Multifunctional Role of Lipids in Modulating the Tumorigenic Properties of 4T1 Breast Cancer Cells
Abstract
1. Introduction
2. Results
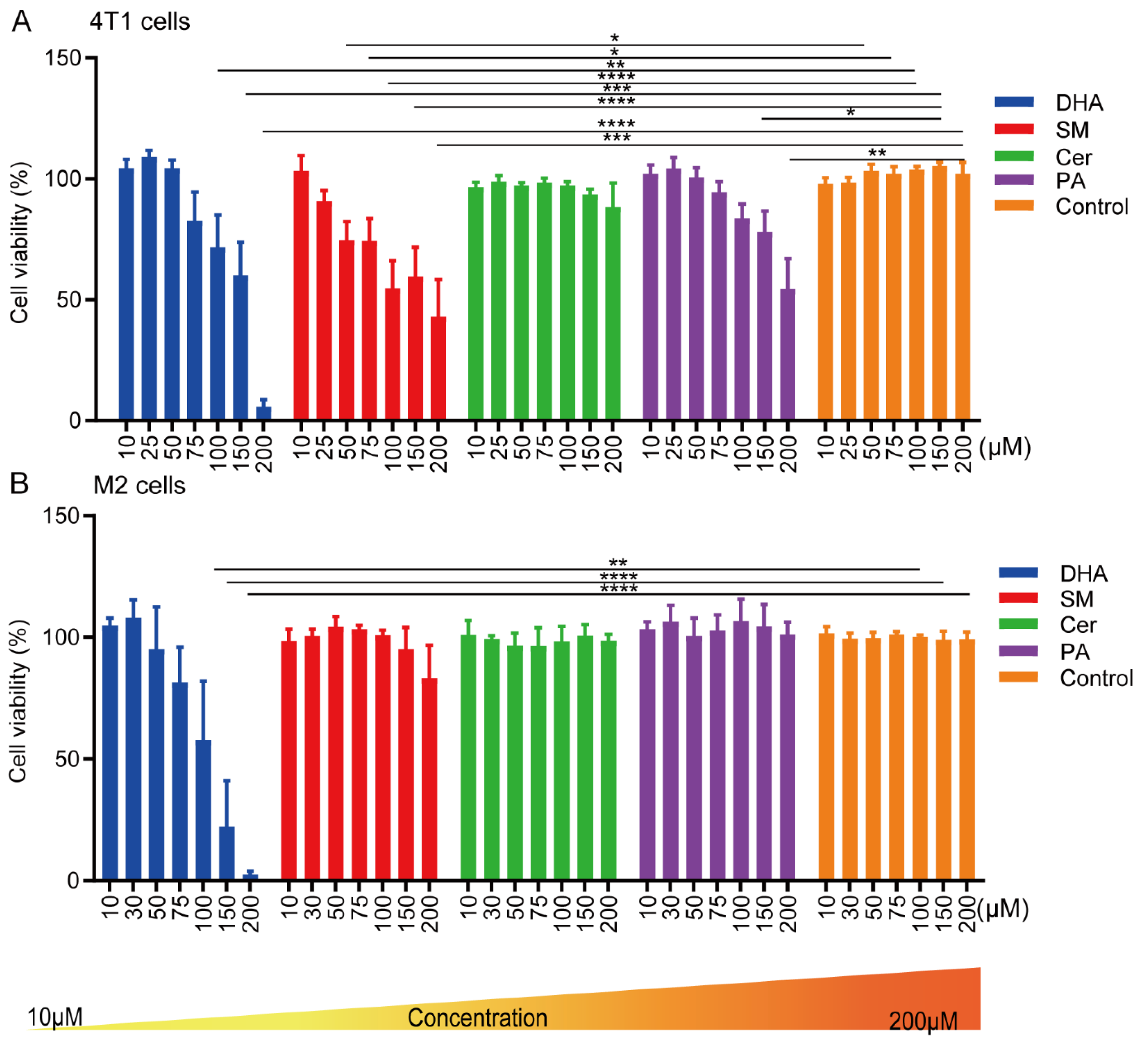
2.1. Effect of Cer, DHA, SM and PA on the Cellular Activity of 4T1 Breast Cancer Cells and Macrophages
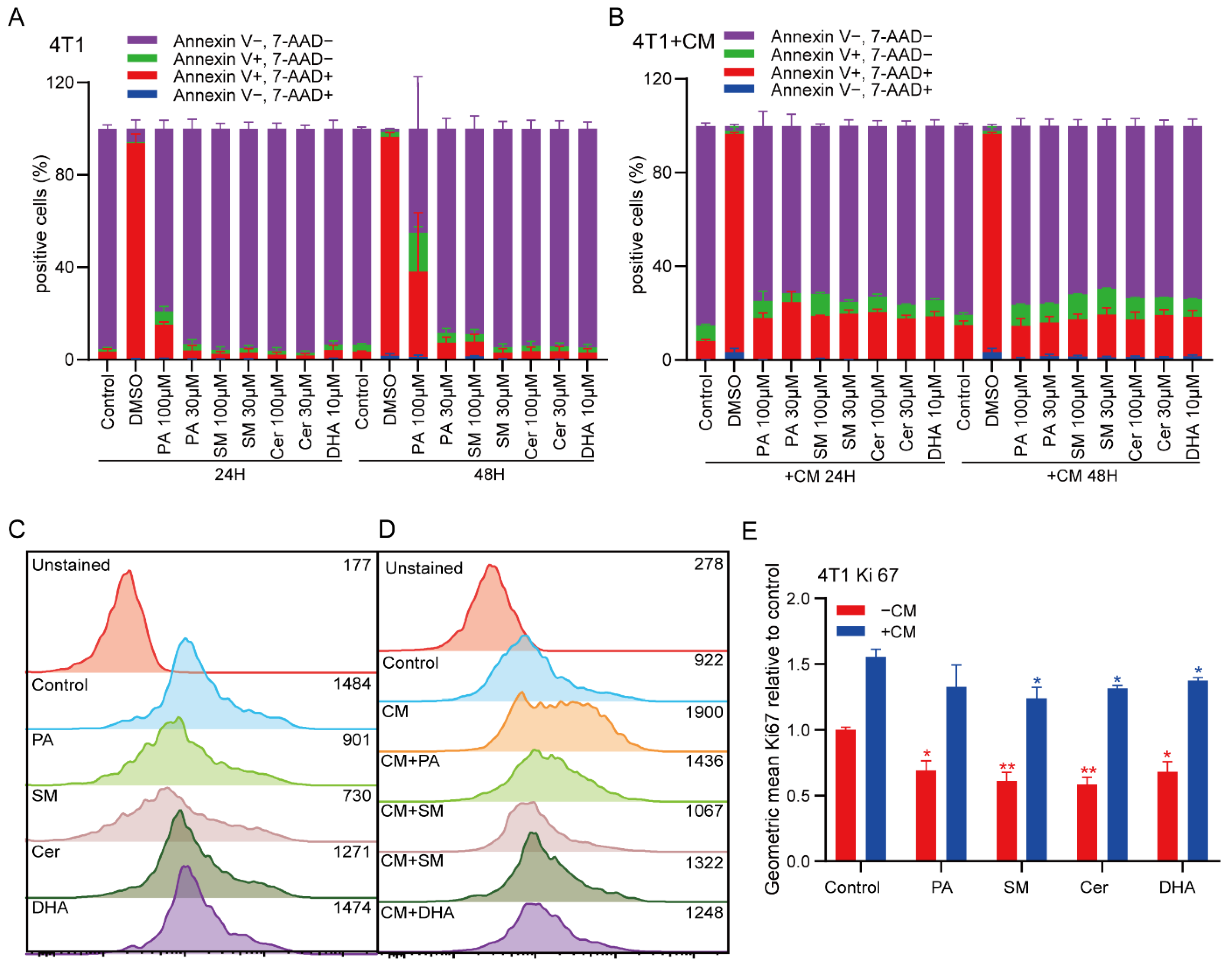
2.2. PA and SM Induce Apoptosis in Breast Cancer Cells
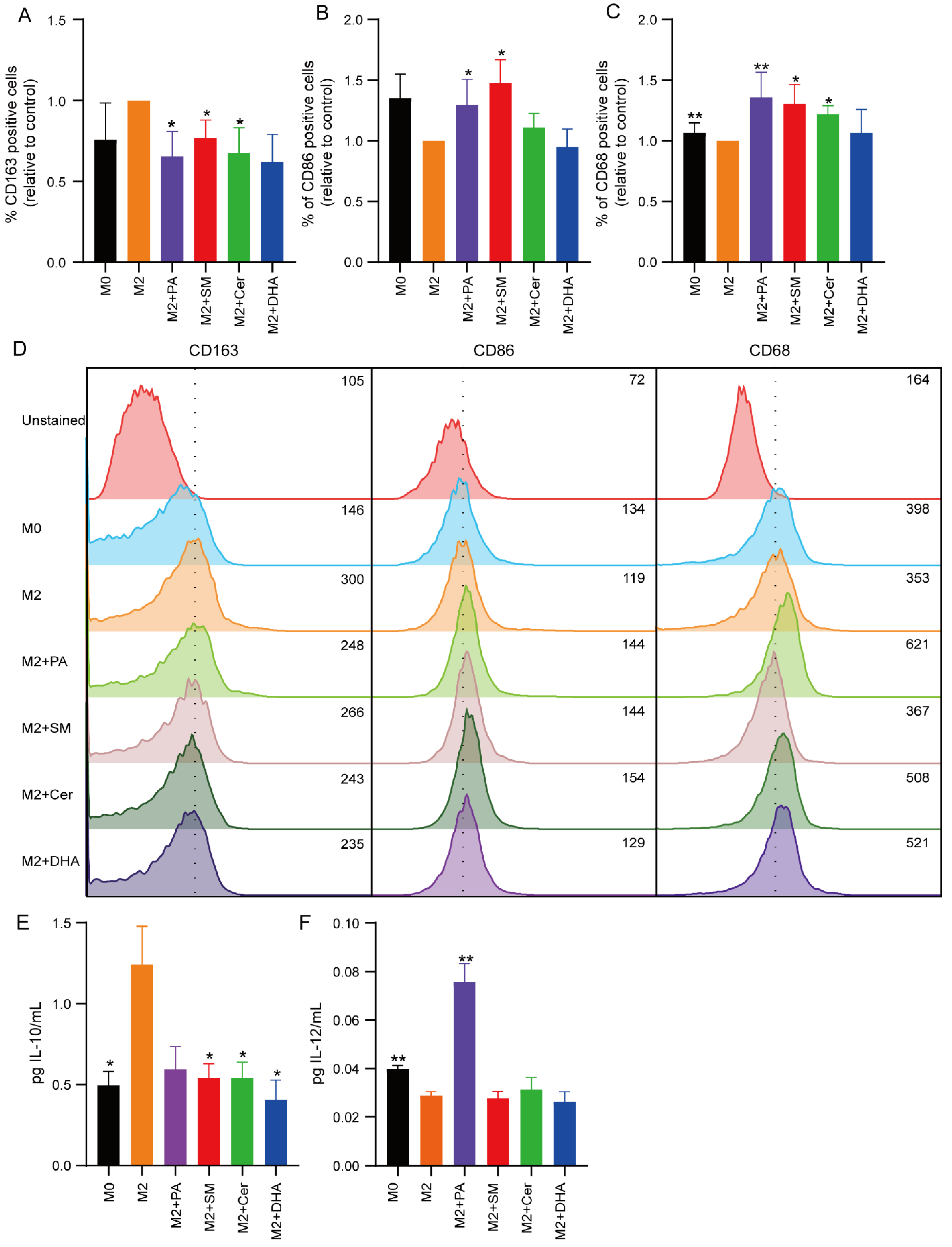
2.3. Effect of Lipids on the Polarization State of M1 and M2 Macrophages
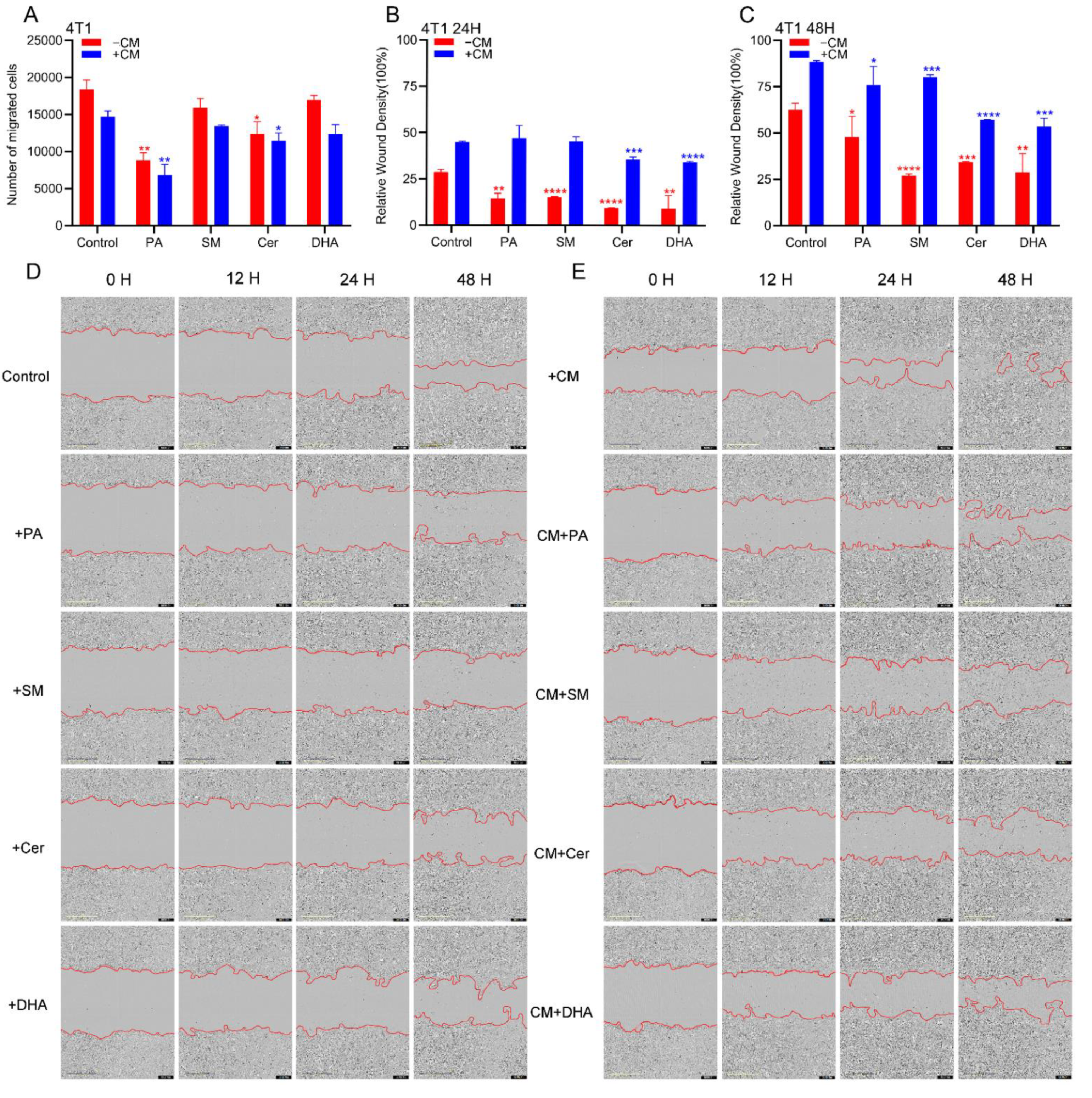
2.4. PA, DHA, SM and Cer Reduce the Migratory Properties of 4T1 Cells
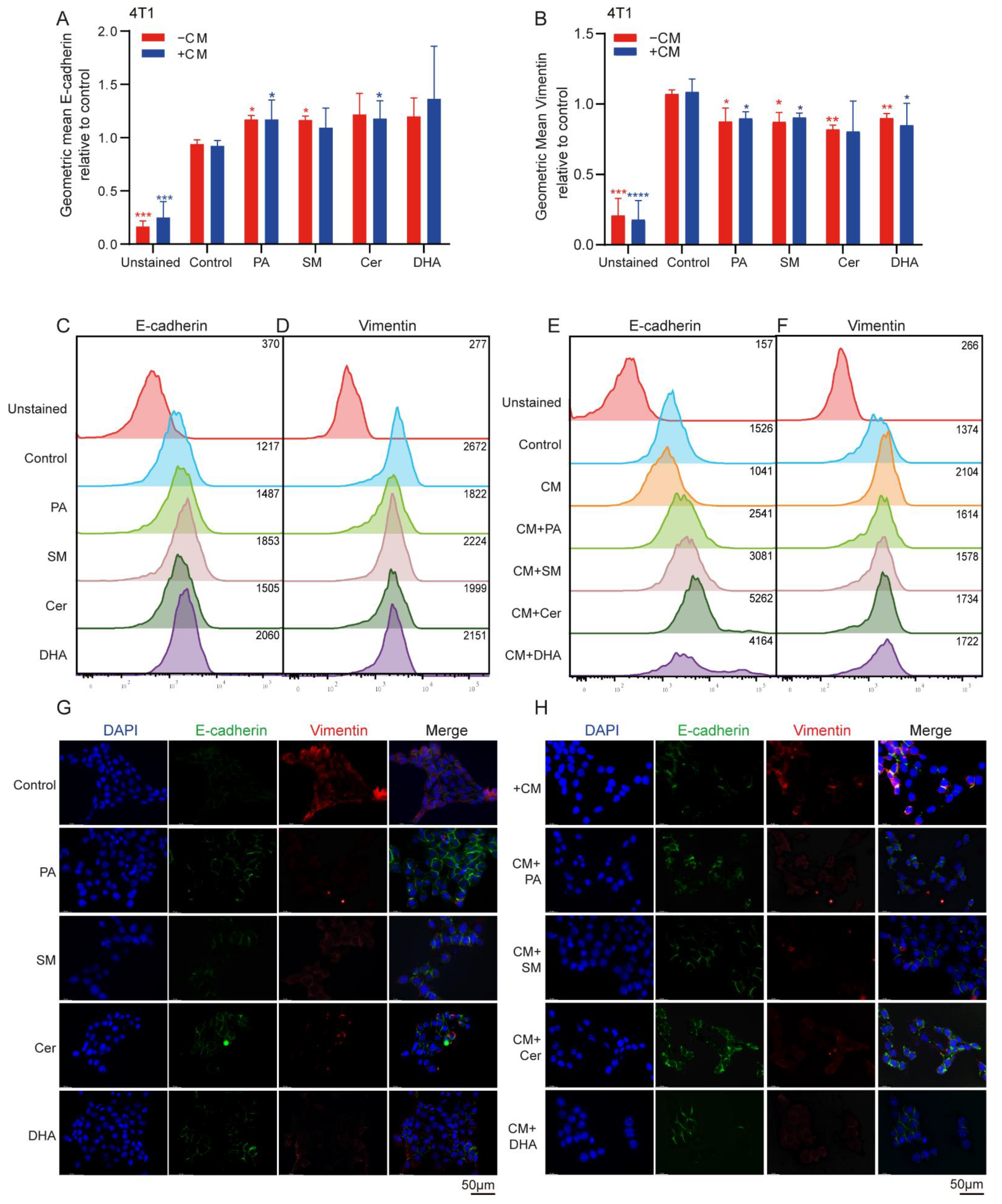
2.5. Lipids Decreased the Mesenchymal Phenotype in Breast Cancer Cells
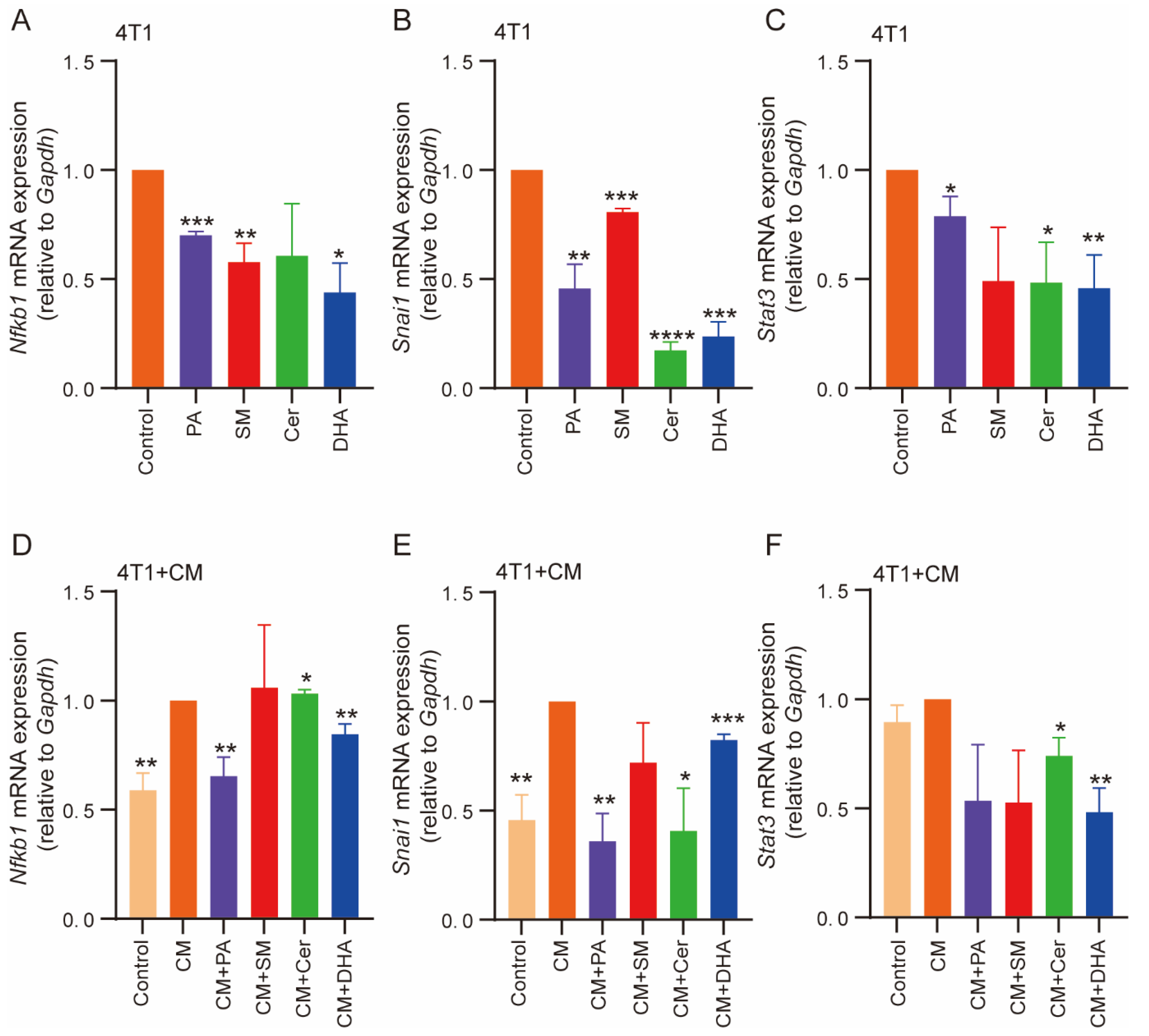
2.6. PA, Cer, SM and DHA Decreased the Mesenchymal Phenotype of 4T1 Cells at the Molecular Level
3. Discussion
4. Materials and Methods
4.1. Materials and Reagents
4.2. Cells
4.3. Preparation, Culture of Bone Marrow-Derived Macrophages (BMDMs)
4.4. Preparation of Lipids
4.5. Polarization of RAW 264.7 Cells towards M1 and M2-Macrophages
4.6. Cell Viability Assays
4.7. Flow Cytometry
4.8. Immunofluorescence
4.9. In Vitro Migration Assays
4.10. Transwell Migration Assays
4.11. Enzyme-Linked Immunosorbent Assay (ELISA)
4.12. Quantitative Real-Time PCR
4.13. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO. Breast Cancer; World Health Organization: Geneva, Switzerland, 2021. [Google Scholar]
- Munoz, D.; Near, A.M.; van Ravesteyn, N.T.; Lee, S.J.; Schechter, C.B.; Alagoz, O.; Berry, D.A.; Burnside, E.S.; Chang, Y.; Chisholm, G.; et al. Effects of screening and systemic adjuvant therapy on ER-specific US breast cancer mortality. JNCI J. Natl. Cancer Inst. 2014, 106, dju289. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, C.G.; Peters, G.J.; Ossendorp, F.; Cruz, L.J. The potential of multi-compound nanoparticles to bypass drug resistance in cancer. Cancer Chemother. Pharmacol. 2017, 80, 881–894. [Google Scholar] [CrossRef] [PubMed]
- Wild, C.P.; Weiderpass, E.; Stewart, B.W. World Cancer Report Cancer Research for Cancer Prevention; IARC Press: Lyon, France, 2020. [Google Scholar]
- Choi, J.; Gyamfi, J.; Jang, H.; Koo, J.S. The Role of Tumor-Associated Macrophage in Breast Cancer Biology. Histol. Histopathol. 2018, 33, 133–145. [Google Scholar] [CrossRef] [PubMed]
- Tang, X. Tumor-associated macrophages as potential diagnostic and prognostic biomarkers in breast cancer. Cancer Lett. 2013, 332, 3–10. [Google Scholar] [CrossRef]
- Tariq, M.; Zhang, J.; Liang, G.; Ding, L.; He, Q.; Yang, B. Macrophage Polarization: Anti-Cancer Strategies to Target Tumor-Associated Macrophage in Breast Cancer. J. Cell. Biochem. 2017, 118, 2484–2501. [Google Scholar] [CrossRef]
- Chanmee, T.; Ontong, P.; Konno, K.; Itano, N. Tumor-associated macrophages as major players in the tumor microenvironment. Cancers 2014, 6, 1670–1690. [Google Scholar] [CrossRef]
- Qiu, S.Q.; Waaijer, S.J.H.; Zwager, M.C.; de Vries, E.G.E.; van der Vegt, B.; Schroder, C.P. Tumor-associated macrophages in breast cancer: Innocent bystander or important player? Cancer Treat. Rev. 2018, 70, 178–189. [Google Scholar] [CrossRef]
