Fine-Tuning Modulation of Oxidation-Mediated Posttranslational Control of Bradyrhizobium diazoefficiens FixK2 Transcription Factor
Abstract
:1. Introduction
2. Results
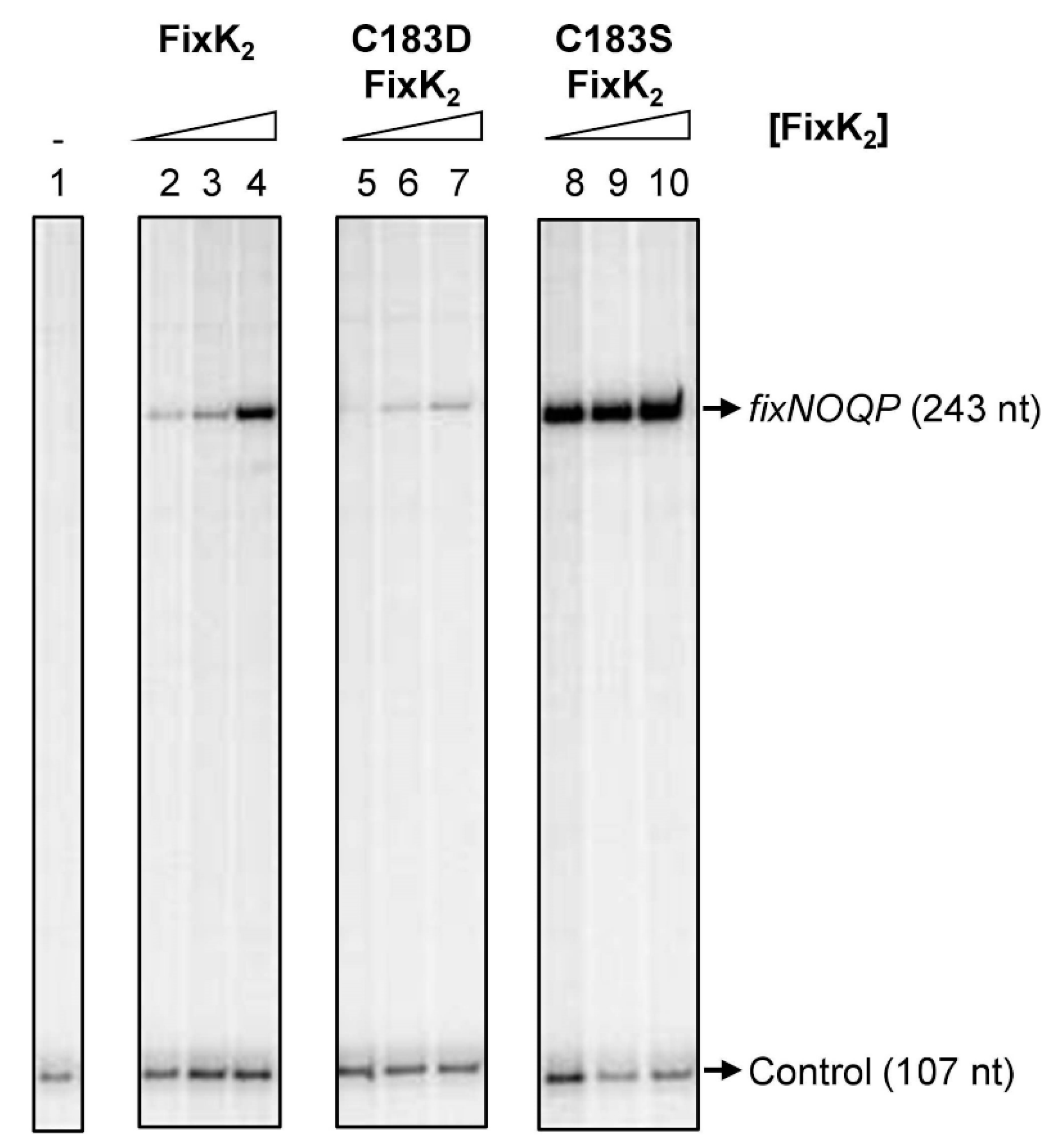
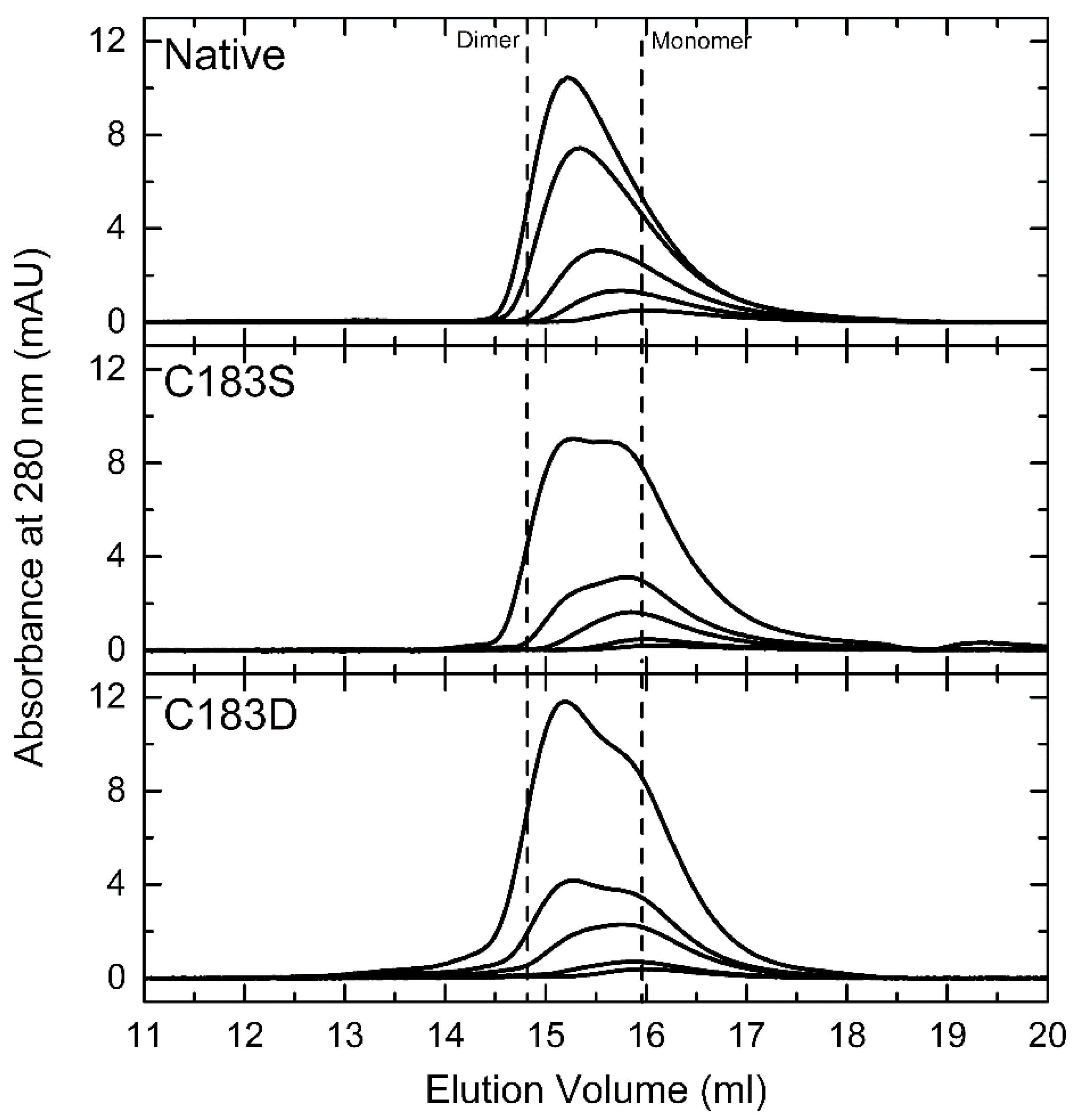
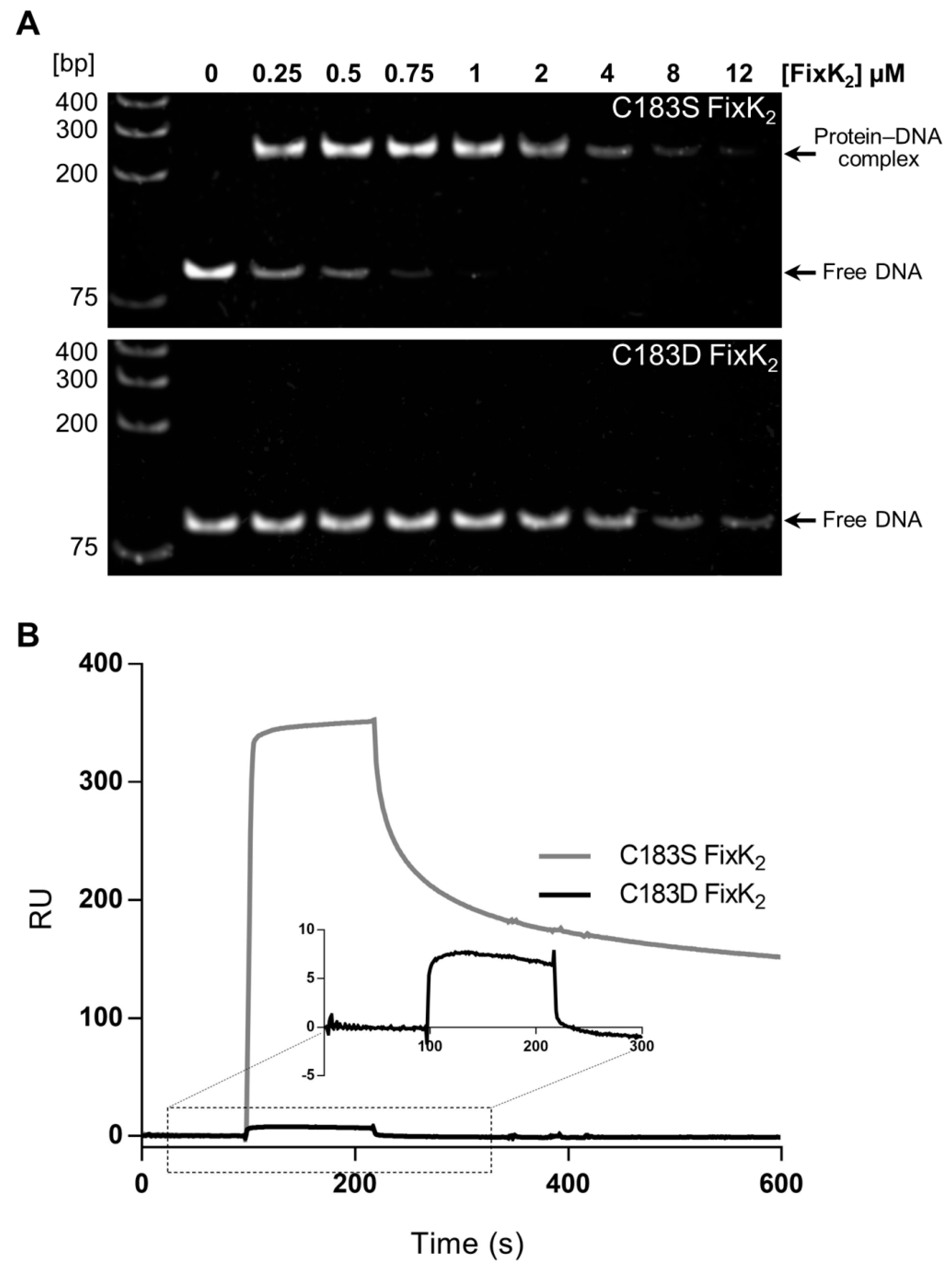
2.1. Assessing the Impact of C183D Exchange in FixK2 on In Vitro Transcription Activation Activity and Protein–DNA Interaction Ability
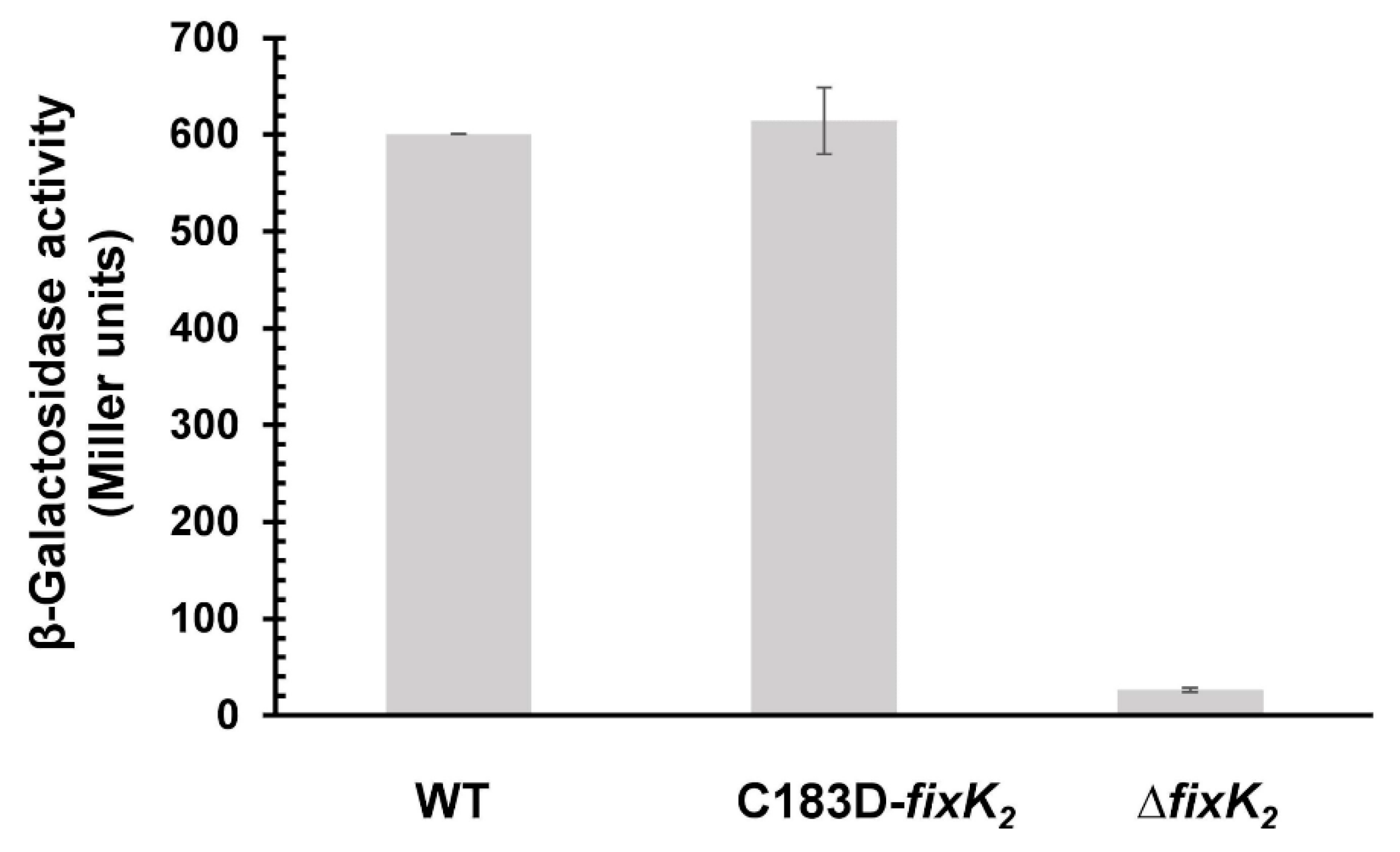
2.2. In Vivo Effects of C183 to Aspartic Acid Replacement in FixK2
2.3. Appraisal of the Impact of the C183D Mutation on a Wider FixK2-Mediated Control Landscape
3. Discussion
4. Materials and Methods
4.1. Strains, Plasmids, and Primers
4.2. Media and Growth Conditions
4.3. Strain and Plasmid Construction
4.4. β-Galactosidase Activity Assays
4.5. Plant Infection Test and Physiological Analyses
4.6. Overexpression and Purification of Non-Tagged FixK2 Protein Variants
4.7. In Vitro Transcription Activation Assay
4.8. Size-Exclusion Chromatography Experiments
4.9. Electrophoretic Mobility Shift DNA Assays
4.10. Surface Plasmon Resonance Analyses
4.11. Immunoblot Detection of FixK2
4.12. Determination of Protein Concentration
4.13. Microarray Sample Preparation and Data Analyses
4.14. Biocomputing Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Galloway, J.N.; Townsend, A.R.; Erisman, J.W.; Bekunda, M.; Cai, Z.; Freney, J.R.; Martinelli, L.A.; Seitzinger, S.P.; Sutton, M.A. Transformation of the nitrogen cycle: Recent trends, questions, and potential solutions. Science 2008, 320, 889–892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galloway, J.N.; Leach, A.M.; Bleeker, A.; Erisman, J.W. A chronology of human understanding of the nitrogen cycle. Philos. Trans. R. Soc. B Biol. Sci. 2013, 368, 20130120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez-Espinosa, R.M.; Cole, J.A.; Richardson, D.J.; Watmough, N.J. Enzymology and ecology of the nitrogen cycle. Biochem. Soc. Trans. 2011, 39, 175–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sprent, J.I.; Ardley, J.; James, E.K. Biogeography of nodulated legumes and their nitrogen-fixing symbionts. New Phytol. 2017, 215, 40–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dixon, R.; Kahn, D. Genetic regulation of biological nitrogen fixation. Nat. Rev. Microbiol. 2004, 2, 621–631. [Google Scholar] [CrossRef] [PubMed]
- Terpolilli, J.J.; Hood, G.A.; Poole, P.S. What determines the efficiency of N2-fixing Rhizobium-legume symbioses? Adv. Microb. Physiol. 2012, 60, 325–389. [Google Scholar] [PubMed]
- Chang, C.; Damiani, I.; Puppo, A.; Frendo, P. Redox changes during the legume-Rhizobium symbiosis. Mol. Plant 2009, 2, 370–377. [Google Scholar] [CrossRef]
- Damiani, I.; Pauly, N.; Puppo, A.; Brouquisse, R.; Boscari, A. Reactive oxygen species and nitric oxide control early steps of the legume—Rhizobium symbiotic interaction. Front. Plant Sci. 2016, 7, 454. [Google Scholar] [CrossRef]
- Poole, P.; Ramachandran, V.; Terpolilli, J. Rhizobia: From saprophytes to endosymbionts. Nat. Rev. Microbiol. 2018, 16, 291–303. [Google Scholar] [CrossRef]
