High Expression of the Lysosomal Protease Cathepsin D Confers Better Prognosis in Neuroblastoma Patients by Contrasting EGF-Induced Neuroblastoma Cell Growth
Abstract
1. Introduction
2. Results
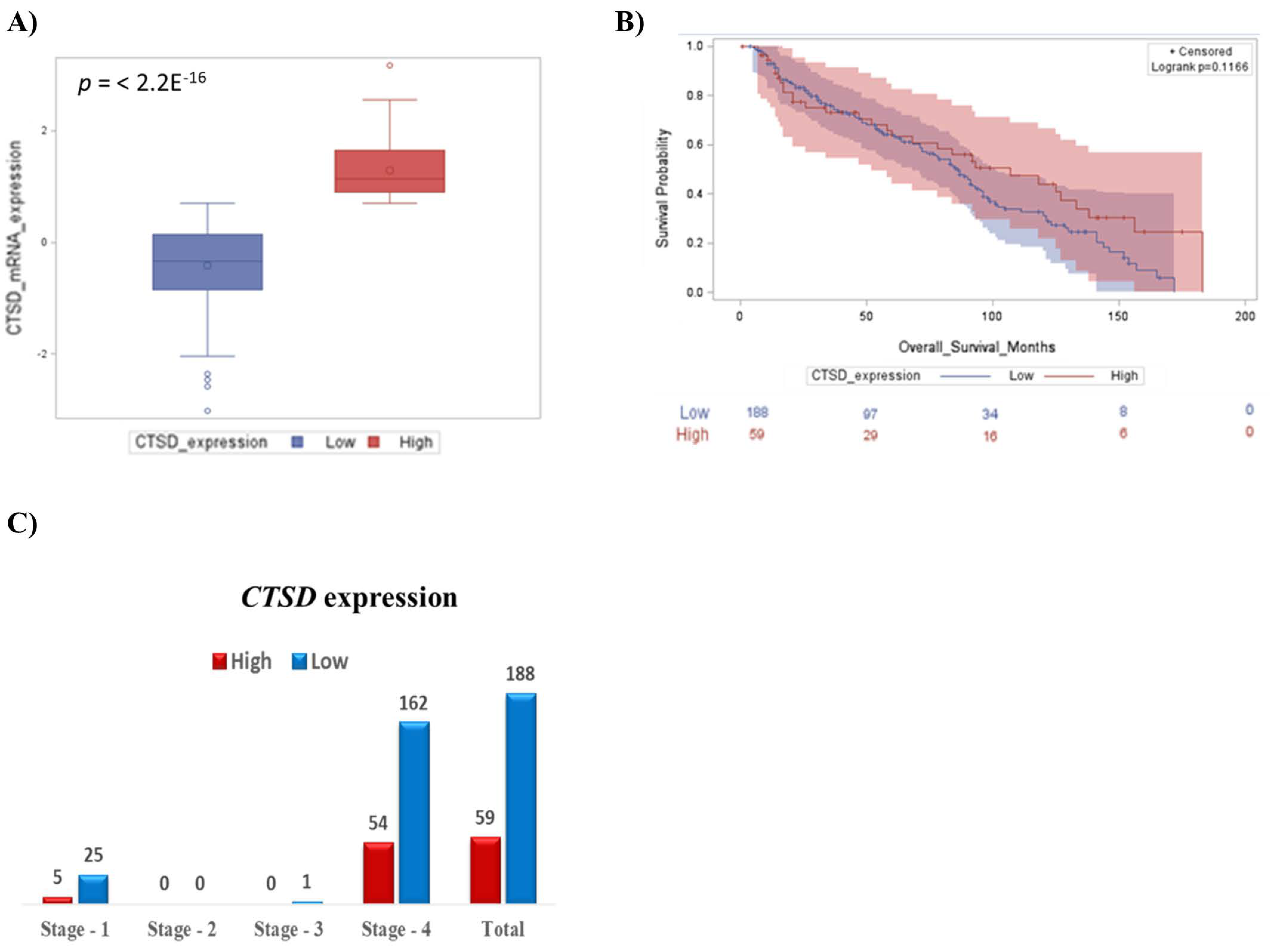
2.1. High CTSD Expression Correlates with Better Prognosis in Pediatric Neuroblastoma Patients
2.2. High CTSD Expression Increases the Overall Survival of Neuroblastoma Patients Highly Expressing EGFR Transcript
2.3. CTSD Gene Expression Negatively Correlates with Genes Involved in Cell Division
2.4. Generation of Transgenic SH-SY5Y Clones Stably Over-Expressing or Silenced for Cathepsin D
2.4.1. Construction of pENTR™/H1/TO Plasmids for Cathepsin D Knockdown
2.4.2. Construction of pcDNA™4/TO Plasmid for Cathepsin D Overexpression
2.5. Overexpression of Cathepsin D Reduces, While Downregulation of Cathepsin D Enhances, the Proliferative Potential of Transgenic SH-SY5Y Clones
2.6. Epidermal Growth Factor Stimulates the Proliferation of KD-CD SH-SY5Y Transgenic Cells While Overexpression of CD Contrasts Its Activity
2.7. EGF Reduces Cathepsin D Protein Level and Increases ERK 1/2 Phosphorylation in SH-SY5Y Neuroblastoma Cells
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Treatment
4.2. Cell Counting, Doubling Time and Cell Cycle Analysis
4.3. Clonogenic Assay
4.4. Antibodies
4.5. Western Blotting
4.6. Immunofluorescence
4.7. Bioinformatic Analysis
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Davidoff, A.M. Neonatal Neuroblastoma. Clin. Perinatol. 2021, 48, 101–115. [Google Scholar] [CrossRef] [PubMed]
- Monclair, T.; Brodeur, G.M.; Ambros, P.F.; Brisse, H.J.; Cecchetto, G.; Holmes, K.; Kaneko, M.; London, W.B.; Matthay, K.K.; Nuchtern, J.G.; et al. The international neuroblastoma risk group (INRG) staging system: An INRG task force report. J. Clin. Oncol. 2009, 27, 298–303. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Kling, M.J.; Griggs, C.N.; McIntyre, E.M.; Alexander, G.; Ray, S.; Challagundla, K.B.; Joshi, S.S.; Coulter, D.W.; Chaturvedi, N.K. Synergistic efficacy of inhibiting MYCN and mTOR signaling against neuroblastoma. BMC Cancer 2021, 21, 1–13. [Google Scholar] [CrossRef]
- Sagulenko, V.; Muth, D.; Sagulenko, E.; Paffhausen, T.; Schwab, M.; Westermann, F. Cathepsin D protects human neuroblastoma cells from doxorubicin-induced cell death. Carcinogenesis 2008, 29, 1869–1877. [Google Scholar] [CrossRef]