- Zhang, S.C.; Hu, Z.Q.; Long, J.H.; Zhu, G.M.; Wang, Y.; Jia, Y.; Zhou, J.; Ouyang, Y.; Zeng, Z. Clinical Implications of Tumor-Infiltrating Immune Cells in Breast Cancer. J. Cancer 2019, 10, 6175–6184. [Google Scholar] [CrossRef]
- Alavi, M.; Hamidi, M. Passive and active targeting in cancer therapy by liposomes and lipid nanoparticles. Drug Metab. Pers. Ther. 2019, 34. [Google Scholar] [CrossRef]
- Kester, M.; Bassler, J.; Fox, T.E.; Carter, C.J.; Davidson, J.A.; Parette, M.R. Preclinical development of a C6-ceramide NanoLiposome, a novel sphingolipid therapeutic. Biol. Chem. 2015, 396, 737–747. [Google Scholar] [CrossRef]
- Lewis, C.E.; Harney, A.S.; Pollard, J.W. The Multifaceted Role of Perivascular Macrophages in Tumors. Cancer Cell 2016, 30, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Goswami, K.K.; Ghosh, T.; Ghosh, S.; Sarkar, M.; Bose, A.; Baral, R. Tumor promoting role of anti-tumor macrophages in tumor microenvironment. Cell Immunol. 2017, 316, 1–10. [Google Scholar] [CrossRef]
- Lee, H.W.; Choi, H.J.; Ha, S.J.; Lee, K.T.; Kwon, Y.G. Recruitment of monocytes/macrophages in different tumor microenvironments. Biochim. Biophys. Acta 2013, 1835, 170–179. [Google Scholar] [CrossRef] [PubMed]
- Martinez, F.O.; Gordon, S. The M1 and M2 paradigm of macrophage activation: Time for reassessment. F1000Prime Rep. 2014, 6, 13. [Google Scholar] [CrossRef]
- Keklikoglou, I.; Cianciaruso, C.; Guc, E.; Squadrito, M.L.; Spring, L.M.; Tazzyman, S.; Lambein, L.; Poissonnier, A.; Ferraro, G.B.; Baer, C.; et al. Chemotherapy elicits pro-metastatic extracellular vesicles in breast cancer models. Nat. Cell Biol. 2019, 21, 190–202. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; de Araujo, R.F., Jr.; Cruz, L.J.; Eich, C. Functionalized Nanoparticles Targeting Tumor-Associated Macrophages as Cancer Therapy. Pharmaceutics 2021, 13, 1670. [Google Scholar] [CrossRef]
- Jiang, N.; Zhang, G.; Pan, L.; Yan, C.; Zhang, L.; Weng, Y.; Wang, W.; Chen, X.; Yang, G. Potential plasma lipid biomarkers in early-stage breast cancer. Biotechnol. Lett. 2017, 39, 1657–1666. [Google Scholar] [CrossRef]
- Shaikh, S.; Channa, N.A.; Talpur, F.N.; Younis, M.; Tabassum, N. Radiotherapy improves serum fatty acids and lipid profile in breast cancer. Lipids Health Dis. 2017, 16, 92. [Google Scholar] [CrossRef]
- Cadenas, C.; Vosbeck, S.; Edlund, K.; Grgas, K.; Madjar, K.; Hellwig, B.; Adawy, A.; Glotzbach, A.; Stewart, J.D.; Lesjak, M.S.; et al. LIPG-promoted lipid storage mediates adaptation to oxidative stress in breast cancer. Int. J. Cancer 2019, 145, 901–915. [Google Scholar] [CrossRef]
- Germain, N.; Dhayer, M.; Boileau, M.; Fovez, Q.; Kluza, J.; Marchetti, P. Lipid Metabolism and Resistance to Anticancer Treatment. Biology 2020, 9, 474. [Google Scholar] [CrossRef]
- Menendez, J.A.; Lupu, R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat. Rev. Cancer 2007, 7, 763–777. [Google Scholar] [CrossRef]
