Oxidative Stress and Nuclear Reprogramming: A Pilot Study of the Effects of Reactive Oxygen Species on Architectural and Epigenetic Landscapes
Abstract
1. Introduction
2. Results
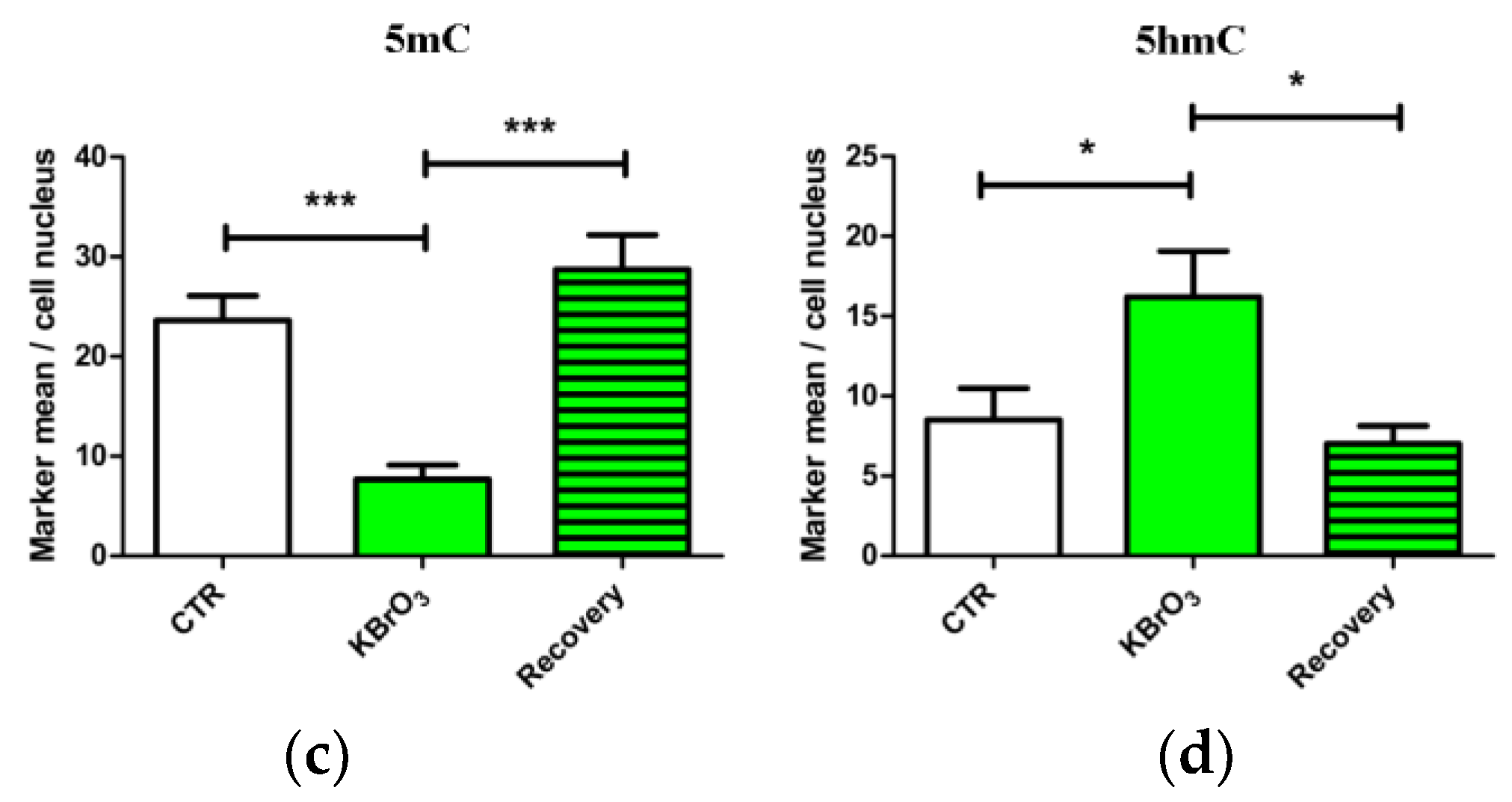
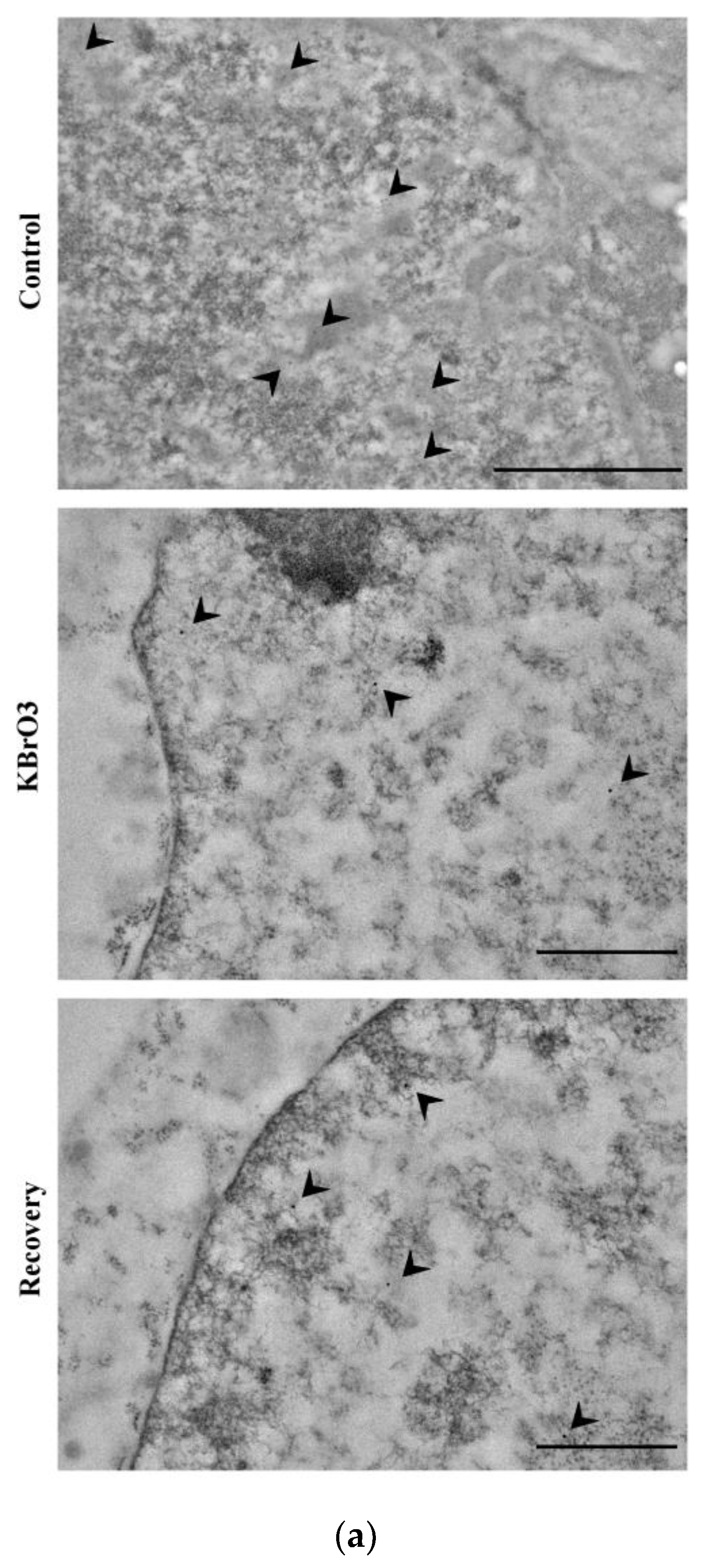
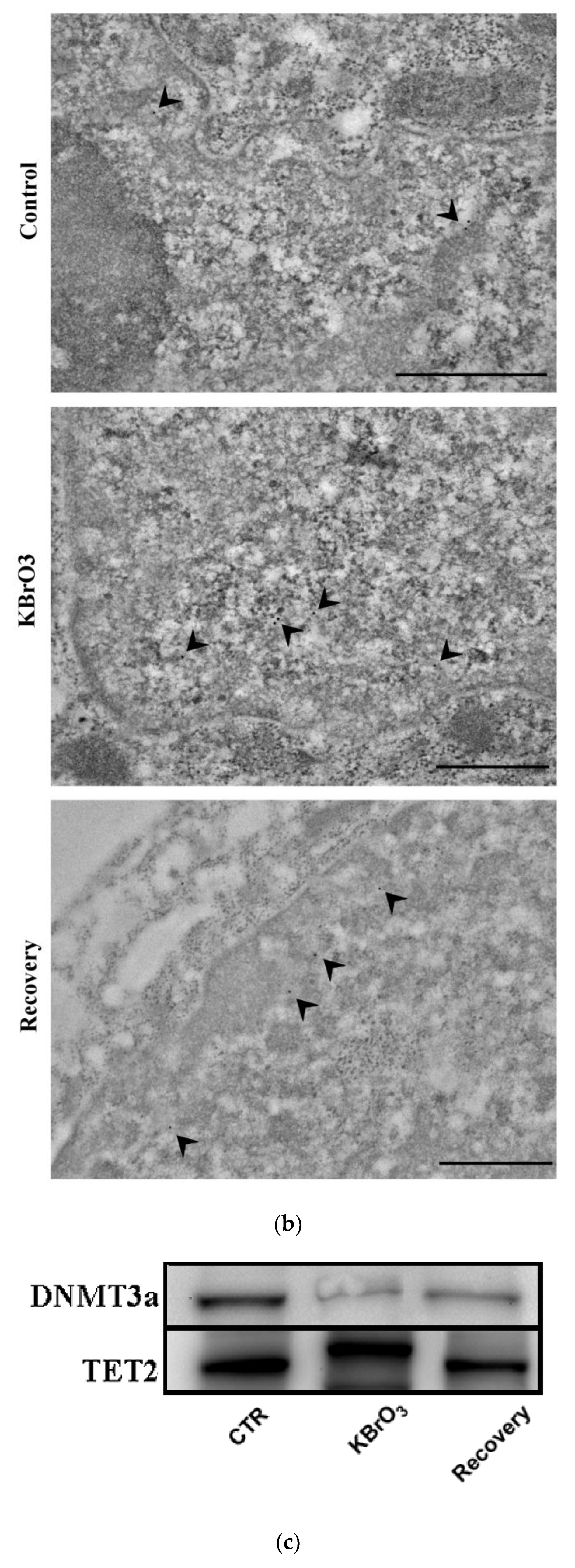
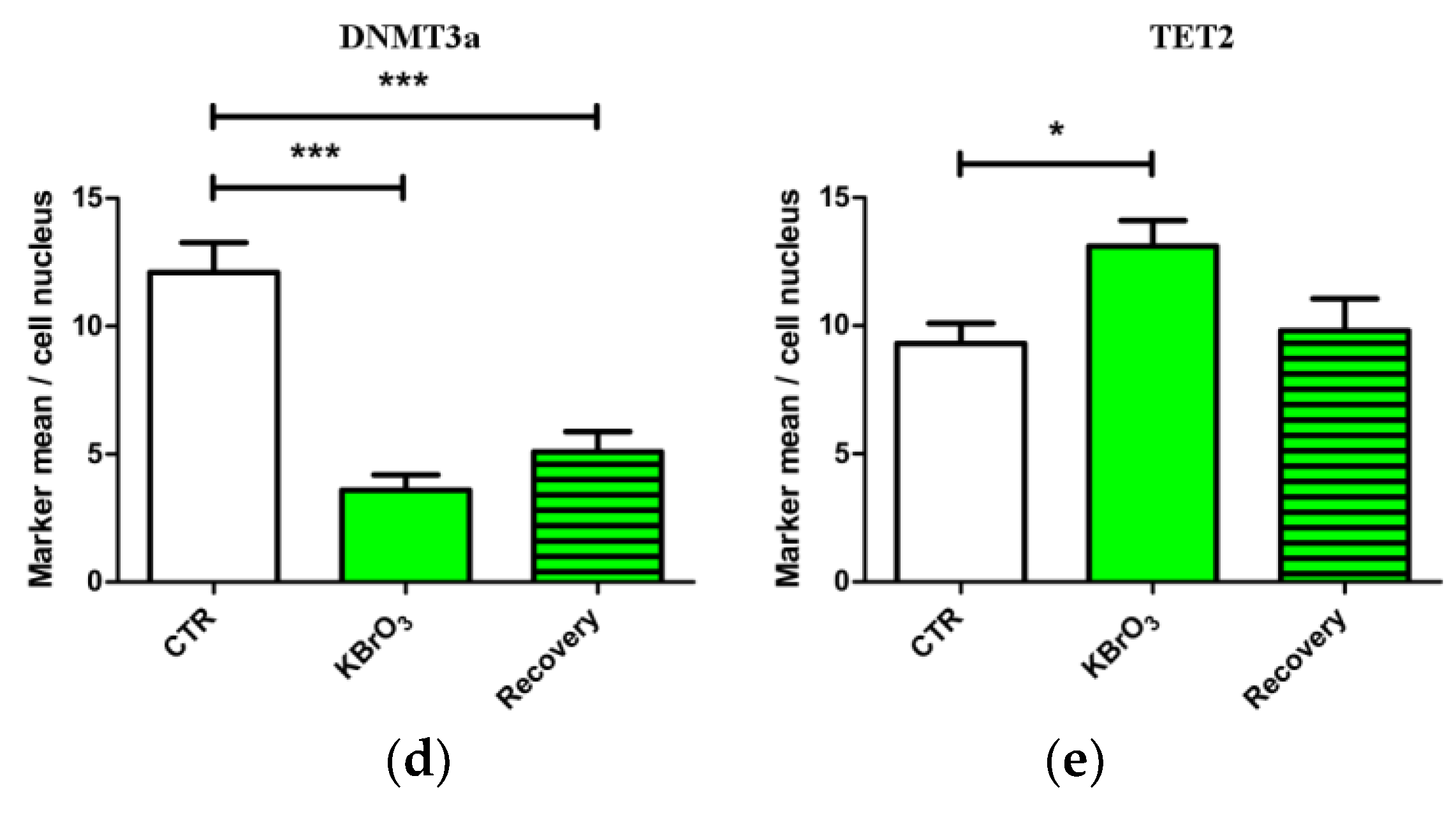
2.1. DNA Methylation and Demethylation Pattern Alteration following KBrO3 Treatment
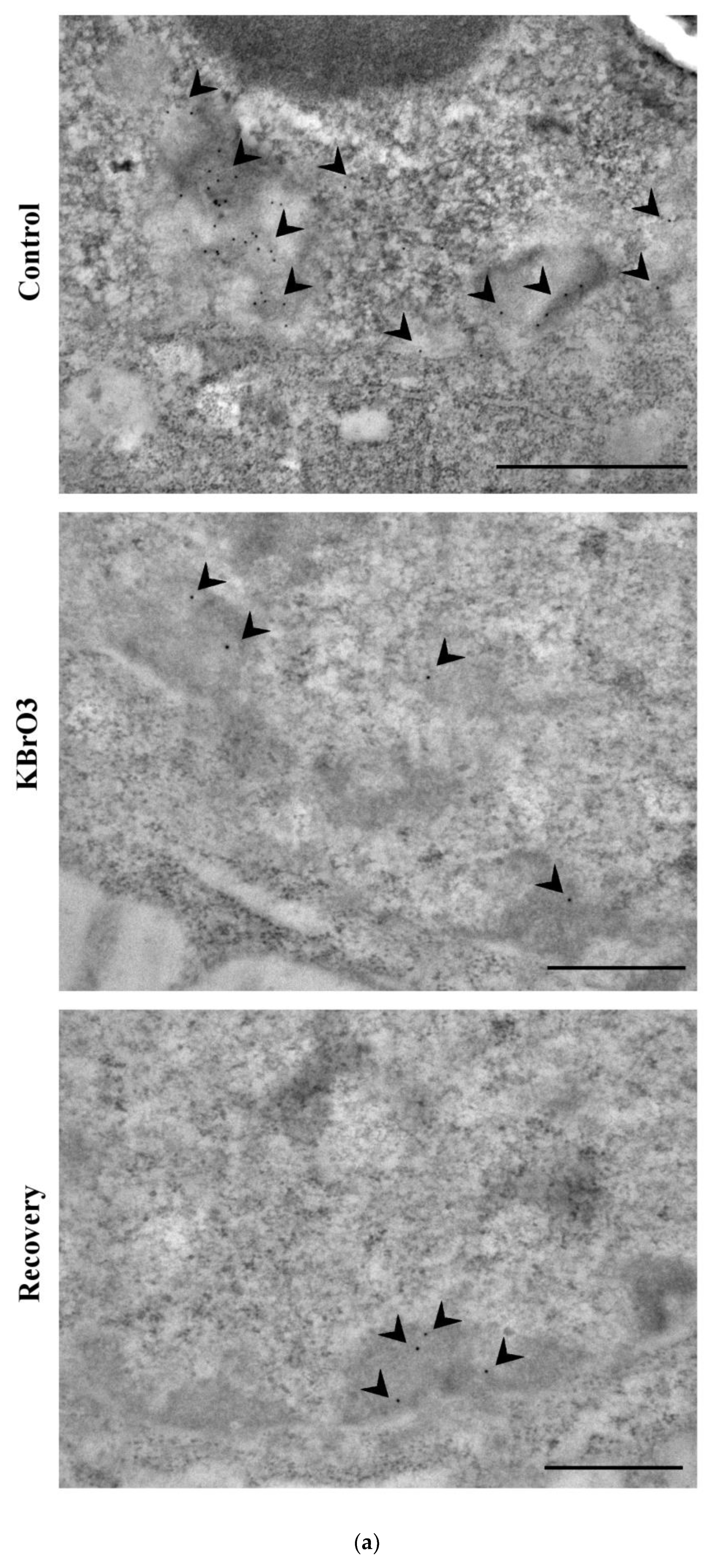
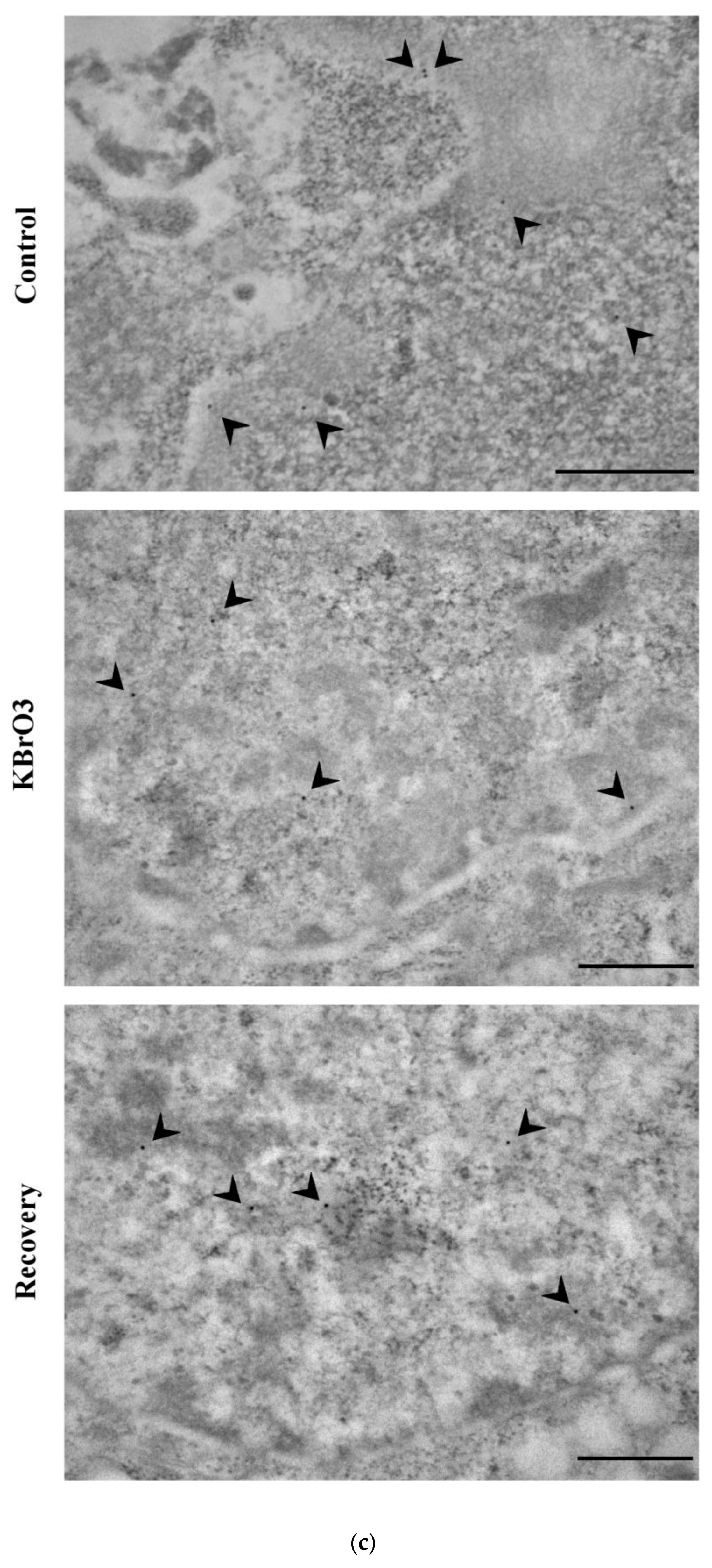
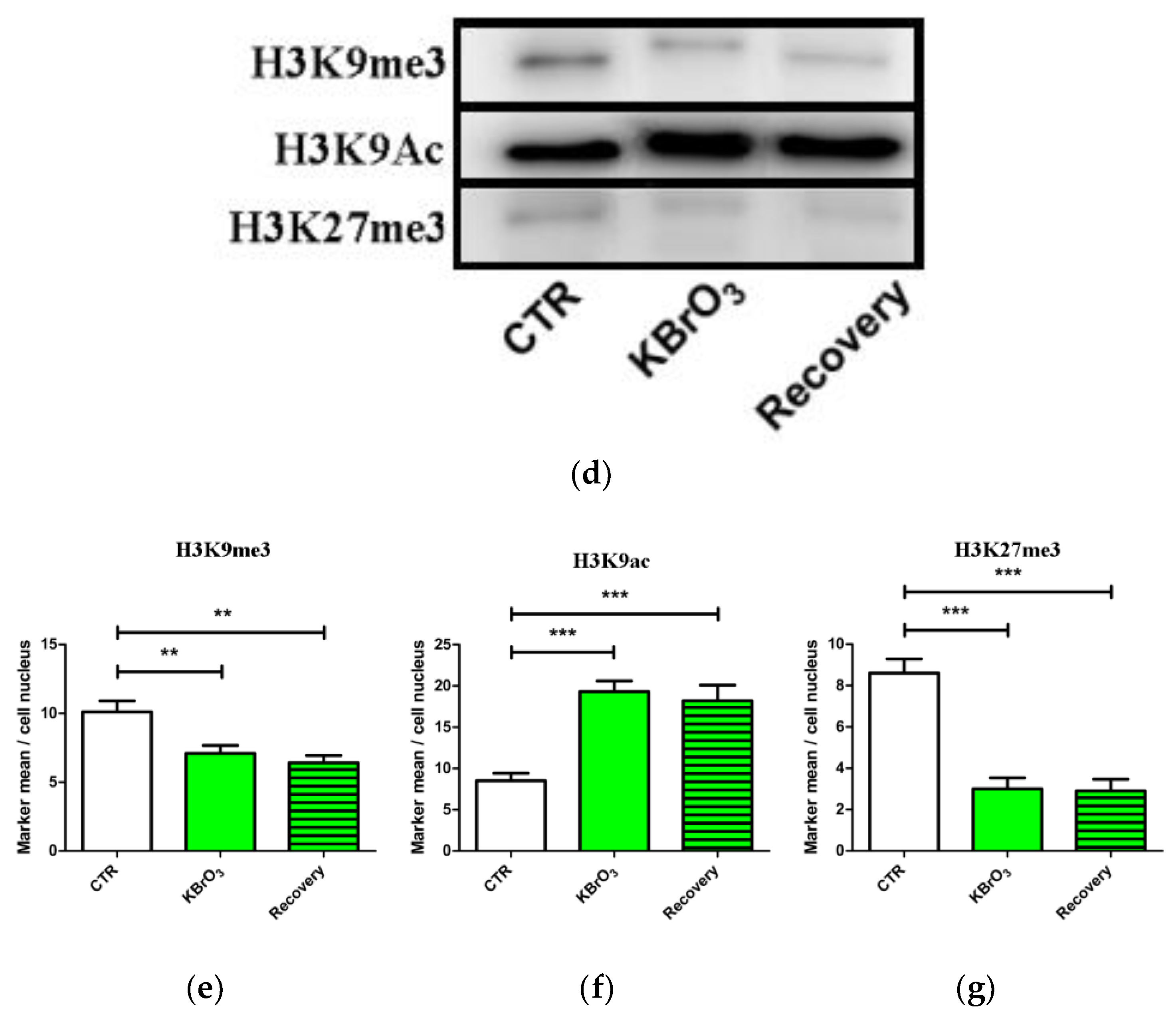
2.2. The Histone Code Reflects Cell Transcriptional Activity
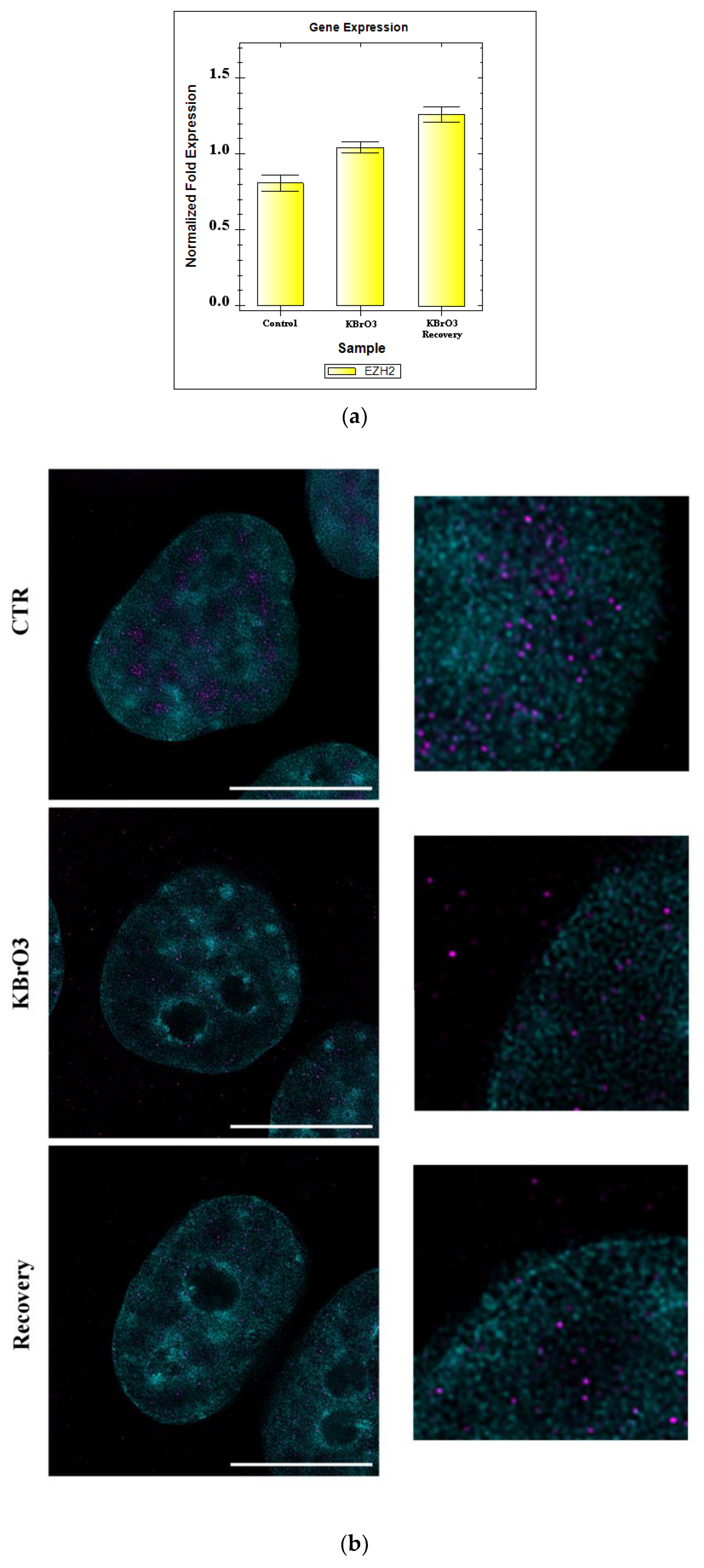
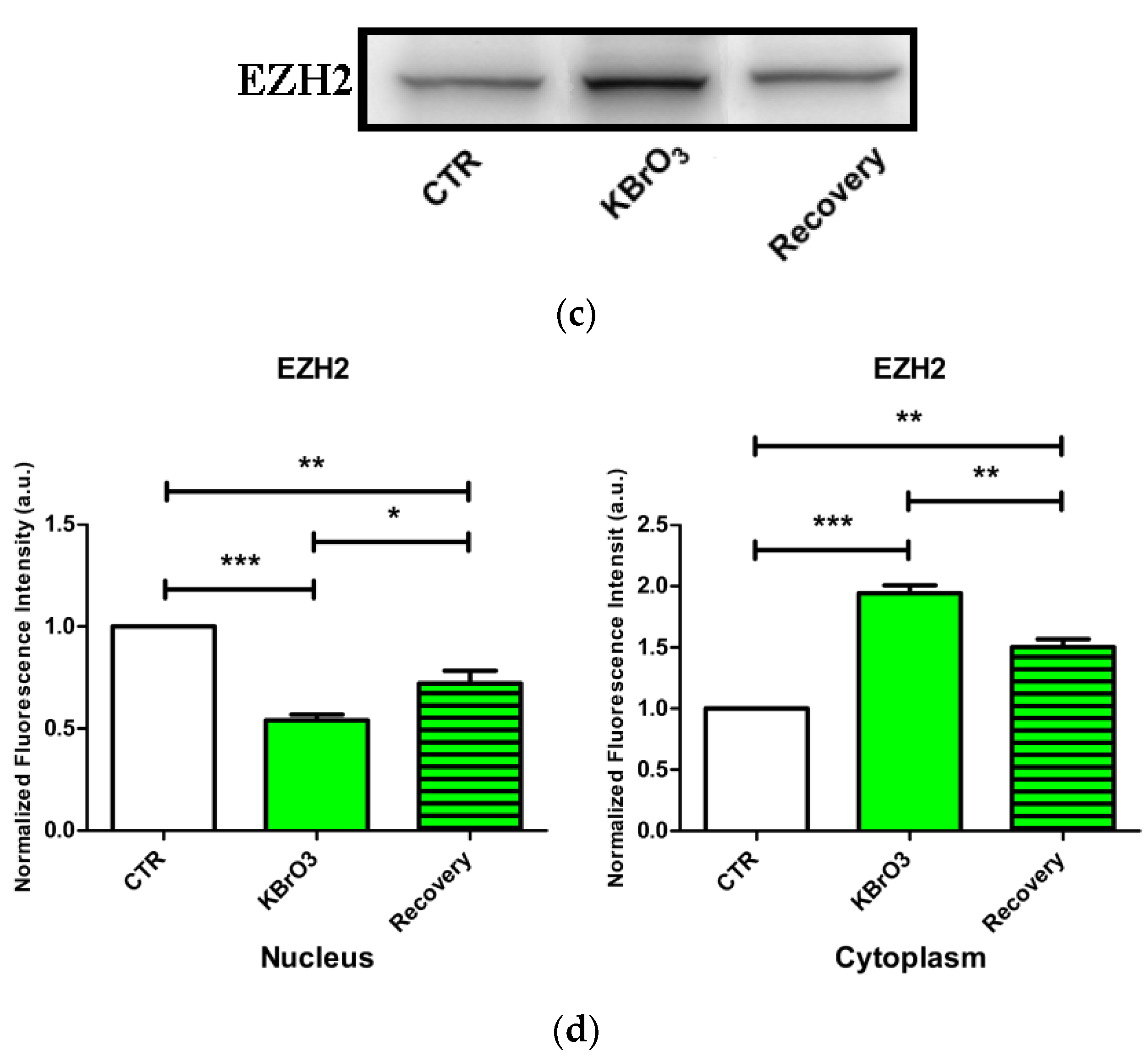
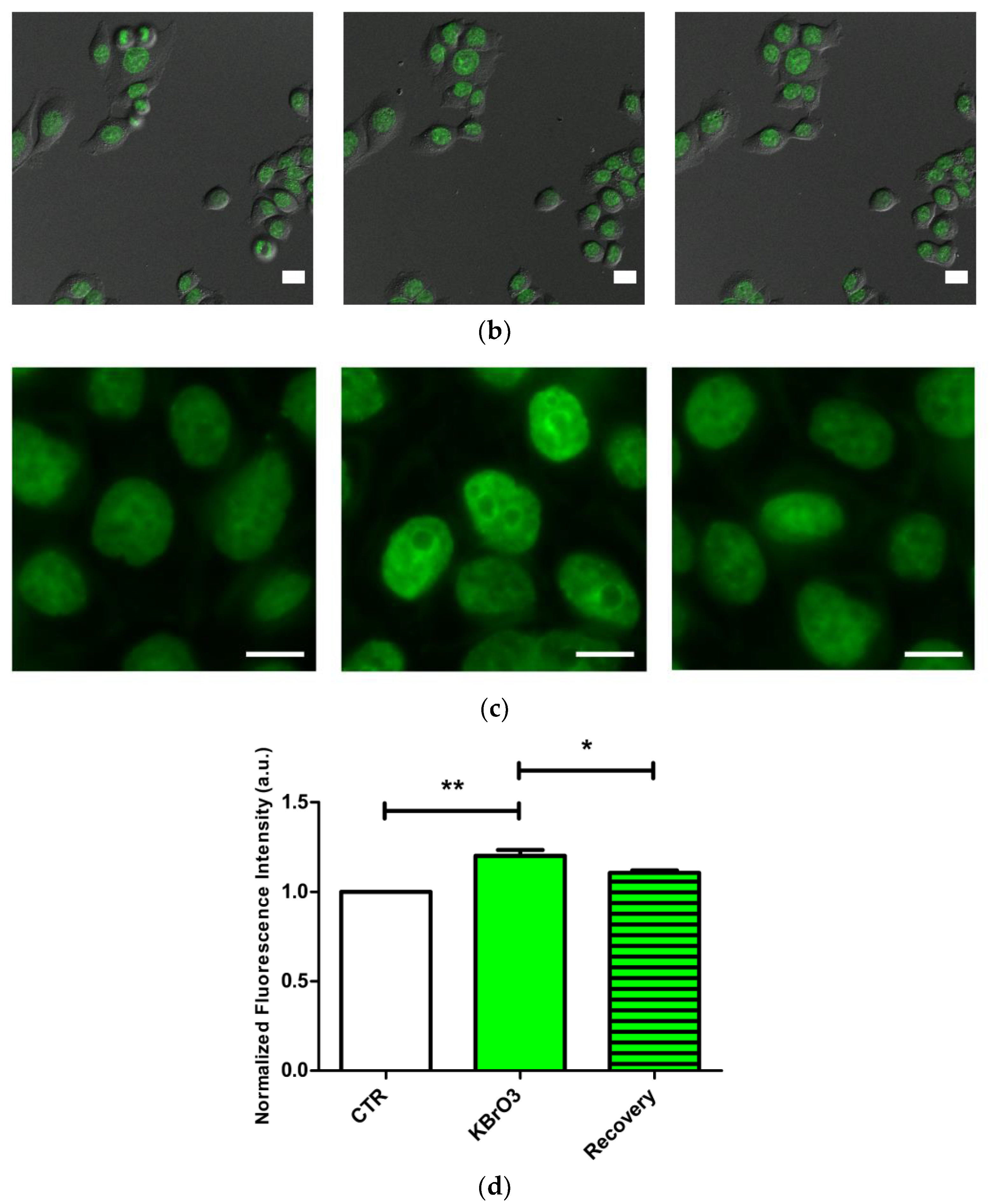
2.3. EZH2 Localization Is Altered following Oxidative Insult
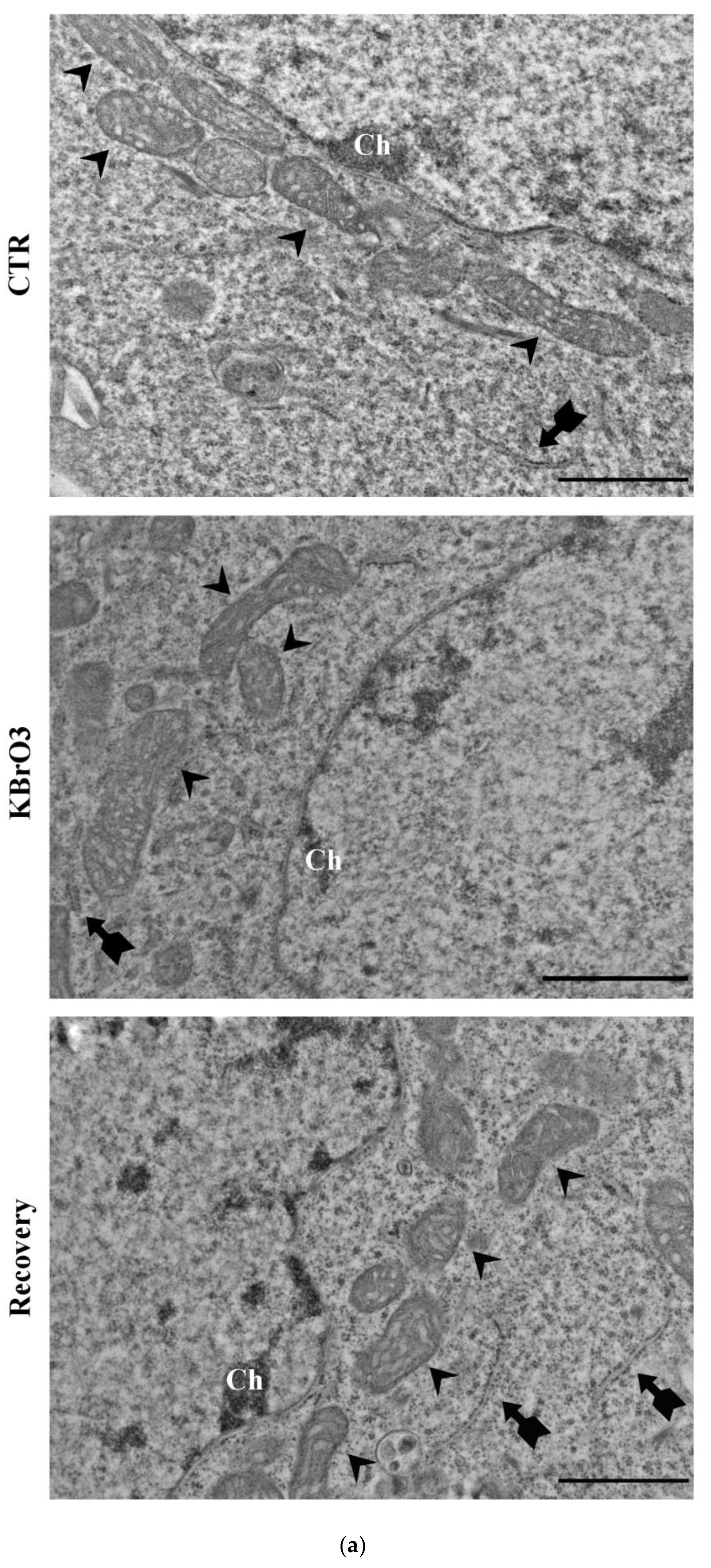
2.4. Oxidative Stress and Chromatin Reorganization
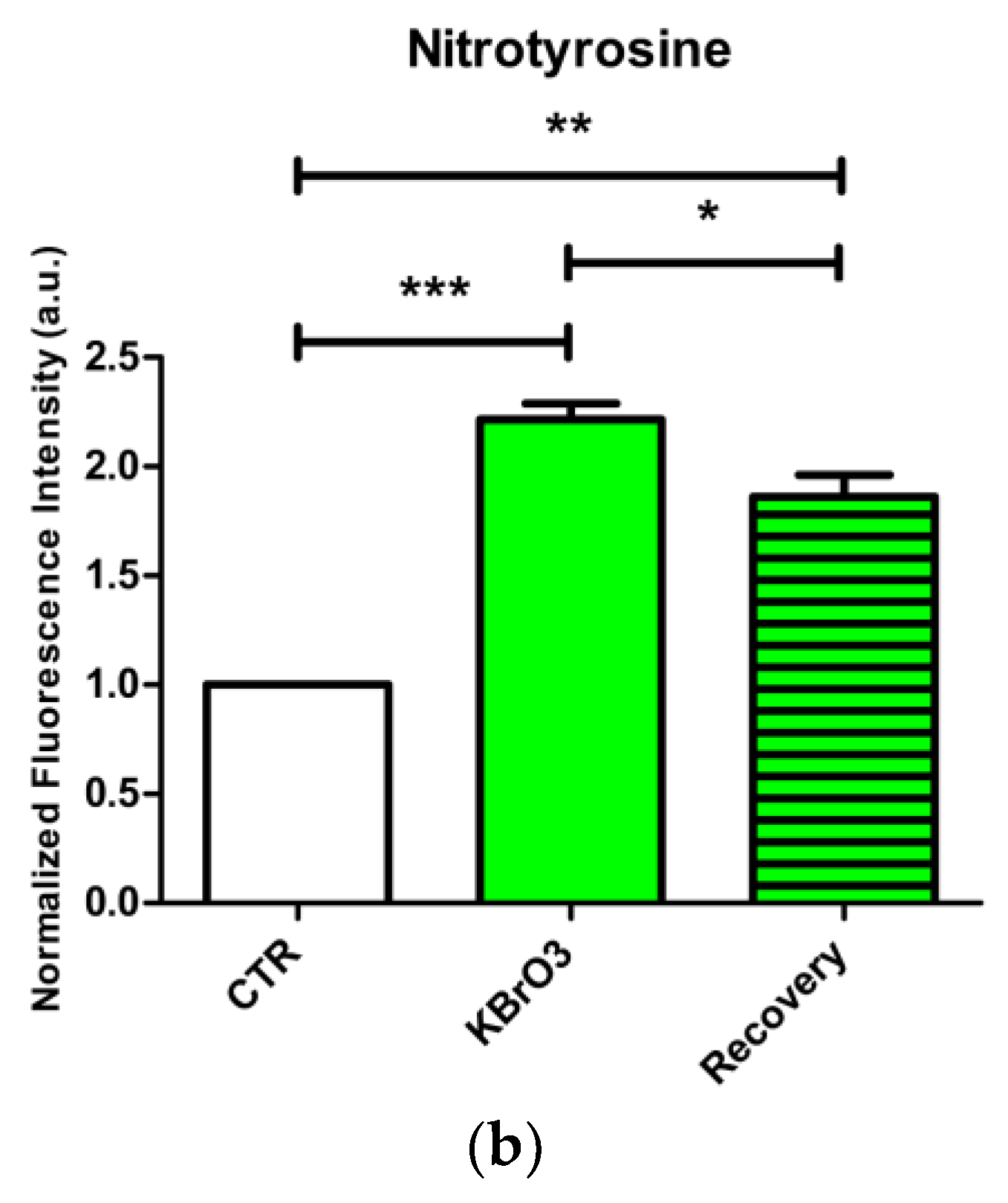
2.5. Oxidative Stress Induction Assessment
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Treatment
4.2. Transmission Electron Microscopy Sample Preparation
Transmission Electron Microscopy Immunocytochemistry and Sample Staining
4.3. Widefield and Confocal Microscopy Sample Preparation and Immunocytochemistry
4.4. Live Cell Imaging
4.5. Stimulated Emission Depletion Microscopy Sample Preparation and Immunocytochemistry
4.6. Western Blotting
4.7. Real Time PCR (qPCR)
4.8. Data and Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Goffart, S.; Tikkanen, P.; Michell, C.; Wilson, T.; Pohjoismäki, J.L.O. The Type and Source of Reactive Oxygen Species Influences the Outcome of Oxidative Stress in Cultured Cells. Cells 2021, 10, 1075. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, A.C.; Alves, R.; Baldeiras, I.; Marques, B.; Oliveiros, B.; Pereira, A.; Nascimento Costa, J.M.; Cortesão, E.; Mota Vieira, L.; Sarmento Ribeiro, A.B. DNA Methylation Is Correlated with Oxidative Stress in Myelodysplastic Syndrome—Relevance as Complementary Prognostic Biomarkers. Cancers 2021, 13, 3138. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, C.; Sueyoshi, Y.; Ema, M.; Mori, Y.; Takaishi, K.; Hisatomi, H. Induction of Oxidative Stress by Anticancer Drugs in the Presence and Absence of Cells. Oncol. Lett. 2017, 14, 6066–6070. [Google Scholar] [CrossRef]
- Totten, S.P.; Im, Y.K.; Cepeda Cañedo, E.; Najyb, O.; Nguyen, A.; Hébert, S.; Ahn, R.; Lewis, K.; Lebeau, B.; La Selva, R.; et al. STAT1 Potentiates Oxidative Stress Revealing a Targetable Vulnerability that Increases Phenformin Efficacy in Breast Cancer. Nat. Commun. 2021, 12, 3299. [Google Scholar] [CrossRef]
- Mancini, A.; Di Segni, C.; Raimondo, S.; Olivieri, G.; Silvestrini, A.; Meucci, E.; Currò, D. Thyroid Hormones, Oxidative Stress, and Inflammation. Mediat. Inflamm. 2016, 2016, 6757154. [Google Scholar] [CrossRef]