- Torres, M.J.; Simon, J.; Rowley, G.; Bedmar, E.J.; Richardson, D.J.; Gates, A.J.; Delgado, M.J. Nitrous oxide metabolism in nitrate-reducing bacteria: Physiology and regulatory mechanisms. Adv. Microb. Physiol. 2016, 68, 353–432. [Google Scholar]
- Rutten, P.J.; Poole, P.S. Oxygen regulatory mechanisms of nitrogen fixation in rhizobia. Adv. Microb. Physiol. 2019, 75, 325–389. [Google Scholar] [PubMed]
- Santos, M.S.; Nogueira, M.A.; Hungria, M. Microbial inoculants: Reviewing the past, discussing the present and previewing an outstanding future for the use of beneficial bacteria in agriculture. AMB Express 2019, 9, 205. [Google Scholar] [CrossRef] [PubMed]
- Delamuta, J.R.M.; Ribeiro, R.A.; Ormeño-Orrillo, E.; Melo, I.S.; Martínez-Romero, E.; Hungria, M. Polyphasic evidence supporting the reclassification of Bradyrhizobium japonicum group Ia strains as Bradyrhizobium diazoefficiens sp. nov. Int. J. Syst. Evol. Microbiol. 2013, 63, 3342–3351. [Google Scholar] [CrossRef] [PubMed]
- Mesa, S.; Alché, J.D.; Bedmar, E.J.; Delgado, M.J. Expression of nir, nor and nos denitrification genes from Bradyrhizobium japonicum in soybean root nodules. Physiol. Plant. 2004, 120, 205–211. [Google Scholar] [CrossRef]
- Bedmar, E.J.; Robles, E.F.; Delgado, M.J. The complete denitrification pathway of the symbiotic, nitrogen-fixing bacterium Bradyrhizobium japonicum. Biochem. Soc. Trans. 2005, 33, 141–144. [Google Scholar] [CrossRef] [Green Version]
- Bedmar, E.J.; Correa, D.; Torres, M.J.; Delgado, M.J.; Mesa, S. Ecology of Denitrification in Soils and Plant-Associated Bacteria. In Beneficial Plant-Microbial Interactions: Ecology and Applications, 1st ed.; Rodelas González, M.B., González-López, J., Eds.; CRC Press: Boca Raton, FL, USA, 2014; pp. 164–182. [Google Scholar]
- Sciotti, M.A.; Chanfon, A.; Hennecke, H.; Fischer, H.M. Disparate oxygen responsiveness of two regulatory cascades that control expression of symbiotic genes in Bradyrhizobium japonicum. J. Bacteriol. 2003, 185, 5639–5642. [Google Scholar] [CrossRef] [Green Version]
- Fernández, N.; Cabrera, J.J.; Salazar, S.; Parejo, S.; Rodríguez, M.C.; Lindemann, A.; Bonnet, M.; Hennecke, H.; Bedmar, E.J.; Mesa, S. Molecular Determinants of Negative Regulation of the Bradyrhizobium diazoefficiens Transcription Factor FixK2. In Biological Nitrogen Fixation and Beneficial Plant-Microbe Interactions, 1st ed.; González-Andrés, F., James, E.K., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 57–72. [Google Scholar]
- Mesa, S.; Hauser, F.; Friberg, M.; Malaguti, E.; Fischer, H.M.; Hennecke, H. Comprehensive assessment of the regulons controlled by the FixLJ-FixK2-FixK1 cascade in Bradyrhizobium japonicum. J. Bacteriol. 2008, 190, 6568–6579. [Google Scholar] [CrossRef] [Green Version]
- Dufour, Y.S.; Kiley, P.J.; Donohue, T.J. Reconstruction of the core and extended regulons of global transcription factors. PLoS Genet. 2010, 6, e1001027. [Google Scholar] [CrossRef] [Green Version]
- Körner, H.; Sofia, H.J.; Zumft, W.G. Phylogeny of the bacterial superfamily of Crp-Fnr transcription regulators: Exploiting the metabolic spectrum by controlling alternative gene programs. FEMS Microbiol. Rev. 2003, 27, 559–592. [Google Scholar] [CrossRef] [Green Version]