- Luo, X.; Zhao, X.; Cheng, C.; Li, N.; Liu, Y.; Cao, Y. The implications of signaling lipids in cancer metastasis. Exp. Mol. Med. 2018, 50, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Marino, N.; German, R.; Rao, X.; Simpson, E.; Liu, S.; Wan, J.; Liu, Y.; Sandusky, G.; Jacobsen, M.; Stoval, M.; et al. Upregulation of lipid metabolism genes in the breast prior to cancer diagnosis. NPJ Breast Cancer 2020, 6, 50. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.; Lee, M.Y.; Park, S.W.; Moon, J.S.; Koh, Y.K.; Ahn, Y.H.; Park, B.W.; Kim, K.S. Up-regulation of acetyl-CoA carboxylase alpha and fatty acid synthase by human epidermal growth factor receptor 2 at the translational level in breast cancer cells. J. Biol. Chem. 2007, 282, 26122–26131. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Liu, Y.; Zhang, J.T. A new mechanism of drug resistance in breast cancer cells: Fatty acid synthase overexpression-mediated palmitate overproduction. Mol. Cancer Ther. 2008, 7, 263–270. [Google Scholar] [CrossRef] [PubMed]
- Guan, X.; Liu, Z.; Zhao, Z.; Zhang, X.; Tao, S.; Yuan, B.; Zhang, J.; Wang, D.; Liu, Q.; Ding, Y. Emerging roles of low-density lipoprotein in the development and treatment of breast cancer. Lipids Health Dis. 2019, 18, 137. [Google Scholar] [CrossRef] [PubMed]
- Nganga, R.; Oleinik, N.; Ogretmen, B. Mechanisms of Ceramide-Dependent Cancer Cell Death. Adv. Cancer Res. 2018, 140, 1–25. [Google Scholar] [CrossRef]
- Salustiano, E.J.; da Costa, K.M.; Freire-de-Lima, L.; Mendonca-Previato, L.; Previato, J.O. Inhibition of glycosphingolipid biosynthesis reverts multidrug resistance by differentially modulating ABC transporters in chronic myeloid leukemias. J. Biol. Chem. 2020, 295, 6457–6471. [Google Scholar] [CrossRef]
- Moro, K.; Nagahashi, M.; Gabriel, E.; Takabe, K.; Wakai, T. Clinical application of ceramide in cancer treatment. Breast Cancer 2019, 26, 407–415. [Google Scholar] [CrossRef]
- Colombini, M. Membrane channels formed by ceramide. In Sphingolipids: Basic Science and Drug Development; Handbook of Experimental Pharmacology; Springer: Vienna, Autria, 2013; Volume 215, pp. 109–126. [Google Scholar] [CrossRef]
- Taniguchi, M.; Okazaki, T. The role of sphingomyelin and sphingomyelin synthases in cell death, proliferation and migration-from cell and animal models to human disorders. Biochim. Biophys. Acta 2014, 1841, 692–703. [Google Scholar] [CrossRef]
- Xiang, H.; Jin, S.; Tan, F.; Xu, Y.; Lu, Y.; Wu, T. Physiological functions and therapeutic applications of neutral sphingomyelinase and acid sphingomyelinase. Biomed. Pharmacother. 2021, 139, 111610. [Google Scholar] [CrossRef] [PubMed]
- Kong, J.N.; He, Q.; Wang, G.; Dasgupta, S.; Dinkins, M.B.; Zhu, G.; Kim, A.; Spassieva, S.; Bieberich, E. Guggulsterone and bexarotene induce secretion of exosome-associated breast cancer resistance protein and reduce doxorubicin resistance in MDA-MB-231 cells. Int. J. Cancer 2015, 137, 1610–1620. [Google Scholar] [CrossRef] [PubMed]
- Krishnamurthy, K.; Wang, G.; Rokhfeld, D.; Bieberich, E. Deoxycholate promotes survival of breast cancer cells by reducing the level of pro-apoptotic ceramide. Breast Cancer Res. 2008, 10, R106. [Google Scholar] [CrossRef] [PubMed]
- Nagahashi, M.; Tsuchida, J.; Moro, K.; Hasegawa, M.; Tatsuda, K.; Woelfel, I.A.; Takabe, K.; Wakai, T. High levels of sphingolipids in human breast cancer. J. Surg. Res. 2016, 204, 435–444. [Google Scholar] [CrossRef]
- Ledet, M.M.; Harman, R.M.; Fan, J.C.; Schmitt-Matzen, E.; Diaz-Rubio, M.E.; Zhang, S.; Van de Walle, G.R. Secreted sphingomyelins modulate low mammary cancer incidence observed in certain mammals. Sci. Rep. 2020, 10, 20580. [Google Scholar] [CrossRef]