- Resmini, E.; Santos, A.; Aulinas, A.; Webb, S.M.; Vives-Gilabert, Y.; Cox, O.H.; Wand, G.; Lee, R.S. Reduced DNA Methylation of FKBP5 in Cushing’s Syndrome. Endocrine 2016, 54, 768–777. [Google Scholar] [CrossRef]
- Gillberg, L.; Hjort, L. Chapter 37—Epigenetics of Metabolic Diseases. In Handbook of Epigenetics, 2nd ed.; Tollefsbol, T.O., Ed.; Academic Press: Cambridge, MA, USA, 2017; pp. 569–580. ISBN 978-0-12-805388-1. [Google Scholar]
- Coppedè, F. Mutations Involved in Premature-Ageing Syndromes. Appl. Clin. Genet. 2021, 14, 279–295. [Google Scholar] [CrossRef]
- Wang, C.; Sun, Y.; Yin, X.; Feng, R.; Feng, R.; Xu, M.; Liang, K.; Zhao, R.; Gu, G.; Jiang, X.; et al. Alterations of DNA Methylation Were Associated with the Rapid Growth of Cortisol-Producing Adrenocortical Adenoma during Pregnancy. Clin. Epigenetics 2021, 13, 213. [Google Scholar] [CrossRef]
- Lima, T.R.R.; Sales, B.C.P.; Pereira, L.C. Oxidative Stress Monitoring in In Vitro and In Vivo Models. In Role of Oxidative Stress in Pathophysiology of Diseases; Maurya, P.K., Dua, K., Eds.; Springer: Singapore, 2020; pp. 163–178. ISBN 9789811515682. [Google Scholar]
- Ballmaier, D.; Epe, B. DNA Damage by Bromate: Mechanism and Consequences. Toxicology 2006, 221, 166–171. [Google Scholar] [CrossRef] [PubMed]
- Campalans, A.; Kortulewski, T.; Amouroux, R.; Menoni, H.; Vermeulen, W.; Radicella, J.P. Distinct Spatiotemporal Patterns and PARP Dependence of XRCC1 Recruitment to Single-Strand Break and Base Excision Repair. Nucleic Acids Res. 2013, 41, 3115–3129. [Google Scholar] [CrossRef]
- Müller, N.; Warwick, T.; Noack, K.; Malacarne, P.F.; Cooper, A.J.L.; Weissmann, N.; Schröder, K.; Brandes, R.P.; Rezende, F. Reactive Oxygen Species Differentially Modulate the Metabolic and Transcriptomic Response of Endothelial Cells. Antioxidants 2022, 11, 434. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, M.K.; Mahmood, R. Protective Effect of Taurine against Potassium Bromate-Induced Hemoglobin Oxidation, Oxidative Stress, and Impairment of Antioxidant Defense System in Blood. Environ. Toxicol. 2016, 31, 304–313. [Google Scholar] [CrossRef]
- Kumar, N.; Theil, A.F.; Roginskaya, V.; Ali, Y.; Calderon, M.; Watkins, S.C.; Barnes, R.P.; Opresko, P.L.; Pines, A.; Lans, H.; et al. Global and Transcription-Coupled Repair of 8-OxoG Is Initiated by Nucleotide Excision Repair Proteins. Nat. Commun. 2022, 13, 974. [Google Scholar] [CrossRef] [PubMed]
- Amouroux, R.; Campalans, A.; Epe, B.; Radicella, J.P. Oxidative Stress Triggers the Preferential Assembly of Base Excision Repair Complexes on Open Chromatin Regions. Nucleic Acids Res. 2010, 38, 2878–2890. [Google Scholar] [CrossRef] [PubMed]
- Lebraud, E.; Pinna, G.; Siberchicot, C.; Depagne, J.; Busso, D.; Fantini, D.; Irbah, L.; Robeska, E.; Kratassiouk, G.; Ravanat, J.-L.; et al. Chromatin Recruitment of OGG1 Requires Cohesin and Mediator and Is Essential for Efficient 8-OxoG Removal. Nucleic Acids Res. 2020, 48, 9082–9097. [Google Scholar] [CrossRef]