- Barrett, A.J. Cathepsin D: The Lysosomal Aspartic Proteinase. In Ciba Foundation Symposium-Aetiology of Diabetes Mellitus and Its Complications (Colloquia on Endocrinology); Excerpta Medica: New York, NY, USA, 2008; Volume 15, pp. 37–50. [Google Scholar] [CrossRef]
- Berg, T.; Gjøen, T.; Bakke, O. Physiological functions of endosomal proteolysis. Biochem. J. 1995, 307, 313–326. [Google Scholar] [CrossRef]
- Saftig, P.; Hetman, M.; Schmahl, W.; Weber, K.; Heine, L.; Mossmann, H.; Köster, A.; Hess, B.; Evers, M.; Von Figura, K. Mice deficient for the lysosomal proteinase cathepsin D exhibit progressive atrophy of the intestinal mucosa and profound destruction of lymphoid cells. EMBO J. 1995, 14, 3599–3608. [Google Scholar] [CrossRef]
- Follo, C.; Ozzano, M.; Mugoni, V.; Castino, R.; Santoro, M.; Isidoro, C. Knock-Down of Cathepsin D Affects the Retinal Pigment Epithelium, Impairs Swim-Bladder Ontogenesis and Causes Premature Death in Zebrafish. PLoS ONE 2011, 6, e21908. [Google Scholar] [CrossRef]
- Isidoro, C.; Demoz, M.; De Stefanis, D.; Baccino, F.M.; Bonelli, G. Synthesis, maturation and extracellular release of procathepsin D as influenced by cell proliferation or transformation. Int. J. Cancer 1995, 63, 866–871. [Google Scholar] [CrossRef]
- Isidoro, C.; Grässel, S.; Baccino, F.M.; Hasilik, A. Determination of the Phosphorylation, Uncovering of Mannose 6-Phosphate Groups and Targeting of Lysosomal Enzymes. Clin. Chem. Lab. Med. (CCLM) 1991, 29, 165–171. [Google Scholar] [CrossRef][Green Version]
- Cunat, S.; Hoffmann, P.; Pujol, P. Estrogens and epithelial ovarian cancer. Gynecol. Oncol. 2004, 94, 25–32. [Google Scholar] [CrossRef]
- Nicotra, G.; Castino, R.; Follo, C.; Peracchio, C.; Valente, G.; Isidoro, C. The dilemma: Does tissue expression of cathepsin D reflect tumor malignancy? The question: Does the assay truly mirror cathepsin D mis-function in the tumor? Cancer Biomarkers 2010, 7, 47–64. [Google Scholar] [CrossRef]
- Tamura, S.; Hosoi, H.; Kuwahara, Y.; Kikuchi, K.; Otabe, O.; Izumi, M.; Tsuchiya, K.; Iehara, T.; Gotoh, T.; Sugimoto, T. Induction of apoptosis by an inhibitor of EGFR in neuroblastoma cells. Biochem. Biophys. Res. Commun. 2007, 358, 226–232. [Google Scholar] [CrossRef]
- Meyers, M.B.; Shen, W.P.V.; Spengler, B.A.; Ciccarone, V.; O’Brien, J.P.; Donner, D.B.; Furth, M.E.; Biedler, J.L. Increased epidermal growth factor receptor in multidrug-resistant human neuroblastoma cells. J. Cell. Biochem. 1988, 38, 87–97. [Google Scholar] [CrossRef]