- Companioni, O.; Mir, C.; Garcia-Mayea, Y.; ME, L.L. Targeting Sphingolipids for Cancer Therapy. Front. Oncol. 2021, 11, 745092. [Google Scholar] [CrossRef]
- Bandet, C.L.; Tan-Chen, S.; Bourron, O.; Le Stunff, H.; Hajduch, E. Sphingolipid Metabolism: New Insight into Ceramide-Induced Lipotoxicity in Muscle Cells. Int. J. Mol. Sci. 2019, 20, 479. [Google Scholar] [CrossRef]
- Eich, C.; Manzo, C.; de Keijzer, S.; Bakker, G.J.; Reinieren-Beeren, I.; García-Parajo, M.F.; Cambi, A. Changes in membrane sphingolipid composition modulate dynamics and adhesion of integrin nanoclusters. Sci. Rep. 2016, 6, 20693. [Google Scholar] [CrossRef]
- Kroll, A.; Cho, H.E.; Kang, M.H. Antineoplastic Agents Targeting Sphingolipid Pathways. Front. Oncol. 2020, 10, 833. [Google Scholar] [CrossRef]
- Xu, S.; Chen, T.; Dong, L.; Li, T.; Xue, H.; Gao, B.; Ding, X.; Wang, H.; Li, H. Fatty acid synthase promotes breast cancer metastasis by mediating changes in fatty acid metabolism. Oncol. Lett. 2021, 21, 27. [Google Scholar] [CrossRef]
- De Araujo, R.F., Jr.; Eich, C.; Jorquera, C.; Schomann, T.; Baldazzi, F.; Chan, A.B.; Cruz, L.J. Ceramide and palmitic acid inhibit macrophage-mediated epithelial-mesenchymal transition in colorectal cancer. Mol. Cell. Biochem. 2020, 468, 153–168. [Google Scholar] [CrossRef] [PubMed]
- Zafaryab, M.; Fakhri, K.U.; Khan, M.A.; Hajela, K.; Rizvi, M.M.A. In vitro Assessment of cytotoxic and apoptotic potential of Palmitic acid for Breast cancer Treatment. Int. J. Life Sci. Res. 2019, 7, 166–174. [Google Scholar] [CrossRef]
- Liu, X.-Z.; Rulina, A.; Choi, M.H.; Pedersen, L.; Lepland, J.; Takle, S.T.; Madeleine, N.; Peters, S.D.; Wogsland, C.E.; Grøndal, S.M.; et al. C/EBPB-dependent Adaptation to Palmitic Acid Promotes Stemness in Hormone Receptor Negative Breast Cancer. Nat. Commun. 2022, 13, 69. [Google Scholar] [CrossRef]
- Giordano, C.; Plastina, P.; Barone, I.; Catalano, S.; Bonofiglio, D. n-3 Polyunsaturated Fatty Acid Amides: New Avenues in the Prevention and Treatment of Breast Cancer. Int. J. Mol. Sci. 2020, 21, 2279. [Google Scholar] [CrossRef] [PubMed]
- Rescigno, T.; Capasso, A.; Tecce, M.F. Effect of Docosahexaenoic Acid on Cell Cycle Pathways in Breast Cell Lines with Different Transformation Degree. J. Cell Physiol. 2016, 231, 1226–1236. [Google Scholar] [CrossRef]
- Pogash, T.J.; El-Bayoumy, K.; Amin, S.; Gowda, K.; de Cicco, R.L.; Barton, M.; Su, Y.; Russo, I.H.; Himmelberger, J.A.; Slifker, M.; et al. Oxidized derivative of docosahexaenoic acid preferentially inhibit cell proliferation in triple negative over luminal breast cancer cells. Vitr. Cell. Dev. Biol. Anim. 2015, 51, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Brown, I.; Lee, J.; Sneddon, A.A.; Cascio, M.G.; Pertwee, R.G.; Wahle, K.W.J.; Rotondo, D.; Heys, S.D. Anticancer effects of n-3 EPA and DHA and their endocannabinoid derivatives on breast cancer cell growth and invasion. Prostaglandins Leukot. Essent. Fat. Acids 2020, 156, 102024. [Google Scholar] [CrossRef]
- Sun, H.; Hu, Y.; Gu, Z.; Owens, R.T.; Chen, Y.Q.; Edwards, I.J. Omega-3 fatty acids induce apoptosis in human breast cancer cells and mouse mammary tissue through syndecan-1 inhibition of the MEK-Erk pathway. Carcinogenesis 2011, 32, 1518–1524. [Google Scholar] [CrossRef]