- Casali, C.; Siciliani, S.; Zannino, L.; Biggiogera, M. Histochemistry for Nucleic Acid Research: 60 Years in the European Journal of Histochemistry. Eur. J. Histochem. 2022, 66, 3409. [Google Scholar] [CrossRef]
- Stepanov, A.I.; Besedovskaia, Z.V.; Moshareva, M.A.; Lukyanov, K.A.; Putlyaeva, L.V. Studying Chromatin Epigenetics with Fluorescence Microscopy. Int. J. Mol. Sci. 2022, 23, 8988. [Google Scholar] [CrossRef]
- Buitrago, D.; Labrador, M.; Arcon, J.P.; Lema, R.; Flores, O.; Esteve-Codina, A.; Blanc, J.; Villegas, N.; Bellido, D.; Gut, M.; et al. Impact of DNA Methylation on 3D Genome Structure. Nat. Commun. 2021, 12, 3243. [Google Scholar] [CrossRef]
- Tyagi, S.C.; Stanisic, D.; Singh, M. Epigenetic Memory: Gene Writer, Eraser and Homocysteine. Mol. Cell Biochem. 2021, 476, 507–512. [Google Scholar] [CrossRef]
- Binder, M.; Carr, R.M.; Lasho, T.L.; Finke, C.M.; Mangaonkar, A.A.; Pin, C.L.; Berger, K.R.; Mazzone, A.; Potluri, S.; Ordog, T.; et al. Oncogenic Gene Expression and Epigenetic Remodeling of Cis-Regulatory Elements in ASXL1-Mutant Chronic Myelomonocytic Leukemia. Nat. Commun. 2022, 13, 1434. [Google Scholar] [CrossRef]
- Lejart, A.; Zentout, S.; Chapuis, C.; D’Augustin, O.; Smith, R.; Salbert, G.; Huet, S. The N-Terminal Domain of TET1 Promotes the Formation of Dense Chromatin Regions Refractory to Transcription. Chromosoma 2022, 131, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.-L.; Nie, J.; Ku, J.; Dougherty, U.; West-Szymanski, D.C.; Collin, F.; Ellison, C.K.; Sieh, L.; Ning, Y.; Deng, Z.; et al. A Human Tissue Map of 5-Hydroxymethylcytosines Exhibits Tissue Specificity through Gene and Enhancer Modulation. Nat. Commun. 2020, 11, 6161. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Zhang, C.; Zhang, X.; Fan, Y.; Zeng, H.; Liu, J.; Meng, H.; Bai, D.; Peng, J.; Zhang, Q.; et al. Tissue-Specific 5-Hydroxymethylcytosine Landscape of the Human Genome. Nat. Commun. 2021, 12, 4249. [Google Scholar] [CrossRef] [PubMed]
- Ghoneim, M.; Fuchs, H.A.; Musselman, C.A. Histone Tail Conformations: A Fuzzy Affair with DNA. Trends Biochem. Sci. 2021, 46, 564–578. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, X.D. Integrative Chemical Biology Approaches to Deciphering the Histone Code: A Problem-Driven Journey. Acc. Chem. Res. 2021, 54, 3734–3747. [Google Scholar] [CrossRef]
- Blackledge, N.P.; Fursova, N.A.; Kelley, J.R.; Huseyin, M.K.; Feldmann, A.; Klose, R.J. PRC1 Catalytic Activity Is Central to Polycomb System Function. Mol. Cell 2020, 77, 857–874. [Google Scholar] [CrossRef]
- Torres-Perez, J.V.; Irfan, J.; Febrianto, M.R.; Giovanni, S.D.; Nagy, I. Histone Post-Translational Modifications as Potential Therapeutic Targets for Pain Management. Trends Pharmacol. Sci. 2021, 42, 897–911. [Google Scholar] [CrossRef]