- Huang, C.-C.; Lee, C.-C.; Lin, H.-H.; Chang, J.-Y. Cathepsin S attenuates endosomal EGFR signalling: A mechanical rationale for the combination of cathepsin S and EGFR tyrosine kinase inhibitors. Sci. Rep. 2016, 6, 29256. [Google Scholar] [CrossRef]
- Authier, F.; Métioui, M.; Bell, A.W.; Mort, J.S. Negative Regulation of Epidermal Growth Factor Signaling by Selective Proteolytic Mechanisms in the Endosome Mediated by Cathepsin B. J. Biol. Chem. 1999, 274, 33723–33731. [Google Scholar] [CrossRef]
- Zheng, C.; Shen, R.; Li, K.; Zheng, N.; Zong, Y.; Ye, D.; Wang, Q.; Wang, Z.; Chen, L.; Ma, Y. Epidermal growth factor receptor is overexpressed in neuroblastoma tissues and cells. Acta Biochim. Et Biophys. Sin. 2016, 48, 762–767. [Google Scholar] [CrossRef]
- Hatziagapiou, K.; Braoudaki, M.; Karpusas, M.; Tzortzatou-Stathopoulou, F. Evaluation of antitumor activity of gefitinib in pediatric glioblastoma and neuroblastoma cells. Clin. Lab. 2011, 57, 22029196. [Google Scholar]
- Ho, R.; Minturn, J.E.; Hishiki, T.; Zhao, H.; Wang, Q.; Cnaan, A.; Maris, J.; Evans, A.E.; Brodeur, G.M. Proliferation of Human Neuroblastomas Mediated by the Epidermal Growth Factor Receptor. Cancer Res. 2005, 65, 9868–9875. [Google Scholar] [CrossRef]
- Ohri, S.S.; Vashishta, A.; Proctor, M.; Fusek, M.; Vetvicka, V. Depletion of procathepsin D gene expression by RNA interference – A potential therapeutic target for breast cancer. Cancer Biol. Ther. 2007, 6, 1081–1087. [Google Scholar] [CrossRef]
- Chiu, B.; Mirkin, B.; Madonna, M.B. Mitogenic and Apoptotic Actions of Epidermal Growth Factor on Neuroblastoma Cells Are Concentration-Dependent. J. Surg. Res. 2006, 135, 209–212. [Google Scholar] [CrossRef]
- Carraway, K.L.; Carraway, C.A.C. Signaling, mitogenesis and the cytoskeleton: Where the action is. BioEssays 1995, 17, 171–175. [Google Scholar] [CrossRef] [PubMed]
- Castino, R.; Démoz, M.; Isidoro, C. Destination ‘Lysosome’: A target organelle for tumour cell killing? J. Mol. Recognit. 2003, 16, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Lockwood, T.D.; Shier, W.T. Regulation of acid proteases during growth, quiescence and starvation in normal and transformed cells. Nature 1977, 267, 252–254. [Google Scholar] [CrossRef] [PubMed]
- Emert-Sedlak, L.; Shangary, S.; Rabinovitz, A.; Miranda, M.B.; Delach, S.M.; Johnson, D.E. Involvement of cathepsin D in chemotherapy-induced cytochrome c release, caspase activation, and cell death. Mol. Cancer Ther. 2005, 4, 733–742. [Google Scholar] [CrossRef] [PubMed]