- Rahman, M.M.; Veigas, J.M.; Williams, P.J.; Fernandes, G. DHA is a more potent inhibitor of breast cancer metastasis to bone and related osteolysis than EPA. Breast Cancer Res. Treat. 2013, 141, 341–352. [Google Scholar] [CrossRef]
- Ravacci, G.R.; Brentani, M.M.; Tortelli, T., Jr.; Torrinhas, R.S.; Saldanha, T.; Torres, E.A.; Waitzberg, D.L. Lipid raft disruption by docosahexaenoic acid induces apoptosis in transformed human mammary luminal epithelial cells harboring HER-2 overexpression. J. Nutr. Biochem. 2013, 24, 505–515. [Google Scholar] [CrossRef]
- Su, P.; Wang, Q.; Bi, E.; Ma, X.; Liu, L.; Yang, M.; Qian, J.; Yi, Q. Enhanced Lipid Accumulation and Metabolism Are Required for the Differentiation and Activation of Tumor-Associated Macrophages. Cancer Res. 2020, 80, 1438–1450. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Han, Y.; Rodriguez Sillke, Y.; Deng, H.; Siddiqui, S.; Treese, C.; Schmidt, F.; Friedrich, M.; Keye, J.; Wan, J.; et al. Lipid droplet-dependent fatty acid metabolism controls the immune suppressive phenotype of tumor-associated macrophages. EMBO Mol. Med. 2019, 11, e10698. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Munoz, A.; Kong, J.Y.; Salh, B.; Steinbrecher, U.P. Ceramide-1-phosphate blocks apoptosis through inhibition of acid sphingomyelinase in macrophages. J. Lipid Res. 2004, 45, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Liu, D.; Kimchi, E.T.; Kaifi, J.T.; Qi, X.; Manjunath, Y.; Liu, X.; Deering, T.; Avella, D.M.; Fox, T.; et al. Nanoliposome C6-Ceramide Increases the Anti-tumor Immune Response and Slows Growth of Liver Tumors in Mice. Gastroenterology 2018, 154, 1024–1036.e1029. [Google Scholar] [CrossRef]
- Nixon, G.F. Sphingolipids in inflammation: Pathological implications and potential therapeutic targets. Br. J. Pharmacol. 2009, 158, 982–993. [Google Scholar] [CrossRef]
- Maceyka, M.; Harikumar, K.B.; Milstien, S.; Spiegel, S. Sphingosine-1-phosphate signaling and its role in disease. Trends Cell Biol. 2012, 22, 50–60. [Google Scholar] [CrossRef]
- Wang, S.W.; Hojabrpour, P.; Zhang, P.; Kolesnick, R.N.; Steinbrecher, U.P.; Gomez-Munoz, A.; Duronio, V. Regulation of ceramide generation during macrophage apoptosis by ASMase and de novo synthesis. Biochim. Biophys. Acta 2015, 1851, 1482–1489. [Google Scholar] [CrossRef]
- Jin, J.; Lu, Z.; Li, Y.; Cowart, L.A.; Lopes-Virella, M.F.; Huang, Y. Docosahexaenoic acid antagonizes the boosting effect of palmitic acid on LPS inflammatory signaling by inhibiting gene transcription and ceramide synthesis. PLoS ONE 2018, 13, e0193343. [Google Scholar] [CrossRef]
- Talamonti, E.; Pauter, A.M.; Asadi, A.; Fischer, A.W.; Chiurchiu, V.; Jacobsson, A. Impairment of systemic DHA synthesis affects macrophage plasticity and polarization: Implications for DHA supplementation during inflammation. Cell. Mol. Life Sci. 2017, 74, 2815–2826. [Google Scholar] [CrossRef]
- McWhorter, F.Y.; Wang, T.; Nguyen, P.; Chung, T.; Liu, W.F. Modulation of macrophage phenotype by cell shape. Proc. Natl. Acad. Sci. USA 2013, 110, 17253–17258. [Google Scholar] [CrossRef]
- Wei, C.; Yang, C.; Wang, S.; Shi, D.; Zhang, C.; Lin, X.; Liu, Q.; Dou, R.; Xiong, B. Crosstalk between cancer cells and tumor associated macrophages is required for mesenchymal circulating tumor cell-mediated colorectal cancer metastasis. Mol. Cancer 2019, 18, 64. [Google Scholar] [CrossRef]