- Liao, Y.; Chen, C.-H.; Xiao, T.; de la Peña Avalos, B.; Dray, E.V.; Cai, C.; Gao, S.; Shah, N.; Zhang, Z.; Feit, A.; et al. Inhibition of EZH2 Transactivation Function Sensitizes Solid Tumors to Genotoxic Stress. Proc. Natl. Acad. Sci. USA 2022, 119, e2105898119. [Google Scholar] [CrossRef]
- Parreno, V.; Martinez, A.-M.; Cavalli, G. Mechanisms of Polycomb Group Protein Function in Cancer. Cell Res. 2022, 32, 231–253. [Google Scholar] [CrossRef]
- Patel, A.J.; Warda, S.; Maag, J.L.V.; Misra, R.; Miranda-Román, M.A.; Pachai, M.R.; Lee, C.J.; Li, D.; Wang, N.; Bayshtok, G.; et al. PRC2-Inactivating Mutations in Cancer Enhance Cytotoxic Response to DNMT1-Targeted Therapy via Enhanced Viral Mimicry. Cancer Discov. 2022, 12, 2120–2139. [Google Scholar] [CrossRef]
- Zhang, L.; Qu, J.; Qi, Y.; Duan, Y.; Huang, Y.-W.; Zhou, Z.; Li, P.; Yao, J.; Huang, B.; Zhang, S.; et al. EZH2 Engages TGFβ Signaling to Promote Breast Cancer Bone Metastasis via Integrin Β1-FAK Activation. Nat. Commun. 2022, 13, 2543. [Google Scholar] [CrossRef]
- Panchal, S.K.; Brown, L. DNA Methylation in Adipose Tissue and Metabolic Syndrome. J. Clin. Med. 2020, 9, 2699. [Google Scholar] [CrossRef]
- Botta, M.; Audano, M.; Sahebkar, A.; Sirtori, C.R.; Mitro, N.; Ruscica, M. PPAR Agonists and Metabolic Syndrome: An Established Role? Int. J. Mol. Sci. 2018, 19, 1197. [Google Scholar] [CrossRef]
- Castellano-Castillo, D.; Moreno-Indias, I.; Sanchez-Alcoholado, L.; Ramos-Molina, B.; Alcaide-Torres, J.; Morcillo, S.; Ocaña-Wilhelmi, L.; Tinahones, F.; Queipo-Ortuño, M.I.; Cardona, F. Altered Adipose Tissue DNA Methylation Status in Metabolic Syndrome: Relationships Between Global DNA Methylation and Specific Methylation at Adipogenic, Lipid Metabolism and Inflammatory Candidate Genes and Metabolic Variables. J. Clin. Med. 2019, 8, 87. [Google Scholar] [CrossRef]
- Yaribeygi, H.; Farrokhi, F.R.; Butler, A.E.; Sahebkar, A. Insulin Resistance: Review of the Underlying Molecular Mechanisms. J. Cell Physiol. 2019, 234, 8152–8161. [Google Scholar] [CrossRef]
- Zhang, X.; Ho, S.-M. Epigenetics Meets Endocrinology. J. Mol. Endocrinol. 2011, 46, R11–R32. [Google Scholar] [CrossRef]
- Duvall, E.; Benitez, C.M.; Tellez, K.; Enge, M.; Pauerstein, P.T.; Li, L.; Baek, S.; Quake, S.R.; Smith, J.P.; Sheffield, N.C.; et al. Single-Cell Transcriptome and Accessible Chromatin Dynamics during Endocrine Pancreas Development. Proc. Natl. Acad. Sci. USA 2022, 119, e2201267119. [Google Scholar] [CrossRef]
- Ahsan, H. 3-Nitrotyrosine: A Biomarker of Nitrogen Free Radical Species Modified Proteins in Systemic Autoimmunogenic Conditions. Hum. Immunol. 2013, 74, 1392–1399. [Google Scholar] [CrossRef]
- Bandookwala, M.; Sengupta, P. 3-Nitrotyrosine: A Versatile Oxidative Stress Biomarker for Major Neurodegenerative Diseases. Int. J. Neurosci. 2020, 130, 1047–1062. [Google Scholar] [CrossRef]