- Matsui, M.; Tomita, M.; Kanai, A. Comprehensive computational analysis of bacterial CRP/FNR superfamily and its target motifs reveals stepwise evolution of transcriptional networks. Genome Biol. Evol. 2013, 5, 267–282. [Google Scholar] [CrossRef]
- Bonnet, M.; Kurz, M.; Mesa, S.; Briand, C.; Hennecke, H.; Grutter, M.G. The structure of Bradyrhizobium japonicum transcription factor FixK2 unveils sites of DNA binding and oxidation. J. Biol. Chem. 2013, 288, 14238–14246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cabrera, J.J.; Jiménez-Leiva, A.; Tomás-Gallardo, L.; Parejo, S.; Casado, S.; Torres, M.J.; Bedmar, E.J.; Delgado, M.J.; Mesa, S. Dissection of FixK2 protein-DNA interaction unveils new insights into Bradyrhizobium diazoefficiens lifestyles control. Environ. Microbiol. 2021, 23, 6194–6209. [Google Scholar] [CrossRef] [PubMed]
- Browning, D.F.; Busby, S.J. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2004, 2, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Nellen-Anthamatten, D.; Rossi, P.; Preisig, O.; Kullik, I.; Babst, M.; Fischer, H.M.; Hennecke, H. Bradyrhizobium japonicum FixK2, a crucial distributor in the FixLJ-dependent regulatory cascade for control of genes inducible by low oxygen levels. J. Bacteriol. 1998, 180, 5251–5255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reutimann, L.; Mesa, S.; Hennecke, H. Autoregulation of fixK2 gene expression in Bradyrhizobium japonicum. Mol. Genet. Genom. 2010, 284, 25–32. [Google Scholar] [CrossRef] [Green Version]
- Mesa, S.; Reutimann, L.; Fischer, H.M.; Hennecke, H. Posttranslational control of transcription factor FixK2, a key regulator for the Bradyrhizobium japonicum-soybean symbiosis. Proc. Natl. Acad. Sci. USA. 2009, 106, 21860–21865. [Google Scholar] [CrossRef] [Green Version]
- Bonnet, M.; Stegmann, M.; Maglica, Ž.; Stiegeler, E.; Weber-Ban, E.; Hennecke, H.; Mesa, S. FixK2, a key regulator in Bradyrhizobium japonicum, is a substrate for the protease ClpAP in vitro. FEBS Lett. 2013, 587, 88–93. [Google Scholar] [CrossRef] [Green Version]
- Fernández, N.; Cabrera, J.J.; Varadarajan, A.R.; Lutz, S.; Ledermann, R.; Roschitzki, B.; Eberl, L.; Bedmar, E.J.; Fischer, H.M.; Pessi, G.; et al. An integrated systems approach unveils new aspects of microoxia-mediated regulation in Bradyrhizobium diazoefficiens. Front. Microbiol. 2019, 10, 924. [Google Scholar] [CrossRef] [Green Version]
- Matamoros, M.A.; Dalton, D.A.; Ramos, J.; Clemente, M.R.; Rubio, M.C.; Becana, M. Biochemistry and molecular biology of antioxidants in the rhizobia-legume symbiosis. Plant Physiol. 2003, 133, 499–509. [Google Scholar] [CrossRef] [Green Version]
- Bonnet, M. Biochemical Studies on FixK2, a Global Regulatory Protein from Bradyrhizobium japonicum: Proteolytic Control and Attempts at Crystallization. Ph.D. Thesis, ETH-Zürich, Zürich, Switzerland, 2011. [Google Scholar] [CrossRef]
- Mesa, S.; Ucurum, Z.; Hennecke, H.; Fischer, H.M. Transcription activation in vitro by the Bradyrhizobium japonicum regulatory protein FixK2. J. Bacteriol. 2005, 187, 3329–3338. [Google Scholar] [CrossRef] [Green Version]