- Yu, H.; Pardoll, D.; Jove, R. STATs in cancer inflammation and immunity: A leading role for STAT3. Nat. Rev. Cancer 2009, 9, 798–809. [Google Scholar] [CrossRef]
- Grivennikov, S.I.; Karin, M. Dangerous liaisons: STAT3 and NF-kappaB collaboration and crosstalk in cancer. Cytokine Growth Factor Rev. 2010, 21, 11–19. [Google Scholar] [CrossRef]
- Cheng, C.; Geng, F.; Cheng, X.; Guo, D. Lipid metabolism reprogramming and its potential targets in cancer. Cancer Commun. 2018, 38, 27. [Google Scholar] [CrossRef] [PubMed]
- Corn, K.C.; Windham, M.A.; Rafat, M. Lipids in the tumor microenvironment: From cancer progression to treatment. Prog. Lipid Res. 2020, 80, 101055. [Google Scholar] [CrossRef]
- Batista-Gonzalez, A.; Vidal, R.; Criollo, A.; Carreno, L.J. New Insights on the Role of Lipid Metabolism in the Metabolic Reprogramming of Macrophages. Front. Immunol. 2019, 10, 2993. [Google Scholar] [CrossRef]
- Ip, W.K.E.; Hoshi, N.; Shouval, D.S.; Snapper, S.; Medzhitov, R. Anti-inflammatory effect of IL-10 mediated by metabolic reprogramming of macrophages. Science 2017, 356, 513–519. [Google Scholar] [CrossRef]
- Baumann, J.; Wong, J.; Sun, Y.; Conklin, D.S. Palmitate-induced ER stress increases trastuzumab sensitivity in HER2/neu-positive breast cancer cells. BMC Cancer 2016, 16, 551. [Google Scholar] [CrossRef]
- Morad, S.A.; Cabot, M.C. Ceramide-orchestrated signalling in cancer cells. Nat. Rev. Cancer 2013, 13, 51–65. [Google Scholar] [CrossRef]
- Struckhoff, A.P.; Patel, B.; Beckman, B.S. Inhibition of p53 sensitizes MCF-7 cells to ceramide treatment. Int. J. Oncol. 2010, 37, 21–30. [Google Scholar] [CrossRef][Green Version]
- Pozuelo-Rubio, M. Regulation of autophagic activity by 14-3-3zeta proteins associated with class III phosphatidylinositol-3-kinase. Cell Death Differ. 2011, 18, 479–492. [Google Scholar] [CrossRef] [PubMed]
- Moro, K.; Kawaguchi, T.; Tsuchida, J.; Gabriel, E.; Qi, Q.; Yan, L.; Wakai, T.; Takabe, K.; Nagahashi, M. Ceramide species are elevated in human breast cancer and are associated with less aggressiveness. Oncotarget 2018, 9, 19874–19890. [Google Scholar] [CrossRef]
- D’Angelo, G.; Moorthi, S.; Luberto, C. Role and Function of Sphingomyelin Biosynthesis in the Development of Cancer. Adv. Cancer Res. 2018, 140, 61–96. [Google Scholar] [CrossRef]
- Mazzei, J.C.; Zhou, H.; Brayfield, B.P.; Hontecillas, R.; Bassaganya-Riera, J.; Schmelz, E.M. Suppression of intestinal inflammation and inflammation-driven colon cancer in mice by dietary sphingomyelin: Importance of peroxisome proliferator-activated receptor gamma expression. J. Nutr. Biochem. 2011, 22, 1160–1171. [Google Scholar] [CrossRef]
- Ogretmen, B. Sphingolipid metabolism in cancer signalling and therapy. Nat. Rev. Cancer 2018, 18, 33–50. [Google Scholar] [CrossRef]
- Deng, Y.; Hu, J.C.; He, S.H.; Lou, B.; Ding, T.B.; Yang, J.T.; Mo, M.G.; Ye, D.Y.; Zhou, L.; Jiang, X.C.; et al. Sphingomyelin synthase 2 facilitates M2-like macrophage polarization and tumor progression in a mouse model of triple-negative breast cancer. Acta Pharmacol. Sin. 2021, 42, 149–159. [Google Scholar] [CrossRef]