- Castino, R.; Peracchio, C.; Salini, A.; Nicotra, G.; Trincheri, N.F.; Démoz, M.; Valente, G.; Isidoro, C. Chemotherapy drug response in ovarian cancer cells strictly depends on a cathepsin D-Bax activation loop. J. Cell. Mol. Med. 2009, 13, 1096–1109. [Google Scholar] [CrossRef] [PubMed]
- Demoz, M.; Castino, R.; Cesaro, P.; Baccino, F.M.; Bonelli, G.; Isidoro, C. Endosomal-Lysosomal Proteolysis Mediates Death Signalling by TNFα, Not by Etoposide, in L929 Fibrosarcoma Cells: Evidence for an Active Role of Cathepsin D. Biol. Chem. 2002, 383, 1237–1248. [Google Scholar] [CrossRef] [PubMed]
- Trincheri, N.F.; Nicotra, G.; Follo, C.; Castino, R.; Isidoro, C. Resveratrol induces cell death in colorectal cancer cells by a novel pathway involving lysosomal cathepsin D. Carcinogenesis 2006, 28, 922–931. [Google Scholar] [CrossRef]
- Castino, R.; Bellio, N.; Nicotra, G.; Follo, C.; Trincheri, N.F.; Isidoro, C. Cathepsin D–Bax death pathway in oxidative stressed neuroblastoma cells. Free Radic. Biol. Med. 2007, 42, 1305–1316. [Google Scholar] [CrossRef]
- Roberg, K.; Ollinger, K. Oxidative stress causes relocation of the lysosomal enzyme cathepsin D with ensuing apoptosis in neonatal rat cardiomyocytes. Am. J. Pathol. 1998, 152, 1151–1156. [Google Scholar]
- Shibata, M.; Kanamori, S.; Isaharaa, K.; Ohsawaa, Y.; Konishia, A.; Kametakaa, S.; Watanabe, T.; Ebisub, S.; Ishidoc, K.; Kominamic, E.; et al. Participation of Cathepsins B and D in Apoptosis of PC12 Cells Following Serum Deprivation. Biochem. Biophys. Res. Commun. 1998, 251, 199–203. [Google Scholar] [CrossRef]
- Vignon, F.; Capony, F.; Ghambon, M.; Freiss, G.; Garcia, M.; Rochefort, H. Autocrine Growth Stimulation of the MCF 7 Breast Cancer Cells by the Estrogen-Regulated 52 K Protein. Endocrinology 1986, 118, 1537–1545. [Google Scholar] [CrossRef]
- Isidoro, C.; Demoz, M.; De Stefanis, D.; Mainferme, F.; Wattiaux, R.; Baccino, F.M. Altered intracellular processing and enhanced secretion of procathepsin D in a highly deviated rat hepatoma. Int. J. Cancer 1995, 60, 61–64. [Google Scholar] [CrossRef]
- Isidoro, C.; Démoz, M.; De Stefanis, D.; Baccino, F.M.; Bonelli, G. High levels of proteolytic enzymes in the ascitic fluid and plasma of rats bearing the Yoshida AH-130 hepatoma. Invasion Metastasis 1995, 15, 8621267. [Google Scholar]
- A Reid, W.; Valler, M.J.; Kay, J. Immunolocalization of cathepsin D in normal and neoplastic human tissues. J. Clin. Pathol. 1986, 39, 1323–1330. [Google Scholar] [CrossRef]