- Bueno, E.; Robles, E.F.; Torres, M.J.; Krell, T.; Bedmar, E.J.; Delgado, M.J.; Mesa, S. Disparate response to microoxia and nitrogen oxides of the Bradyrhizobium japonicum napEDABC, nirK and norCBQD denitrification genes. Nitric Oxide 2017, 68, 137–149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torres, M.J.; Bueno, E.; Jiménez-Leiva, A.; Cabrera, J.J.; Bedmar, E.J.; Mesa, S.; Delgado, M.J. FixK2 is the main transcriptional activator of Bradyrhizobium diazoefficiens nosRZDYFLX genes in response to low oxygen. Front. Microbiol. 2017, 8, 1621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pessi, G.; Ahrens, C.H.; Rehrauer, H.; Lindemann, A.; Hauser, F.; Fischer, H.M.; Hennecke, H. Genome-wide transcript analysis of Bradyrhizobium japonicum bacteroids in soybean root nodules. Mol. Plant-Microbe Interact. 2007, 20, 1353–1363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hauser, F.; Pessi, G.; Friberg, M.; Weber, C.; Rusca, N.; Lindemann, A.; Fischer, H.M.; Hennecke, H. Dissection of the Bradyrhizobium japonicum NifA+σ54 regulon, and identification of a ferredoxin gene (fdxN) for symbiotic nitrogen fixation. Mol. Genet. Genom. 2007, 278, 255–271. [Google Scholar] [CrossRef]
- Kaneko, T.; Nakamura, Y.; Sato, S.; Minamisawa, K.; Uchiumi, T.; Sasamoto, S.; Watanabe, A.; Idesawa, K.; Iriguchi, M.; Kawashima, K.; et al. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA 110. DNA Res. 2002, 9, 189–197. [Google Scholar] [CrossRef]
- Mesa, S.; Hennecke, H.; Fischer, H.M. A multitude of CRP/FNR-like transcription proteins in Bradyrhizobium japonicum. Biochem. Soc. Trans. 2006, 34, 156–159. [Google Scholar] [CrossRef] [Green Version]
- Agari, Y.; Kashihara, A.; Yokoyama, S.; Kuramitsu, S.; Shinkai, A. Global gene expression mediated by Thermus thermophilus SdrP, a CRP/FNR family transcriptional regulator. Mol. Microbiol. 2008, 70, 60–75. [Google Scholar] [CrossRef]
- Eiting, M.; Hagelüken, G.; Schubert, W.D.; Heinz, D.W. The mutation G145S in PrfA, a key virulence regulator of Listeria monocytogenes, increases DNA-binding affinity by stabilizing the HTH motif. Mol. Microbiol. 2005, 56, 433–446. [Google Scholar] [CrossRef]
- Freitag, N.E.; Port, G.C.; Miner, M.D. Listeria monocytogenes—From saprophyte to intracellular pathogen. Nat. Rev. Microbiol. 2009, 7, 623–628. [Google Scholar] [CrossRef]
- Reniere, M.L.; Whiteley, A.T.; Hamilton, K.L.; John, S.M.; Lauer, P.; Brennan, R.G.; Portnoy, D.A. Glutathione activates virulence gene expression of an intracellular pathogen. Nature 2015, 517, 170–173. [Google Scholar] [CrossRef] [Green Version]
- Hall, M.; Grundström, C.; Begum, A.; Lindberg, M.J.; Sauer, U.H.; Almqvist, F.; Johansson, J.; Sauer-Eriksson, A.E. Structural basis for glutathione-mediated activation of the virulence regulatory protein PrfA in Listeria. Proc. Natl. Acad. Sci. USA. 2016, 113, 14733–14738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osorio, H. Caracterización de la proteína FNR de Acidithiobacillus ferrooxidans, estructural y funcionalmente. Ph.D Thesis, University Andres Bello, Santiago de Chile, Chile, 2012. Available online: http://repositorio.unab.cl/xmlui/handle/ria/1225 (accessed on 21 July 2021).