- Modrak, D.E.; Cardillo, T.M.; Newsome, G.A.; Goldenberg, D.M.; Gold, D.V. Synergistic Interaction between Sphingomyelin and Gemcitabine Potentiates Ceramide-Mediated Apoptosis in Pancreatic Cancer. Cancer Res. 2004, 64, 8405–8410. [Google Scholar] [CrossRef]
- Xue, M.; Wang, Q.; Zhao, J.; Dong, L.; Ge, Y.; Hou, L.; Liu, Y.; Zheng, Z. Docosahexaenoic acid inhibited the Wnt/beta-catenin pathway and suppressed breast cancer cells in vitro and in vivo. J. Nutr. Biochem. 2014, 25, 104–110. [Google Scholar] [CrossRef]
- Newell, M.; Baker, K.; Postovit, L.M.; Field, C.J. A Critical Review on the Effect of Docosahexaenoic Acid (DHA) on Cancer Cell Cycle Progression. Int. J. Mol. Sci. 2017, 18, 1784. [Google Scholar] [CrossRef]
- Blanckaert, V.; Ulmann, L.; Mimouni, V.; Antol, J.; Brancquart, L.; Chénais, B. Docosahexaenoic acid intake decreases proliferation, increases apoptosis and decreases the invasive potential of the human breast carcinoma cell line MDA-MB-231. Int. J. Oncol. 2010, 36, 737–742. [Google Scholar] [CrossRef]
- Ding, W.-Q.; Vaught, J.L.; Yamauchi, H.; Lind, S.E. Differential sensitivity of cancer cells to docosahexaenoic acid–induced cytotoxicity: The potential importance of down-regulation of superoxide dismutase 1 expression. Mol. Cancer Ther. 2004, 3, 1109–1117. [Google Scholar]
- Kawano, A.; Ariyoshi, W.; Yoshioka, Y.; Hikiji, H.; Nishihara, T.; Okinaga, T. Docosahexaenoic acid enhances M2 macrophage polarization via the p38 signaling pathway and autophagy. J. Cell. Biochem. 2019, 120, 12604–12617. [Google Scholar] [CrossRef] [PubMed]
- Williams-Bey, Y.; Boularan, C.; Vural, A.; Huang, N.-N.; Hwang, I.-Y.; Shan-Shi, C.; Kehrl, J.H. Omega-3 Free Fatty Acids Suppress Macrophage Inflammasome Activation by Inhibiting NF-κB Activation and Enhancing Autophagy. PLoS ONE 2014, 9, e97957. [Google Scholar] [CrossRef] [PubMed]
- Gladine, C.; Zmojdzian, M.; Joumard-Cubizolles, L.; Verny, M.-A.; Comte, B.; Mazur, A. The omega-3 fatty acid docosahexaenoic acid favorably modulates the inflammatory pathways and macrophage polarization within aorta of LDLR−/− mice. Genes Nutr. 2014, 9, 424. [Google Scholar] [CrossRef]
- Rajasinghe, L.D.; Chauhan, P.S.; Wierenga, K.A.; Evered, A.O.; Harris, S.N.; Bates, M.A.; Gavrilin, M.A.; Pestka, J.J. Omega-3 Docosahexaenoic Acid (DHA) Impedes Silica-Induced Macrophage Corpse Accumulation by Attenuating Cell Death and Potentiating Efferocytosis. Front. Immunol. 2020, 11, 2179. [Google Scholar] [CrossRef] [PubMed]
- Nieto, M.A.; Huang, R.Y.; Jackson, R.A.; Thiery, J.P. Emt: 2016. Cell 2016, 166, 21–45. [Google Scholar] [CrossRef]
- Tam, W.L.; Weinberg, R.A. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat. Med. 2013, 19, 1438–1449. [Google Scholar] [CrossRef]
- Gordon-Weeks, A.N.; Lim, S.Y.; Yuzhalin, A.E.; Jones, K.; Markelc, B.; Kim, K.J.; Buzzelli, J.N.; Fokas, E.; Cao, Y.; Smart, S.; et al. Neutrophils promote hepatic metastasis growth through fibroblast growth factor 2-dependent angiogenesis in mice. Hepatology 2017, 65, 1920–1935. [Google Scholar] [CrossRef]
- Toh, B.; Wang, X.; Keeble, J.; Sim, W.J.; Khoo, K.; Wong, W.C.; Kato, M.; Prevost-Blondel, A.; Thiery, J.P.; Abastado, J.P. Mesenchymal transition and dissemination of cancer cells is driven by myeloid-derived suppressor cells infiltrating the primary tumor. PLoS Biol. 2011, 9, e1001162. [Google Scholar] [CrossRef]