- Laurent-Matha, V.; Maruani-Herrmann, S.; Prébois, C.; Beaujouin, M.; Glondu, M.; Noel, A.; Alvarez-Gonzalez, M.L.; Blacher, S.; Coopman, P.; Baghdiguian, S.; et al. Catalytically inactive human cathepsin D triggers fibroblast invasive growth. J. Cell Biol. 2005, 168, 489–499. [Google Scholar] [CrossRef]
- Pranjol, Z.I.; Gutowski, N.J.; Hannemann, M.; Whatmore, J.L. Cathepsin D non-proteolytically induces proliferation and migration in human omental microvascular endothelial cells via activation of the ERK1/2 and PI3K/AKT pathways. Biochim. Et Biophys. Acta 2018, 1865, 25–33. [Google Scholar] [CrossRef]
- Benes, P.; Vetvicka, V.; Fusek, M. Cathepsin D—Many functions of one aspartic protease. Crit. Rev. Oncol. Hematol. 2008, 68, 12–28. [Google Scholar] [CrossRef]
- Jha, S.K.; Rauniyar, K.; Chronowska, E.; Mattonet, K.; Maina, E.W.; Koistinen, H.; Stenman, U.-H.; Alitalo, K.; Jeltsch, M. KLK3/PSA and cathepsin D activate VEGF-C and VEGF-D. eLife 2019, 8, e44478. [Google Scholar] [CrossRef]
- Sasaki, T.; Hiroki, K.; Yamashita, Y. The Role of Epidermal Growth Factor Receptor in Cancer Metastasis and Microenvironment. BioMed Res. Int. 2013, 2013, 1–8. [Google Scholar] [CrossRef]
- Ongusaha, P.P.; Kwak, J.C.; Zwible, A.J.; Macip, S.; Higashiyama, S.; Taniguchi, N.; Fang, L.; Lee, S.W. HB-EGF Is a Potent Inducer of Tumor Growth and Angiogenesis. Cancer Res. 2004, 64, 5283–5290. [Google Scholar] [CrossRef]
- Yagi, H.; Yotsumoto, F.; Miyamoto, S. Heparin-binding epidermal growth factor–like growth factor promotes transcoelomic metastasis in ovarian cancer through epithelial-mesenchymal transition. Mol. Cancer Ther. 2008, 7, 3441–3451. [Google Scholar] [CrossRef] [PubMed]
- Rao, L.; Giannico, D.; Leone, P.; Solimando, A.G.; Maiorano, E.; Caporusso, C.; Duda, L.; Tamma, R.; Mallamaci, R.; Susca, N.; et al. HB-EGF–EGFR Signaling in Bone Marrow Endothelial Cells Mediates Angiogenesis Associated with Multiple Myeloma. Cancers 2020, 12, 173. [Google Scholar] [CrossRef]
- Braig, F.; Kriegs, M.; Voigtlaender, M.; Habel, B.; Grob, T.; Biskup, K.; Blanchard, V.; Sack, M.; Thalhammer, A.; Ben Batalla, I.; et al. Cetuximab Resistance in Head and Neck Cancer Is Mediated by EGFR-K521 Polymorphism. Cancer Res. 2016, 77, 1188–1199. [Google Scholar] [CrossRef] [PubMed]
- Sok, J.C.; Coppelli, F.M.; Thomas, S.; Lango, M.N.; Xi, S.; Hunt, J.L.; Freilino, M.L.; Graner, M.W.; Wikstrand, C.J.; Bigner, D.D.; et al. Mutant Epidermal Growth Factor Receptor (EGFRvIII) Contributes to Head and Neck Cancer Growth and Resistance to EGFR Targeting. Clin. Cancer Res. 2006, 12, 5064–5073. [Google Scholar] [CrossRef]