- Li, Q.; Sun, X.; Tang, Y.; Qu, Y.; Zhou, Y.; Zhang, Y. EZH2 Reduction Is an Essential Mechanoresponse for the Maintenance of Super-Enhancer Polarization against Compressive Stress in Human Periodontal Ligament Stem Cells. Cell Death Dis. 2020, 11, 757. [Google Scholar] [CrossRef]
- Liu, Y.; Tu, C.; Guo, X.; Wu, C.; Gu, C.; Lai, Q.; Fang, Y.; Huang, J.; Wang, Z.; Li, A.; et al. Tumor-Suppressive Function of EZH2 Is through Inhibiting Glutaminase. Cell Death Dis. 2021, 12, 975. [Google Scholar] [CrossRef] [PubMed]
- Anwar, T.; Arellano-Garcia, C.; Ropa, J.; Chen, Y.-C.; Kim, H.S.; Yoon, E.; Grigsby, S.; Basrur, V.; Nesvizhskii, A.I.; Muntean, A.; et al. P38-Mediated Phosphorylation at T367 Induces EZH2 Cytoplasmic Localization to Promote Breast Cancer Metastasis. Nat. Commun. 2018, 9, 2801. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, M.; Wang, D.; Hou, P.; Chen, X.; Chu, S.; Chai, D.; Zheng, J.; Bai, J. Post-Translational Modifications of EZH2 in Cancer. Cell Biosci. 2020, 10, 143. [Google Scholar] [CrossRef] [PubMed]
- Madugundu, G.S.; Cadet, J.; Wagner, J.R. Hydroxyl-Radical-Induced Oxidation of 5-Methylcytosine in Isolated and Cellular DNA. Nucleic Acids Res. 2014, 42, 7450–7460. [Google Scholar] [CrossRef] [PubMed]
- Kietzmann, T.; Petry, A.; Shvetsova, A.; Gerhold, J.M.; Görlach, A. The Epigenetic Landscape Related to Reactive Oxygen Species Formation in the Cardiovascular System: ROS and Epigenetic Modifications. Br. J. Pharmacol. 2017, 174, 1533–1554. [Google Scholar] [CrossRef] [PubMed]
- Khurana, V.; Lu, Y.; Steinhilb, M.L.; Oldham, S.; Shulman, J.M.; Feany, M.B. TOR-Mediated Cell-Cycle Activation Causes Neurodegeneration in a Drosophila Tauopathy Model. Curr. Biol. 2006, 16, 230–241. [Google Scholar] [CrossRef] [PubMed]
- Frost, B.; Hemberg, M.; Lewis, J.; Feany, M.B. Tau Promotes Neurodegeneration through Global Chromatin Relaxation. Nat. Neurosci. 2014, 17, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Kreuz, S.; Fischle, W. Oxidative Stress Signaling to Chromatin in Health and Disease. Epigenomics 2016, 8, 843–862. [Google Scholar] [CrossRef]
- García-Giménez, J.L.; Olaso, G.; Hake, S.B.; Bönisch, C.; Wiedemann, S.M.; Markovic, J.; Dasí, F.; Gimeno, A.; Pérez-Quilis, C.; Palacios, Ò.; et al. Histone H3 Glutathionylation in Proliferating Mammalian Cells Destabilizes Nucleosomal Structure. Antioxid. Redox Signal. 2013, 19, 1305. [Google Scholar] [CrossRef]
- Agudelo, M.; Gandhi, N.; Saiyed, Z.; Pichili, V.; Thangavel, S.; Khatavkar, P.; Yndart-Arias, A.; Nair, M. Effects of Alcohol on Histone Deacetylase 2 (HDAC2) and the Neuroprotective Role of Trichostatin A (TSA). Alcohol Clin. Exp. Res. 2011, 35, 1550–1556. [Google Scholar] [CrossRef]
- Hwang, J.; Yao, H.; Caito, S.; Sundar, I.K.; Rahman, I. Redox Regulation of SIRT1 in Inflammation and Cellular Senescence. Free Radic. Biol. Med. 2013, 61, 95–110. [Google Scholar] [CrossRef] [PubMed]