- Soberón-Chávez, G.; Alcaraz, L.D.; Morales, E.; Ponce-Soto, G.Y.; Servín-González, L. The transcriptional regulators of the CRP family regulate different essential bacterial functions and can be inherited vertically and horizontally. Front. Microbiol. 2017, 8, 959. [Google Scholar] [CrossRef] [PubMed]
- Scortti, M.; Monzo, H.J.; Lacharme-Lora, L.; Lewis, D.A.; Vazquez-Boland, J.A. The PrfA virulence regulon. Microbes Infect. 2007, 9, 1196–1207. [Google Scholar] [CrossRef] [PubMed]
- Mettert, E.L.; Kiley, P.J. ClpXP-dependent proteolysis of FNR upon loss of its O2-sensing [4Fe-4S] cluster. J. Mol. Biol. 2005, 354, 220–232. [Google Scholar] [CrossRef]
- Pan, Q.; Shan, Y.; Yan, A. A region at the C-terminus of the Escherichia coli global transcription factor FNR negatively mediates its degradation by the ClpXP protease. Biochemistry 2012, 51, 5061–5071. [Google Scholar] [CrossRef] [PubMed]
- Simon, R.; Priefer, U.; Pühler, A. Vector plasmids for in-vivo and in-vitro manipulations of Gram-negative bacteria. In Molecular Genetics of the Bacteria–Plant Interaction, 1st ed.; Pühler, A., Ed.; Springer: Berlin/Heidelberg, Germany, 1983; pp. 98–106. [Google Scholar]
- Regensburger, B.; Hennecke, H. RNA polymerase from Rhizobium japonicum. Arch. Microbiol. 1983, 135, 103–109. [Google Scholar] [CrossRef]
- Zufferey, R.; Preisig, O.; Hennecke, H.; Thöny-Meyer, L. Assembly and function of the cytochrome cbb3 oxidase subunits in Bradyrhizobium japonicum. J. Biol. Chem. 1996, 271, 9114–9119. [Google Scholar] [CrossRef] [Green Version]
- Kovach, M.E.; Elzer, P.H.; Hill, D.S.; Robertson, G.T.; Farris, M.A.; Roop, R.M., II; Peterson, K.M. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 1995, 166, 175–176. [Google Scholar] [CrossRef]
- Schäfer, A.; Tauch, A.; Jager, W.; Kalinowski, J.; Thierbach, G.; Pühler, A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: Selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 1994, 145, 69–73. [Google Scholar] [CrossRef]
- Miller, J.H. Experiments in Molecular Genetics, 1st ed.; Cold Spring Harbor Laboratory: New York, NY, USA, 1972. [Google Scholar]
- Daniel, R.M.; Appleby, C.A. Anaerobic-nitrate, symbiotic and aerobic growth of Rhizobium japonicum: Effects on cytochrome P450, other haemoproteins, nitrate and nitrite reductases. Biochim. Biophys. Acta. 1972, 275, 347–354. [Google Scholar] [CrossRef]
- Cabrera, J.J.; Salas, A.; Torres, M.J.; Bedmar, E.J.; Richardson, D.J.; Gates, A.J.; Delgado, M.J. An integrated biochemical system for nitrate assimilation and nitric oxide detoxification in Bradyrhizobium japonicum. Biochem. J. 2016, 473, 297–309. [Google Scholar] [CrossRef] [Green Version]
- Tortosa, G.; Parejo, S.; Cabrera, J.J.; Bedmar, E.J.; Mesa, S. Oxidative stress produced by paraquat reduces nitrogen fixation in soybean-Bradyrhizobium diazoefficiens symbiosis by decreasing nodule functionality. Nitrogen 2021, 2, 30–40. [Google Scholar] [CrossRef]
- Tortosa, G.; Pacheco, P.J.; Hidalgo-García, A.; Granados, A.; Delgado, A.; Mesa, S.; Bedmar, E.J.; Delgado, M.J. Copper modulates nitrous oxide emissions from soybean root nodules. Environ. Exp. Bot. 2020, 180, 104262. [Google Scholar] [CrossRef]
