis, G., Ridge, B., Hewitt, A. W., Vote, B. J., Verma, N., Craig, J. E., & Burdon, K. P. (2022). Identifying Genetic Biomarkers Predicting Response to Anti-Vascular Endothelial Growth Factor Injections in Diabetic Macular Edema. International Journal of Molecular Sciences, 23(7), 4042. https://doi.org/10.3390/ijms23074042