- Berby, B.; Bichara, C.; Rives-Feraille, A.; Jumeau, F.; Pizio, P.D.; Sétif, V.; Sibert, L.; Dumont, L.; Rondanino, C.; Rives, N. Oxidative Stress Is Associated with Telomere Interaction Impairment and Chromatin Condensation Defects in Spermatozoa of Infertile Males. Antioxidants 2021, 10, 593. [Google Scholar] [CrossRef] [PubMed]
- Vasileva, B.; Staneva, D.; Krasteva, N.; Miloshev, G.; Georgieva, M. Changes in Chromatin Organization Eradicate Cellular Stress Resilience to UVA/B Light and Induce Premature Aging. Cells 2021, 10, 1755. [Google Scholar] [CrossRef] [PubMed]
- Guillaumet-Adkins, A.; Yañez, Y.; Peris-Diaz, M.D.; Calabria, I.; Palanca-Ballester, C.; Sandoval, J. Epigenetics and Oxidative Stress in Aging. Oxid. Med. Cell Longev. 2017, 2017, 9175806. [Google Scholar] [CrossRef] [PubMed]
- Bazopoulou, D.; Knoefler, D.; Zheng, Y.; Ulrich, K.; Oleson, B.J.; Xie, L.; Kim, M.; Kaufmann, A.; Lee, Y.-T.; Dou, Y.; et al. Developmental ROS Individualizes Organismal Stress Resistance and Lifespan. Nature 2019, 576, 301–305. [Google Scholar] [CrossRef] [PubMed]
- Mongioì, L.M.; La Vignera, S.; Cannarella, R.; Cimino, L.; Compagnone, M.; Condorelli, R.A.; Calogero, A.E. The Role of Resveratrol Administration in Human Obesity. Int. J. Mol. Sci. 2021, 22, 4362. [Google Scholar] [CrossRef] [PubMed]
- Pérusse, L.; Jacob, R.; Drapeau, V.; Llewellyn, C.; Arsenault, B.J.; Bureau, A.; Labonté, M.-È.; Tremblay, A.; Vohl, M.-C. Understanding Gene-Lifestyle Interaction in Obesity: The Role of Mediation versus Moderation. Lifestyle Genom. 2022, 15, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Mancini, A.; Martorana, G.E.; Magini, M.; Festa, R.; Raimondo, S.; Silvestrini, A.; Nicolotti, N.; Mordente, A.; Mele, M.C.; Miggiano, G.A.D.; et al. Oxidative Stress and Metabolic Syndrome: Effects of a Natural Antioxidants Enriched Diet on Insulin Resistance. Clin. Nutr. ESPEN 2015, 10, e52–e60. [Google Scholar] [CrossRef] [PubMed]
- Mancini, A.; Silvestrini, A. Oxidative Stress in Metabolic and Endocrine Diseases: Basic and Translational Aspects. Int. J. Mol. Sci. 2022, 23, 4346. [Google Scholar] [CrossRef] [PubMed]
- Butterfield, D.A.; Halliwell, B. Oxidative Stress, Dysfunctional Glucose Metabolism and Alzheimer Disease. Nat. Rev. Neurosci. 2019, 20, 148–160. [Google Scholar] [CrossRef]
- Palierse, E.; Przybylski, C.; Brouri, D.; Jolivalt, C.; Coradin, T. Interactions of Calcium with Chlorogenic and Rosmarinic Acids: An Experimental and Theoretical Approach. Int. J. Mol. Sci. 2020, 21, 4948. [Google Scholar] [CrossRef] [PubMed]
- Ražná, K.; Sawinska, Z.; Ivanišová, E.; Vukovic, N.; Terentjeva, M.; Stričík, M.; Kowalczewski, P.Ł.; Hlavačková, L.; Rovná, K.; Žiarovská, J.; et al. Properties of Ginkgo biloba L.: Antioxidant Characterization, Antimicrobial Activities, and Genomic MicroRNA Based Marker Fingerprints. Int. J. Mol. Sci. 2020, 21, 3087. [Google Scholar] [CrossRef] [PubMed]