- Kang, E.; Seo, J.; Yoon, H.; Cho, S. The Post-Translational Regulation of Epithelial-Mesenchymal Transition-Inducing Transcription Factors in Cancer Metastasis. Int. J. Mol. Sci. 2021, 22, 3591. [Google Scholar] [CrossRef]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef]
- Solanas, G.; Porta-de-la-Riva, M.; Agusti, C.; Casagolda, D.; Sanchez-Aguilera, F.; Larriba, M.J.; Pons, F.; Peiro, S.; Escriva, M.; Munoz, A.; et al. E-cadherin controls beta-catenin and NF-kappaB transcriptional activity in mesenchymal gene expression. J. Cell Sci. 2008, 121, 2224–2234. [Google Scholar] [CrossRef]
- Cavalcante, R.S.; Ishikawa, U.; Silva, E.S.; Silva, A.A., Jr.; Araujo, A.A.; Cruz, L.J.; Chan, A.B.; Araujo, R.F., Jr. M2 TAM-associated STAT3/NF-kappaB signalling suppression as major target of immunomodulatory therapy with PLGA-based nanocarriers and anti-PD-L1 in breast cancer. Br. J. Pharmacol. 2021, 178, 2284–2304. [Google Scholar] [CrossRef]
- Ying, W.; Cheruku, P.S.; Bazer, F.W.; Safe, S.H.; Zhou, B. Investigation of macrophage polarization using bone marrow derived macrophages. J. Vis. Exp. 2013, 76, e50323. [Google Scholar] [CrossRef]
- Trouplin, V.; Boucherit, N.; Gorvel, L.; Conti, F.; Mottola, G.; Ghigo, E. Bone marrow-derived macrophage production. J. Vis. Exp. 2013, 81, e50966. [Google Scholar] [CrossRef]
- Lin, L.; Ding, Y.; Wang, Y.; Wang, Z.; Yin, X.; Yan, G.; Zhang, L.; Yang, P.; Shen, H. Functional lipidomics: Palmitic acid impairs hepatocellular carcinoma development by modulating membrane fluidity and glucose metabolism. Hepatology 2017, 66, 432–448. [Google Scholar] [CrossRef]
| Effect on: | 4T1 | M2 TAMs | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Viability | Apoptosis | Proliferation | E-cadherin | Vimentin | NFκB | Snai1 | Stat3 | CD163 | CD86 | CD68 | IL10 | IL12 | |
| PA/CM+PA | + | + | + | +/+ | +/+ | +/+ | +/+ | + | + | + | + | + | |
| SM/CM+SM | + | +/+ | + | +/+ | + | + | + | + | + | + | |||
| Cer/CM+Cer | +/+ | + | + | + | +/+ | +/+ | + | + | + | ||||
| DHA/CM+DHA | +/+ | +/+ | +/+ | +/+ | +/+ | + | + | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, Y.; Rezaei, S.; Júnior, R.F.d.A.; Cruz, L.J.; Eich, C. Multifunctional Role of Lipids in Modulating the Tumorigenic Properties of 4T1 Breast Cancer Cells. Int. J. Mol. Sci. 2022, 23, 4240. https://doi.org/10.3390/ijms23084240
He Y, Rezaei S, Júnior RFdA, Cruz LJ, Eich C. Multifunctional Role of Lipids in Modulating the Tumorigenic Properties of 4T1 Breast Cancer Cells. International Journal of Molecular Sciences. 2022; 23(8):4240. https://doi.org/10.3390/ijms23084240
Chicago/Turabian StyleHe, Yuanyuan, Somayeh Rezaei, Raimundo Fernandes de Araújo Júnior, Luis J. Cruz, and Christina Eich. 2022. "Multifunctional Role of Lipids in Modulating the Tumorigenic Properties of 4T1 Breast Cancer Cells" International Journal of Molecular Sciences 23, no. 8: 4240. https://doi.org/10.3390/ijms23084240
APA StyleHe, Y., Rezaei, S., Júnior, R. F. d. A., Cruz, L. J., & Eich, C. (2022). Multifunctional Role of Lipids in Modulating the Tumorigenic Properties of 4T1 Breast Cancer Cells. International Journal of Molecular Sciences, 23(8), 4240. https://doi.org/10.3390/ijms23084240