- Liao, H.-W.; Hsu, J.-M.; Xia, W.; Wang, H.-L.; Wang, Y.-N.; Chang, W.-C.; Arold, S.T.; Chou, C.-K.; Tsou, P.-H.; Yamaguchi, H.; et al. PRMT1-mediated methylation of the EGF receptor regulates signaling and cetuximab response. J. Clin. Investig. 2015, 125, 4529–4543. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Chen, Y.; Ren, W.; Hu, S.; Tan, Z.; Wang, Y.; Chen, Y.; Zhang, J.; Wu, J.; Li, T.; et al. Transcriptome Alterations in Liver Metastases of Colorectal Cancer After Acquired Resistance to Cetuximab. Cancer Genom. Proteom. 2019, 16, 207–219. [Google Scholar] [CrossRef] [PubMed]
- Vidoni, C.; Ferraresi, A.; Secomandi, E.; Vallino, L.; Dhanasekaran, D.N.; Isidoro, C. Epigenetic targeting of autophagy for cancer prevention and treatment by natural compounds. Semin. Cancer Biol. 2020, 66, 34–44. [Google Scholar] [CrossRef]
- Thongchot, S.; Ferraresi, A.; Vidoni, C.; Loilome, W.; Yongvanit, P.; Namwat, N.; Isidoro, C. Erratum to “Resveratrol interrupts the pro-invasive communication between Cancer associated Fibroblasts and Cholangiocarcinoma cells” [Cancer Letters 430C (2018) 160–171]. Cancer Lett. 2018, 434, 206–207. [Google Scholar] [CrossRef]
- Buhrmann, C.; Shayan, P.; Brockmueller, A.; Shakibaei, M. Resveratrol Suppresses Cross-Talk between Colorectal Cancer Cells and Stromal Cells in Multicellular Tumor Microenvironment: A Bridge between In Vitro and In Vivo Tumor Microenvironment Study. Molecules 2020, 25, 4292. [Google Scholar] [CrossRef]
- Brâkenhielm, E.; Cao, R.; Cao, Y. Suppression of angiogenesis, tumor growth, and wound healing by resveratrol, a natural compound in red wine and grapes. FASEB J. 2001, 15, 1798–1800. [Google Scholar] [CrossRef]
- Wu, H.; He, L.; Shi, J.; Hou, X.; Zhang, H.; Zhang, X.; An, Q.; Fan, F. Resveratrol inhibits VEGF-induced angiogenesis in human endothelial cells associated with suppression of aerobic glycolysis via modulation of PKM2 nuclear translocation. Clin. Exp. Pharmacol. Physiol. 2018, 45, 1265–1273. [Google Scholar] [CrossRef]
- Cottart, C.-H.; Nivet-Antoine, V.; Laguillier-Morizot, C.; Beaudeux, J.-L. Resveratrol bioavailability and toxicity in humans. Mol. Nutr. Food Res. 2009, 54, 7–16. [Google Scholar] [CrossRef]
- Rössler, J.; Odenthal, E.; Geoerger, B.; Gerstenmeyer, A.; Lagodny, J.; Niemeyer, C.M.; Vassal, G. EGFR inhibition using gefitinib is not active in neuroblastoma cell lines. Anticancer. Res. 2009, 29, 1327–1333. [Google Scholar]