- Bruno, C.; Basile, U.; Vergani, E.; Napodano, C.; Oliva, A.; Gulli, F.; Meucci, E.; Silvestrini, A.; Orlando, P.; Silvestri, S.; et al. Inflammation and Oxidative Stress in Seminal Plasma: Search for Biomarkers in Diagnostic Approach to Male Infertility. J. Pers. Med. 2022, 12, 857. [Google Scholar] [CrossRef]
- Kaur, J.; Daoud, A.; Eblen, S.T. Targeting Chromatin Remodeling for Cancer Therapy. Curr. Mol. Pharmacol. 2019, 12, 215–229. [Google Scholar] [CrossRef]
- Lima, D.G.; do Amaral, G.C.L.S.; Planello, A.C.; Borgato, G.B.; Guimarães, G.N.; de Souza, A.P. Combined Therapy with Cisplatin and 5-AZA-2CdR Modifies Methylation and Expression of DNA Repair Genes in Oral Squamous Cell Carcinoma. Int. J. Clin. Exp. Pathol. 2022, 15, 131–144. [Google Scholar]
- Traube, F.R.; Brás, N.F.; Roos, W.P.; Sommermann, C.C.; Diehl, T.; Mayer, R.J.; Ofial, A.R.; Müller, M.; Zipse, H.; Carell, T. Epigenetic Anti-Cancer Treatment with a Stabilized Carbocyclic Decitabine Analogue. Chemistry 2022, 28, e202200640. [Google Scholar] [CrossRef]
- Norbury, J.W.; Slaba, T.C.; Aghara, S.; Badavi, F.F.; Blattnig, S.R.; Clowdsley, M.S.; Heilbronn, L.H.; Lee, K.; Maung, K.M.; Mertens, C.J.; et al. Advances in Space Radiation Physics and Transport at NASA. Life Sci. Space Res. 2019, 22, 98–124. [Google Scholar] [CrossRef]
- da Silveira, W.A.; Fazelinia, H.; Rosenthal, S.B.; Laiakis, E.C.; Kim, M.S.; Meydan, C.; Kidane, Y.; Rathi, K.S.; Smith, S.M.; Stear, B.; et al. Comprehensive Multi-Omics Analysis Reveals Mitochondrial Stress as a Central Biological Hub for Spaceflight Impact. Cell 2020, 183, 1185–1201. [Google Scholar] [CrossRef]
- Slaba, T.C.; Wilson, J.W.; Werneth, C.M.; Whitman, K. Updated Deterministic Radiation Transport for Future Deep Space Missions. Life Sci. Space Res. 2020, 27, 6–18. [Google Scholar] [CrossRef]
- Fleisch, A.F.; Wright, R.O.; Baccarelli, A.A. Environmental Epigenetics: A Role in Endocrine Disease? J. Mol. Endocrinol. 2012, 49, R61–R67. [Google Scholar] [CrossRef]
- Crafa, A.; Calogero, A.E.; Cannarella, R.; Mongioi’, L.M.; Condorelli, R.A.; Greco, E.A.; Aversa, A.; La Vignera, S. The Burden of Hormonal Disorders: A Worldwide Overview with a Particular Look in Italy. Front. Endocrinol. 2021, 12, 694325. [Google Scholar] [CrossRef] [PubMed]
- Bernhard, W. A New Staining Procedure for Electron Microscopical Cytology. J. Ultrastruct. Res. 1969, 27, 250–265. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, E.S. The Use of Lead Citrate at High pH as an Electron-Opaque Stain in Electron Microscopy. J. Cell Biol. 1963, 17, 208–212. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Rueden, C.T.; Schindelin, J.; Hiner, M.C.; DeZonia, B.E.; Walter, A.E.; Arena, E.T.; Eliceiri, K.W. ImageJ2: ImageJ for the next Generation of Scientific Image Data. BMC Bioinform. 2017, 18, 529. [Google Scholar] [CrossRef] [PubMed]