- LaRue, T.A.; Child, J.J. Sensitive fluorometric assay for leghemoglobin. Anal. Biochem. 1979, 92, 11–15. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Jiménez-Leiva, A.; Cabrera, J.J.; Bueno, E.; Torres, M.J.; Salazar, S.; Bedmar, E.J.; Delgado, M.J.; Mesa, S. Expanding the regulon of the Bradyrhizobium diazoefficiens NnrR transcription factor: New insights into the denitrification pathway. Front. Microbiol. 2019, 10, 1926. [Google Scholar] [CrossRef] [Green Version]
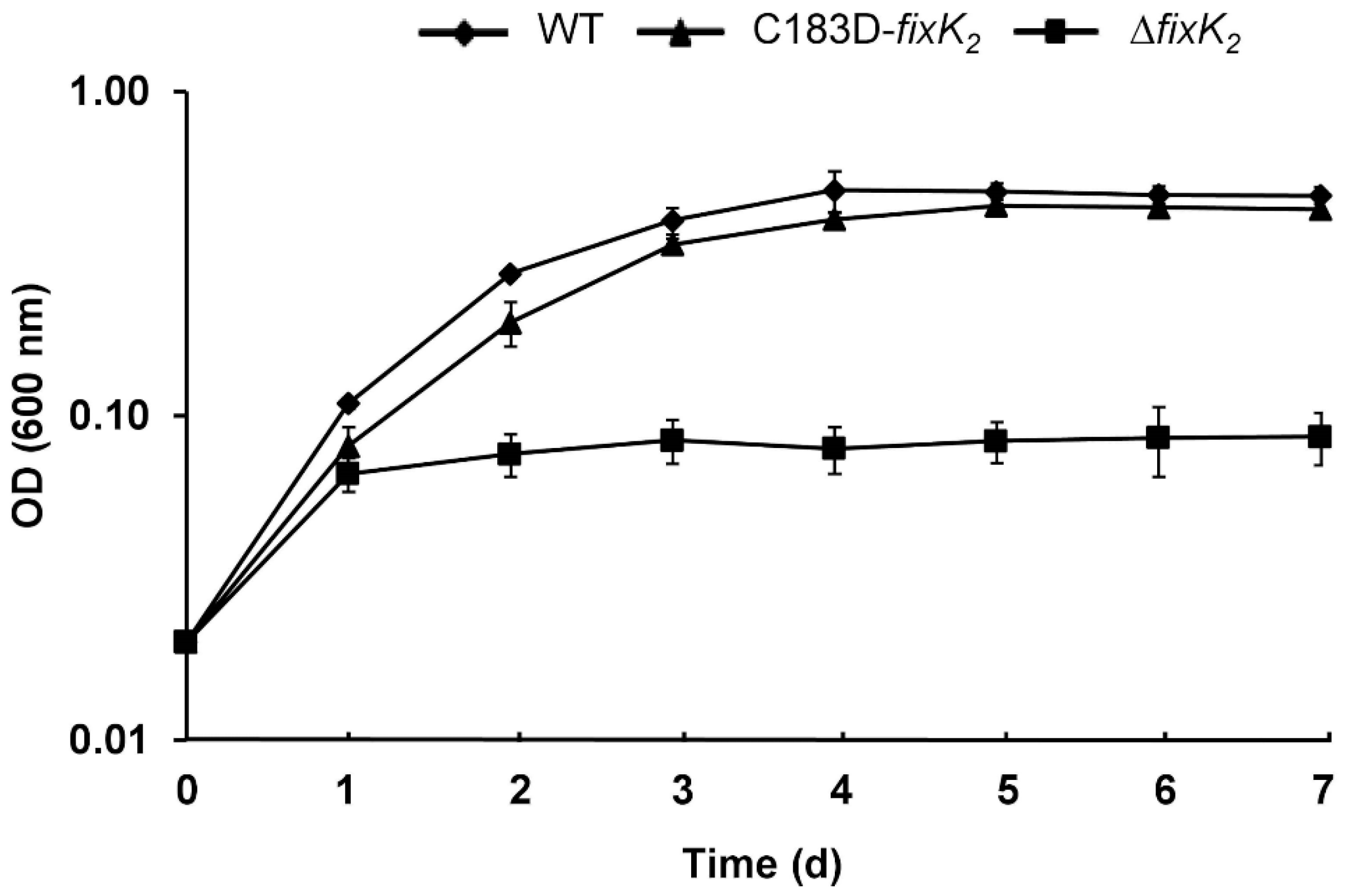
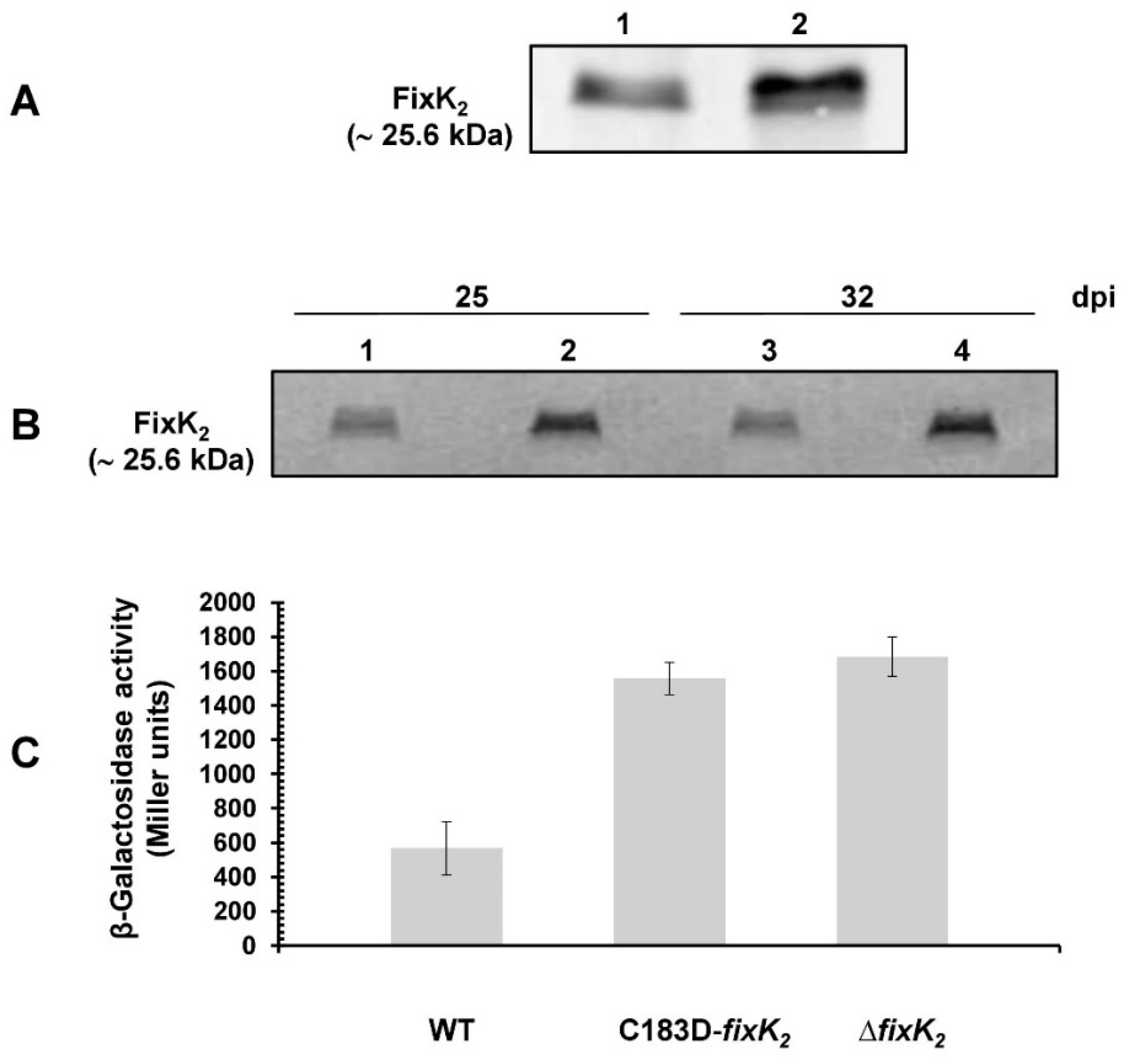
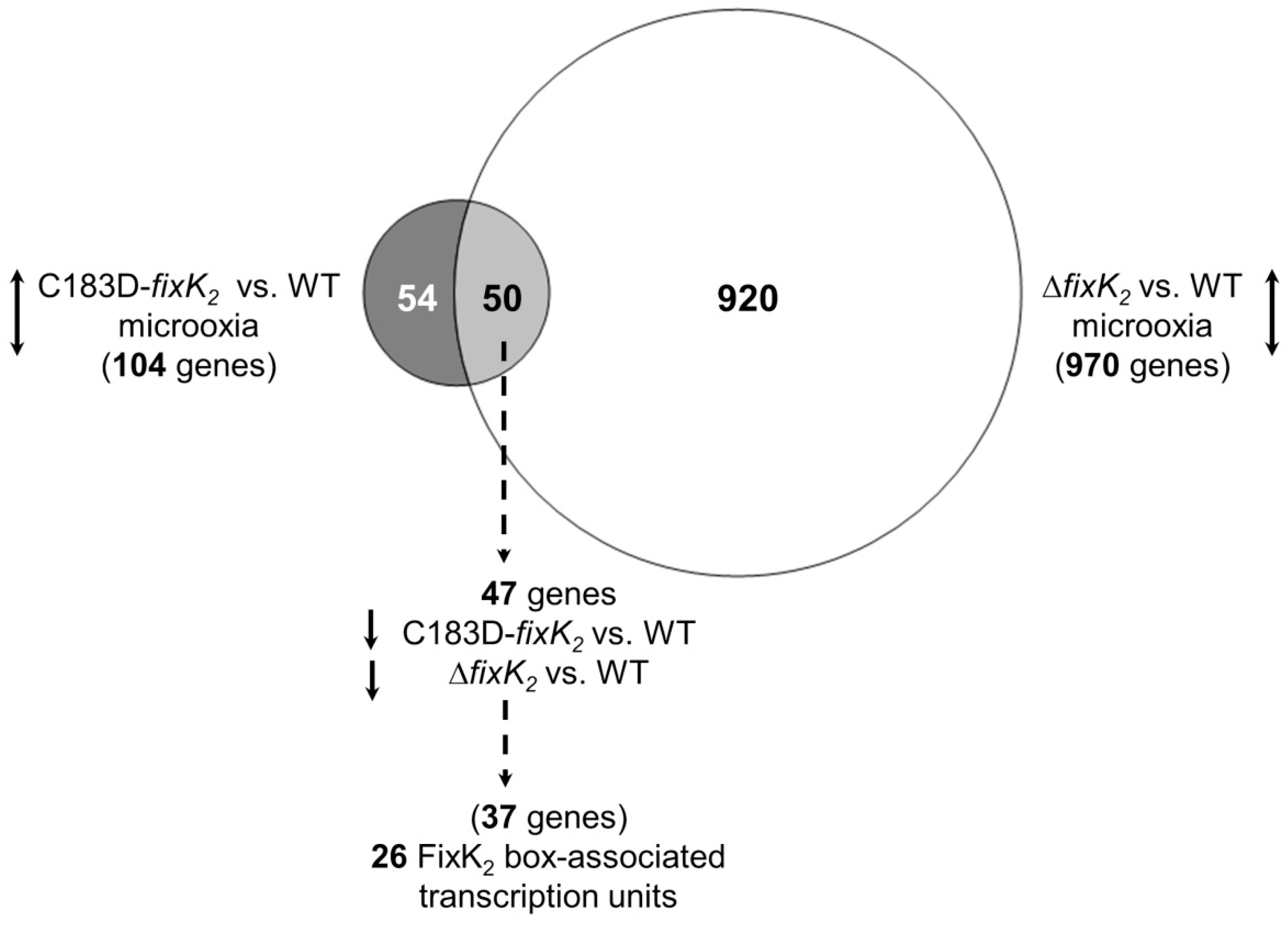

| Parameters | WT | ΔfixK2 | C183D-fixK2 |
|---|---|---|---|
| 25 dpi | |||
| SDW (g) | (0.54 ± 0.13) | (0.59 ± 0.10) | (0.47 ± 0.11) |
| N (mg) | (12.60 ± 4.0) | (4.90 ± 1.2) | (12.20 ± 3.90) |
| NN | (38.30 ± 4.5) | (34.50 ± 3.5) | (32.20 ± 6.70) |
| NDW (mg) | (38.67 ± 7.58) | (16.83 ± 1.94) | (32.50 ± 6.63) |
| NDW/NN (mg) | (1.03 ± 0.24) | (0.49 ± 0.03) | (1.03 ± 0.22) |
| Lb (mg Lb · g NFW−1) | (11.83 ± 0.59) | (0.11 ± 0.02) | (10.08 ± 0.35) |
| 32 dpi | |||
| SDW (g) | (0.92 ± 0.14) | (0.68 ± 0.13) | (0.77 ± 0.01) |
| N (mg) | (18.60 ± 5.90) | (5.00 ± 1.30) | (21.00 ± 5.80) |
| NN | (31.30 ± 13.60) | (50.70 ± 13.60) | (27.80 ± 4.00) |
| NDW (mg) | (38.17 ± 5.04) | (25.33 ± 6.31) | (32.83 ± 4.96) |
| NFW/NN (mg) | (1.44 ± 0.65) | (0.50 ± 0.05) | (1.21 ± 0.28) |
| Lb (mg Lb · g NFW−1) | (11.51 ± 0.24) | (0.15 ± 0.02) | (11.23 ± 0.71) |
| Query a | FC (C183D-fixK2 vs. WT) b | FC (ΔfixK2 vs. WT) c | Locus_Tag d | Gene Name e | Product f | Position g | Motif h | Predicted Operon Structure i |
|---|---|---|---|---|---|---|---|---|
| bll0330 | −2.4 | −11.0 | Bdiaspc4_01315 | - | DNA-binding response regulator | −106 | TTGACCTGGATCAA | - |
| bll0818 | −2.1 | −9.3 | Bdiaspc4_03880 | - | hypothetical protein | −66 | TTGATCCCGGTCAA | - |
| blr1289 | −3.2 | −23.1 | Bdiaspc4_06390 | - | oleate hydratase | −37 | TTGATCCAGCGCAA | - |
| bll2517 | −3.2 | −10.2 | Bdiaspc4_12930 | - | acetate/propionate family kinase | - | ||
| bll2518 | −2.6 | −10.0 | Bdiaspc4_12935 | - | phosphoketolase family protein | −89 | TTGACCTCACGCAA | bll2518-bll2517 |
| bll3115 | −9.6 | −30.6 | Bdiaspc4_16100 | - | MBL fold metallo-hydrolase | - | ||
| bll3117 | −2.4 | −6.6 | Bdiaspc4_16110 | - | thymidine phosphorylase family protein | −74 | ATGATCTGGGTCAA | bll3117-bll3116- bll3115 |
| blr3815 | −2.2 | −7.6 | Bdiaspc4_19720 | - | HAD family hydrolase | −287 | TTGACGTATCGCAA | - |
| blr4240 | −3.1 | −25.1 | Bdiaspc4_22005 | - | pyridoxamine 5’-phosphate oxidase family protein | −69 | TTGAGGTGCATCAA | blr4240-blr4241 |
| blr4241 | −2.9 | −83.3 | Bdiaspc4_22010 | - | cytochrome c | - | ||
| bll4412 | −3.2 | −20.7 | Bdiaspc4_22980 | - | translational machinery protein | −38 | TTGACCTGCGTCAA | - |
| bll4634 | −2.8 | −20.2 | Bdiaspc4_24260 | - | efflux RND transporter periplasmic adaptor subunit | −75 | TTGACCTAGCGCAA | - |
| blr4635 | −2.5 | −29.4 | Bdiaspc4_24265 | groL5, groEL5 | chaperonin GroEL | −150 | TTGCGCTAGGTCAA | - |
| blr4637 | −2.6 | −111.5 | Bdiaspc4_24275 | hspC2 | Hsp20/alpha crystallin family protein | −86 | TTGAGCAAAATCAA | - |
| bll4644 | −3.2 | −20.9 | Bdiaspc4_24320 | - | universal stress protein | −72 | TTGATTTCGGTCAA | - |