- Thongchot, S.; Vidoni, C.; Ferraresi, A.; Loilome, W.; Khuntikeo, N.; Sangkhamanon, S.; Titapun, A.; Isidoro, C.; Namwat, N. Cancer-Associated Fibroblast-Derived IL-6 Determines Unfavorable Prognosis in Cholangiocarcinoma by Affecting Autophagy-Associated Chemoresponse. Cancers 2021, 13, 2134. [Google Scholar] [CrossRef]
- Salwa, A.; Ferraresi, A.; Chinthakindi, M.; Vallino, L.; Vidoni, C.; Dhanasekaran, D.; Isidoro, C. BECN1 and BRCA1 Deficiency Sensitizes Ovarian Cancer to Platinum Therapy and Confers Better Prognosis. Biomedicines 2021, 9, 207. [Google Scholar] [CrossRef]






| shRNA | Oligo Strand | Sequence |
|---|---|---|
| 879 | Top | 5′-caccGCACAGACTCCAAGTATTACACGAATGTAATACTTGGAGTCTGTGC |
| Bottom | 5′-aaaaGCACAGACTCCAAGTATTACATTCGTGTAATACTTGGAGTCTGTGC | |
| 486 | Top | 5′-caccGGATCCACCACAAGTACAACACGAATGTTGTACTTGTGGTGGATCC |
| Bottom | 5′-aaaaGGATCCACCACAAGTACAACATTCGTGTTGTACTTGTGGTGGATCC | |
| Ohri | Top | 5′-caccGGCAAAGGCTACAAGCTGTTTCAAGAGAACAGCTTGTAGCCTTTGCC |
| Bottom | 5′-aaaaGGCAAAGGCTACAAGCTGTTCTCTTGAAACAGCTTGTAGCCTTTGCC |
| Clone | Doubling Time |
|---|---|
| Co Sham | 38.6 ± 4.8 |
| EGF Sham | 29.9 ± 1.9 |
| Co KD-CD | 26.8 ± 1.25 |
| EGF KD-CD | 19.05 ± 1.23 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Secomandi, E.; Salwa, A.; Vidoni, C.; Ferraresi, A.; Follo, C.; Isidoro, C. High Expression of the Lysosomal Protease Cathepsin D Confers Better Prognosis in Neuroblastoma Patients by Contrasting EGF-Induced Neuroblastoma Cell Growth. Int. J. Mol. Sci. 2022, 23, 4782. https://doi.org/10.3390/ijms23094782
Secomandi E, Salwa A, Vidoni C, Ferraresi A, Follo C, Isidoro C. High Expression of the Lysosomal Protease Cathepsin D Confers Better Prognosis in Neuroblastoma Patients by Contrasting EGF-Induced Neuroblastoma Cell Growth. International Journal of Molecular Sciences. 2022; 23(9):4782. https://doi.org/10.3390/ijms23094782
Chicago/Turabian StyleSecomandi, Eleonora, Amreen Salwa, Chiara Vidoni, Alessandra Ferraresi, Carlo Follo, and Ciro Isidoro. 2022. "High Expression of the Lysosomal Protease Cathepsin D Confers Better Prognosis in Neuroblastoma Patients by Contrasting EGF-Induced Neuroblastoma Cell Growth" International Journal of Molecular Sciences 23, no. 9: 4782. https://doi.org/10.3390/ijms23094782
APA StyleSecomandi, E., Salwa, A., Vidoni, C., Ferraresi, A., Follo, C., & Isidoro, C. (2022). High Expression of the Lysosomal Protease Cathepsin D Confers Better Prognosis in Neuroblastoma Patients by Contrasting EGF-Induced Neuroblastoma Cell Growth. International Journal of Molecular Sciences, 23(9), 4782. https://doi.org/10.3390/ijms23094782