| Antibody (Host) | Target | Dilution | Reference |
|---|---|---|---|
| Anti-5mC (mouse) | 5mC | 1:500 in PBS/BSA/TWEEN20 | GTX629448–AB_2888114 (GeneTex, Irvine, CA, USA) |
| Anti-5hmC (mouse) | 5hmC | 1:150 in PBS/BSA/TWEEN20 | GTX629765–AB_2736902 (GeneTex) |
| Anti-DNMT3a (rabbit) | DNMT3a | 1:10 in PBS/BSA/TWEEN20 | GTX129125–AB_2885902 (GeneTex) |
| Anti-TET2 (mouse) | TET2 | 1:20 in PBS/BSA/TWEEN20 | GTX629881–AB_2888171 (GeneTex) |
| Anti-H3K9me3 (rabbit) | H3K9me3 | 1:20 in PBS/BSA/TWEEN20 | GTX121677–AB_10721938 (GeneTex) |
| Anti-H3K9Ac (rabbit) | H3K9Ac | 1:20 in PBS/BSA/TWEEN20 | GTX88007–AB_10731164 (GeneTex) |
| Anti-H3K27me3 (rabbit) | H3K27me3 | 1:20 in PBS/BSA/TWEEN20 | GTX121184–AB_10618572 (GeneTex) |
| Anti-Rabbit 12 nm gold particle conjugated (goat) | Rabbit IgG | 1:20 in PBS | 111-205-144– AB_2338016 (Jackson ImmunoResearch, West Grove, PA, USA) |
| Anti-Mouse 12 nm gold particle conjugated (goat) | Mouse IgG | 1:20 in PBS | 115-205-068– AB_2338730 (Jackson ImmunoResearch) |
| Antibody (Host) | Target | Dilution | Reference |
|---|---|---|---|
| Anti-EZH2 (rabbit) | EZH2 | 1:200 in PBS/BSA 1% | GTX110384–AB_2885283 (GeneTex) |
| Anti-NO-Tyr | Nitrotyrosine | 1:200 in PBS/BSA 1% | sc-32757–AB_628022 (Santa Cruz Biotechnology, Dallas, TX, USA) |
| Alexa 568 (goat) | Rabbit IgG | 1:100 in PBS/BSA 1% | A11036–AB_10563566 (Invitrogen, Waltham, MA, USA) |
| DyLight594 (goat) | Mouse IgG | 1:100 in PBS/BSA 1% | GTX213111-05–AB_2887581 (GeneTex) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Casali, C.; Siciliani, S.; Galgano, L.; Biggiogera, M. Oxidative Stress and Nuclear Reprogramming: A Pilot Study of the Effects of Reactive Oxygen Species on Architectural and Epigenetic Landscapes. Int. J. Mol. Sci. 2023, 24, 153. https://doi.org/10.3390/ijms24010153
Casali C, Siciliani S, Galgano L, Biggiogera M. Oxidative Stress and Nuclear Reprogramming: A Pilot Study of the Effects of Reactive Oxygen Species on Architectural and Epigenetic Landscapes. International Journal of Molecular Sciences. 2023; 24(1):153. https://doi.org/10.3390/ijms24010153
Chicago/Turabian StyleCasali, Claudio, Stella Siciliani, Luca Galgano, and Marco Biggiogera. 2023. "Oxidative Stress and Nuclear Reprogramming: A Pilot Study of the Effects of Reactive Oxygen Species on Architectural and Epigenetic Landscapes" International Journal of Molecular Sciences 24, no. 1: 153. https://doi.org/10.3390/ijms24010153
APA StyleCasali, C., Siciliani, S., Galgano, L., & Biggiogera, M. (2023). Oxidative Stress and Nuclear Reprogramming: A Pilot Study of the Effects of Reactive Oxygen Species on Architectural and Epigenetic Landscapes. International Journal of Molecular Sciences, 24(1), 153. https://doi.org/10.3390/ijms24010153