| bll4645 | −2.8 | −10.6 | Bdiaspc4_24325 | - | host attachment protein | −69 | TTGATCGGGATCAA | - |
| blr4652 | −3.1 | −95.2 | Bdiaspc4_24370 | - | nitroreductase | −48 | TTGATCGACATCAA | blr4652-blr4653- blr4654 |
| blr4653 | −2.8 | −16.8 | Bdiaspc4_24375 | dnaJ | J domain-containing protein | - | ||
| blr4654 | −2.8 | −30.0 | Bdiaspc4_24380 | - | hypothetical protein | - | ||
| blr4655 | −2.5 | −14.2 | Bdiaspc4_24385 | ppsA | phosphoenolpyruvate synthase | −47 | TTGACCTGCCTCAA | - |
| bsr6066 | −4.0 | −92.6 | Bdiaspc4_31980 | - | hypothetical protein | −105 | TTGACCTGTCTCAA | bsr6066-blr6067 |
| blr6067 | −2.7 | −20.9 | Bdiaspc4_31985 | - | phage holin family protein | - | ||
| bll6073 | −3.5 | −27.9 | Bdiaspc4_32015 | phaC2 | probable poly-beta-hydroxybutyrate polymerase | −81 | TTGATGCAGCTCAA | - |
| blr6074 | −2.7 | −90.9 | Bdiaspc4_32020 | - | CBS domain-containing protein | −143 | TTGAGCTGCATCAA | - |
| bll6525 | −2.1 | −7.7 | Bdiaspc4_34395 | - | hypothetical protein | −22 | TTGATCTGCATCAA | - |
| bll7086 | −2.3 | −97.1 | Bdiaspc4_37390 | hemN2 | oxygen-independent coproporphyrinogen III oxidase | −140 | TTGCGCGAGCGCAA | - |
| bsr7087 | −3.2 | −53.8 | Bdiaspc4_37395 | - | hypothetical protein | −115 | TTGCGCTCGCGCAA | bsr7087-blr7088 |
| blr7088 | −2.2 | −8.1 | Bdiaspc4_37400 | - | copper chaperone PCu(A)C | - | ||
| blr7345 | −2.9 | −16.8 | Bdiaspc4_38745 | - | hypothetical protein | −76 | TTGATCCGCATCAA | - |
| bll7986 | −2.1 | −5.6 | Bdiaspc4_42230 | - | HlyD family efflux transporter periplasmic adaptor subunit | - | ||
| bll7987 | −2.5 | −17.4 | Bdiaspc4_42235 | - | ABC transporter permease | - | ||
| bll7988 | −3.3 | −33.1 | Bdiaspc4_42240 | - | ABC transporter ATP-binding protein | −66 | CTGATCTAAATCAA | bll7988-bll7987- bll7986 |
| bll7989 | −2.6 | −5.3 | Bdiaspc4_42245 | mat | methionine adenosyltransferase | −203 | TTGAGCCAATGCAG | - |
| bll7990 | −3.2 | −19.7 | Bdiaspc4_42250 | - | hypothetical protein | - | ||
| bll7991 | −2.8 | −22.8 | Bdiaspc4_42255 | - | isoprenylcysteine carboxylmethyltransferase family protein | - | ||
| bsl7992 | −2.7 | −23.0 | Bdiaspc4_42260 | - | DUF2933 domain-containing protein | −59 | TTGATCTGCGTCAA | bsl7992-bll7991- bll7990 |
| bll7993 | −2.8 | −8.5 | Bdiaspc4_42265 | - | hypothetical protein | −60 | TTGAGGGATTGCAA | - |
| Strain or Plasmid | Description | Resistance | Source or Reference |
|---|---|---|---|
| Strains | |||
| E. coli | |||
| DH5α | supE44 ΔlacU169 (φ80 lacZ ΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Bethesda Research Laboratories Inc., Gaithersburg, MD, USA | |
| S17-1 | thi pro recA hsdR hsdM RP4Tc::Mu Km::Tn7 | Tpr Smr Spcr | [50] |
| ER2566 | fhuA2 lacZ::T7 gene1 [lon] ompT gal sulA11 R(mcr-73::miniTn10-TetS)2 [dcm] R(zgb-210::Tn10-TetS) endA1 Δ(mcrC-mrr)114::IS10 | NEB, USA | |
| B. diazoefficiens | |||
| 110spc4 | Wild type (WT) | Cmr Spcr | [51] |
| 9043 | ΔfixK2 | Cmr Spcr Smr | [26] |
| 1255 | C183D-fixK2 | Cmr Spcr | This work |
| 3604 | WT::fixNOQP’-‘lacZ | Cmr Spcr Tcr | [52] |
| 9043-3603 | ΔfixK2::fixNOQP’-‘lacZ | Cmr Spcr Tcr | This work |
| 1255-3603 | C183D-fixK2::fixNOQP’-‘lacZ | Cmr Spcr Tcr | This work |
| 1109 | WT:: fixK2′-‘lacZ | Cmr Spcr Tcr | [18] |
| 9043-1109 | ΔfixK2:: fixK2′-‘lacZ | Cmr Spcr Tcr | [18] |
| 1255-1109 | C183D-fixK2::fixK2′-‘lacZ | Cmr Spcr Tcr | This work |
| Plasmids | |||
| pTXB1 | Expression vector for the IMPACT protein purification system. It codes for a C-terminal thiol-cleavable Mxe GyrA-intein–chitin-binding domain (CBD) under T7 promoter control | Ampr | NEB, USA |
| pBBR1MCS-2 | lacPOZ mobRP4, low-copy-number cloning vector | Kmr | [53] |
| pK18mobsacB | Mobilizable pUC18 derivative, mob, sacB | Kmr | [54] |
| pRJ0051 | [pTXB1] with a 715-bp NdeI/SpeI fragment encoding C183S FixK2-intein fused in frame with the CBD of the vector | [32] | |
| pRJ0053 | [pTXB1] with a 715-bp NdeI/SpeI fragment encoding FixK2-Intein fused in frame with the CBD of the vector | Ampr | [32] |
| pRJ8848 | [pUC19] with a 2.288-kb SalI fragment encoding C183S FixK2 | Ampr | [23] |
| pMB1250 | [pRJ8848] with a 2.288-kb SalI fragment encoding C183D FixK2 | Ampr | This work |
| pMB1251 | [pBBR1MCS-2] with a 1.843-kb BamHI-XbaI fragment from pMB1250 | Kmr | This work |
| pMB1253 | [pTXB1] with a 715-bp NdeI/SpeI fragment from pMB1251 encoding C183D FixK2 | Ampr | This work |
| pRJ9041 | [pUC19] with a 2.288-kb SalI fragment encoding FixK2 | Ampr | [33] |
| pMB1256 | [pRJ9041] with a 3.965-kb NotI fragment from pMB1251 | Ampr Kmr | This work |
| pMB1254 | [pMB1256] Religation of a 4.974-kb BglII fragment | Ampr | This work |
| pMB1255 | [pK18mobsacB] with a 1.849-kb BamHI fragment from pMB1254 | Kmr | This work |
| pRJ3603 | [pSUP202pol2] ‘blr2761, blr2762 and fixNOQP’-‘lacZ on a 8.261-kb XhoI fragment | Tcr | [52] |
| pRJ9054 | [pSUP202] fixJ, bll2758 and fixK2′-‘lacZ on a 4.434-kb NsiI/DraI fragment | Tcr | [26] |
| pMB1109 | [pRJ9054] fixK2′-‘lacZ with a 136-bp SmaI fragment deletion within the bll2758 coding region | Tcr | [18] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parejo, S.; Cabrera, J.J.; Jiménez-Leiva, A.; Tomás-Gallardo, L.; Bedmar, E.J.; Gates, A.J.; Mesa, S. Fine-Tuning Modulation of Oxidation-Mediated Posttranslational Control of Bradyrhizobium diazoefficiens FixK2 Transcription Factor. Int. J. Mol. Sci. 2022, 23, 5117. https://doi.org/10.3390/ijms23095117
Parejo S, Cabrera JJ, Jiménez-Leiva A, Tomás-Gallardo L, Bedmar EJ, Gates AJ, Mesa S. Fine-Tuning Modulation of Oxidation-Mediated Posttranslational Control of Bradyrhizobium diazoefficiens FixK2 Transcription Factor. International Journal of Molecular Sciences. 2022; 23(9):5117. https://doi.org/10.3390/ijms23095117
Chicago/Turabian StyleParejo, Sergio, Juan J. Cabrera, Andrea Jiménez-Leiva, Laura Tomás-Gallardo, Eulogio J. Bedmar, Andrew J. Gates, and Socorro Mesa. 2022. "Fine-Tuning Modulation of Oxidation-Mediated Posttranslational Control of Bradyrhizobium diazoefficiens FixK2 Transcription Factor" International Journal of Molecular Sciences 23, no. 9: 5117. https://doi.org/10.3390/ijms23095117
APA StyleParejo, S., Cabrera, J. J., Jiménez-Leiva, A., Tomás-Gallardo, L., Bedmar, E. J., Gates, A. J., & Mesa, S. (2022). Fine-Tuning Modulation of Oxidation-Mediated Posttranslational Control of Bradyrhizobium diazoefficiens FixK2 Transcription Factor. International Journal of Molecular Sciences, 23(9), 5117. https://doi.org/10.3390/ijms23095117









